Significance
Accurate DNA replication is a prerequisite for cell proliferation and genetic stability, but obstacles to smooth replication fork progression are frequent. The oncogenic activity of Mdm2 has been largely ascribed to its ability of antagonizing the tumor suppressor p53. This report, however, points out a p53-independent activity of Mdm2 in suppressing R loops, a structure that includes DNA:RNA hybrids and has recently emerged as a key obstacle to DNA replication. Accordingly, Mdm2 is required for sustaining DNA replication. Our results also reveal that Mdm2 and the polycomb repressor complexes act in parallel to not only modify histones but also support DNA replication. Thus, chromatin modifiers that were traditionally implicated in transcription regulation are enabling unperturbed DNA replication as well.
Keywords: Mdm2, p53, RNF2, histone, ubiquitin
Abstract
The p53–Mdm2 system is key to tumor suppression. We have recently reported that p53 as well as Mdm2 are capable of supporting DNA replication fork progression. On the other hand, we found that Mdm2 is a modifier of chromatin, modulating polycomb repressor complex (PRC)-driven histone modifications. Here we show that, similar to Mdm2 knockdown, the depletion of PRC members impairs DNA synthesis, as determined in fiber assays. In particular, the ubiquitin ligase and PRC1 component RNF2/Ring1B is required to support DNA replication, similar to Mdm2. Moreover, the Ring finger domain of Mdm2 is not only essential for its ubiquitin ligase activity, but also for proper DNA replication. Strikingly, Mdm2 overexpression can rescue RNF2 depletion with regard to DNA replication fork progression, and vice versa, strongly suggesting that the two ubiquitin ligases perform overlapping functions in this context. H2A overexpression also rescues fork progression upon depletion of Mdm2 or RNF2, but only when the ubiquitination sites K118/K119 are present. Depleting the H2A deubiquitinating enzyme BAP1 reduces the fork rate, suggesting that both ubiquitination and deubiquitination of H2A are required to support fork progression. The depletion of Mdm2 elicits the accumulation of RNA/DNA hybrids, suggesting R-loop formation as a mechanism of impaired DNA replication. Accordingly, RNase H overexpression or the inhibition of the transcription elongation kinase CDK9 each rescues DNA replication upon depletion of Mdm2 or RNF2. Taken together, our results suggest that chromatin modification by Mdm2 and PRC1 ensures smooth DNA replication through the avoidance of R-loop formation.
The tumor suppressor p53 is unique with regard to its mutation rate in human malignancies, exceeding 50%. Its activity is balanced by the ubiquitin ligase Mdm2, the product of a p53-inducible gene. Some tumors overexpress or activate Mdm2 as part of an oncogenic mechanism, most notably to antagonize p53.
More recently, however, p53-independent activities of Mdm2 were discovered. Our laboratory described a chromatin modifier function of Mdm2 which contributes to histone H2A ubiquitination at K119, as well as to histone H3 trimethylation at K27 (1, 2). Accordingly, Mdm2 physically associates with the protein complexes that confer these chromatin modifications, i.e., the members of the polycomb repressor complexes (PRCs) 1 (3) and 2 (1). These modifications were mostly characterized as mediating transcriptional repression, along with stemness (4, 5).
On the other hand, we have also identified a p53 function that broadens the traditional view on the “guardian of the genome.” On top of eliminating cells with damaged DNA by apoptosis or senescence, p53 is also capable of enhancing the processivity of DNA replication forks (6). Other groups reported similar findings, considering various mechanisms of how p53 might enhance DNA replication, e.g., through avoiding topological stress (7), inducing DNA polymerase-eta and translesion synthesis (8), orchestrating fork restart (9), or enhancing the levels of pCDKN1A/p21 and its association with PCNA (10). In other systems, p53 can also compromise DNA replication through p53-associated exonuclease activity and DNA polymerase-iota (11), and forced CDKN1A/p21 synthesis impairs DNA replication in UV-irradiated cells (12). These findings are at least not easy to reconcile, underscoring the need for further investigations on how the p53–Mdm2 system affects DNA replication. In our hands, the p53 target gene product Mdm2 is supporting DNA replication fork progression, similar to p53 itself (6). This suggests that the induction of Mdm2 constitutes a major mechanism of how p53 supports replication. This notion, however, did not answer the question by what mechanism(s) Mdm2’s impact on DNA replication could be explained.
Conflicts between transcription and DNA replication appear to represent a major cause of replication stress (13). Such conflicts may not simply result from collisions between RNA and DNA polymerases, but rather from the cotranscriptional occurrence of DNA:RNA hybrids with an additional single DNA strand, so-called R loops. R loops seem to hinder DNA replication forks from progressing (14). This raises the possibility that chromatin modifications, e.g., through Mdm2 or PRCs (1), might help to avoid R loops and ease DNA replication fork progression.
Here we show that Mdm2 and PRCs act similarly to sustain DNA replication fork progression. The Ring finger domain of Mdm2, conferring the ubiquitin ligase function, is essential for smooth DNA replication. Furthermore, Mdm2 and the PRC1 component RNF2 can substitute for each other in this function, suggesting that both ubiquitin ligases employ similar mechanisms to facilitate DNA replication through chromatin modification. And indeed, the depletion of either ubiquitin ligase induces R loops, whereas DNA replication is restored by ribonuclease H1 (RNaseH1)-mediated removal of R loops. Integrating these findings, we propose that Mdm2 and PRC1 contribute to DNA replication fork processivity through the avoidance of R loops by chromatin modification.
Results
Depletion of Mdm2 or Polycomb Repressor Complex 2 Members Similarly Decreases DNA Replication Fork Progression.
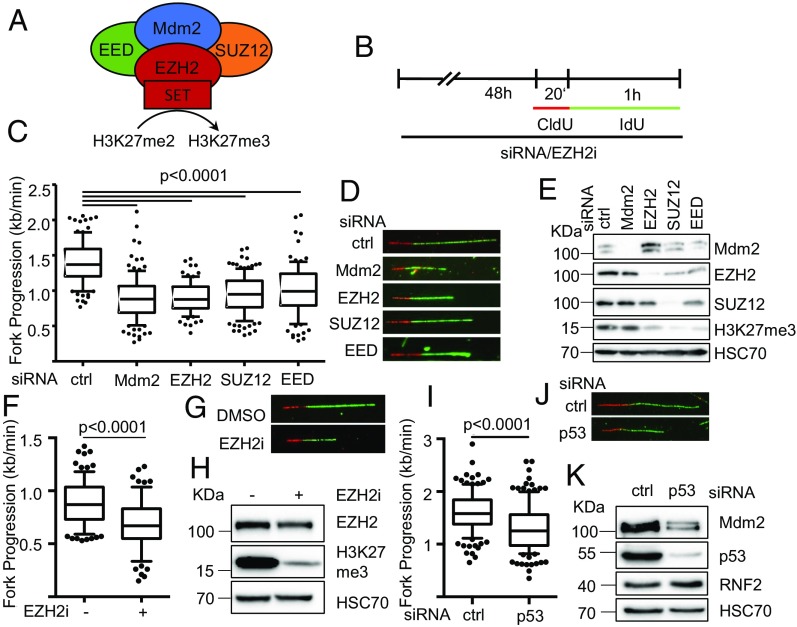
Our previously published results suggested that Mdm2 induces chromatin modifications with a pattern analogous to PRCs, through a mechanism that involves the physical association between Mdm2 and PRC2 (1). On the other hand, we observed that p53 and Mdm2 are required to support DNA replication fork progression (6). To understand whether the chromatin modifier activity of Mdm2 might also be involved in the support of DNA replication, we first compared the depletion of Mdm2 and PRC2 members regarding their impact on DNA replication fork progression (Fig. 1A). To address this, we employed the p53−/− lung large cell carcinoma cell line H1299 (15) to knock down Mdm2 or the PRC2 components EZH2, EED, or SUZ12 by siRNA, followed by fiber assays for DNA replication (Fig. 1B). Strikingly, the depletion of all tested PRC2 components led to a marked decrease in DNA replication fork progression, to a similar extent as the removal of Mdm2 (Fig. 1 C–E and SI Appendix, Fig. S1 A–H). In the case of EZH2, this is consistent with a recently published result indicating that EZH2-depleted cells resume replication to a lesser extent upon temporary fork stalling (16). Similarly, a pharmacological inhibitor of EZH2 catalytic activity as a histone methyl transferase compromised DNA replication (Fig. 1 F–H and SI Appendix, Fig. S1 I–M). The impact of Mdm2 on DNA replication was still observed in a p53-proficient cell line with Mdm2 gene amplification. Here, depleting p53 and concomitant Mdm2 loss led to a reduction in the fork rate as well, arguing that even high amounts of Mdm2 can still be rate limiting for its function in DNA replication (Fig. 1 I–K and SI Appendix, Fig. S1 N and O). Thus, interfering with PRC2 and its activity phenocopies the impact of Mdm2 depletion on DNA replication, providing a first hint that Mdm2 and PRC2 might affect DNA replication by similar pathways.
Fig. 1.
Compromised DNA replication fork progression upon depletion of Mdm2 or PRC2 members. (A) Schematic diagram of the main components of the PRC2 complex and its interaction partner Mdm2 that have been subjected to replication studies. siRNA transfections targeting Mdm2, EZH2, SUZ12, and EED as well as an inhibitor targeting the SET domain of EZH2 were used. (B) H1299 cells were transfected with siRNAs each against the targets Mdm2, EZH2, SUZ12, and EED or treated with the EZH2 inhibitor EPZ-6438 for 48 h and incubated with 5′-chloro-2′-deoxyuridine (25 μM CldU, 20 min) followed by 5′-iodo-2′-deoxyuridine (25 μM IdU, 60 min) as indicated. (C) Fork progression was determined from the track length of the second label (IdU; kilobases per minute) and displayed in a boxplot with 10–90 percentile whiskers. (D) Representative images of tracks of newly synthesized DNA were visualized by immunostaining of CldU (red) and IdU (green). (E) Cells were treated as in B and subjected to immunoblot analysis. Knockdowns were confirmed by staining for the target proteins as well as the target of active PRC2, i.e., trimethylated histone 3 (H3K27me3). (F) H1299 cells were treated with 5 μM of the EZH2 inhibitor EPZ-6438 for 48 h, followed by incubation with CldU and IdU as in B. Boxplot analysis of the fork progression during the time of IdU label with 10–90 percentile whiskers. (G) Representative labeled tracks were immunostained as described in D. (H) Cells were treated as in F and subjected to immunoblot analysis of H3K27me3 as a readout for inhibitor activity. SJSA cells were transfected with siRNA targeting p53 for 48 h followed by incubation with CldU (25 μM; 20 min) and IdU (25 μM; 60 min) for fiber analysis. (I) The fork rate of replication during the IdU label is displayed in boxplots with 10–90 percentile whiskers. (J) Representative images of CldU (red) and IdU (green) labeled tracks. (K) Immunoblot analysis of proteins after siRNA treatment described in I to confirm p53 depletion and protein levels of Mdm2 and RNF2 in response to p53 knockdown. In addition to the DNA fiber assays shown in this figure, replicates with the siRNAs used, alternative siRNAs, as well as in HCT116 p53−/− cells, are shown in SI Appendix, Fig. S1.
Interfering with the Polycomb Repressor Complex 1 Slows Down DNA Replication Forks and Cell Proliferation.
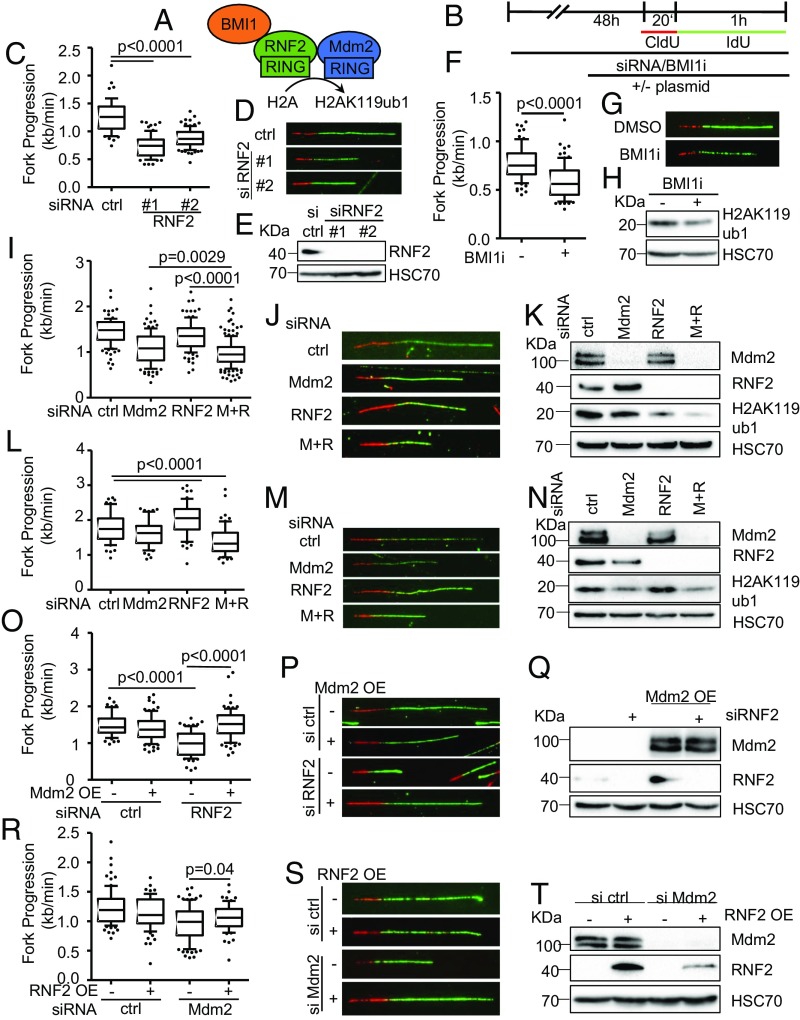
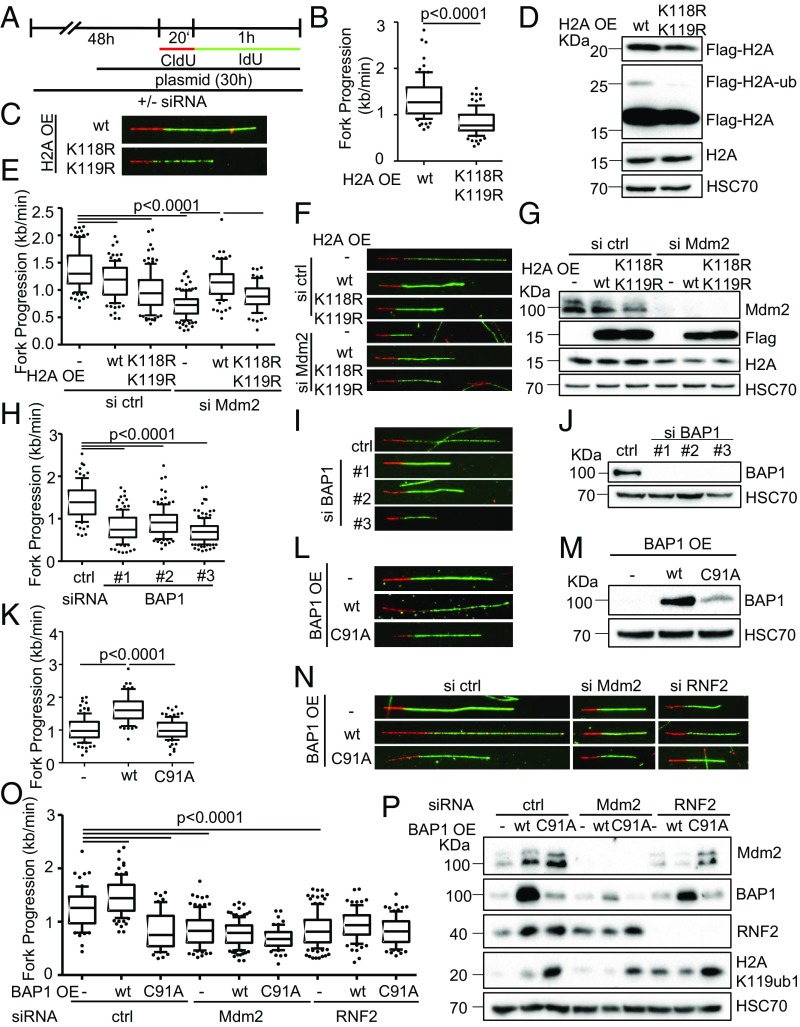
Besides the trimethylation of histone H3 at K27, Mdm2 also supports the ubiquitination of histone H2A at K119 (1), perhaps as a result of direct ubiquitin ligase activity (17). This site is mostly known for its ubiquitination by PRC1, typically through its RNF2 component (4). To test whether RNF2 knockdown might affect DNA replication in a similar manner as Mdm2 depletion, we performed fiber assays and found reduced DNA replication fork progression again (Fig. 2 A–E and SI Appendix, Fig. S2 A and B). Analogous observations were made in the presence of a pharmacological inhibitor of Bmi1, another PRC1 component (Fig. 2 F–H and SI Appendix, Fig. S2 C and D). When depleting Mdm2 and RNF2 simultaneously, an additive effect on the progression of DNA replication forks was observed (Fig. 2 I–K and SI Appendix, Fig. S2 H and I). Similarly, the codepletion of Mdm2 and RNF2 also impaired fork progression and H2A ubiquitination in nontransformed retinal pigment epithelial (RPE) cells with a targeted deletion of TP53; however, the single knockdowns had little effect (Fig. 2 L–N and SI Appendix, Fig. S2 K and L), suggesting that sufficient levels of Mdm2 and RNF2 can at least partially substitute for each other. We propose that Mdm2 and RNF2 might become rate limiting for DNA replication preferably in transformed compared with normal cells, perhaps constituting a therapeutic window when targeting this activity. To further explore whether Mdm2 might support DNA replication by a similar mechanism as PRC2, we performed rescue experiments upon knockdown of Mdm2 or RNF2. As expected, the overexpression of each component reconstituted DNA replication when it had been depleted by siRNA (SI Appendix, Fig. S2 E–G). Although the siRNA could also target the overexpressed Mdm2, the remaining amounts of Mdm2 still exceeded the endogenous levels, explaining the rescue of DNA replication (SI Appendix, Fig. S2E). Of note, however, the overexpression of Mdm2 also rescued DNA replication when RNF2 had been depleted (Fig. 2 L–Q and SI Appendix, Fig. S2M) while overexpression of RNF2 partially restored fork progression upon Mdm2 knockdown (Fig. 2 O–T and SI Appendix, Fig. S2N). Thus, enhanced Mdm2 levels can compensate for a lack of RNF2 and vice versa. Since both proteins are capable of enhancing H2A ubiquitination at K119 (1, 17), this at least suggests that the ubiquitination of this histone enables smooth DNA replication.
Fig. 2.
Decreased fork progression upon RNF2 depletion or Bmi1 inhibition. (A) Diagram of the catalytic subunit of the PRC1 complex mediating H2AK119ub1, RNF2, that was targeted by siRNA transfections as well as another component of the complex, BMI1, targeted by an inhibitor. (B) H1299 and RPE p53−/− were depleted of endogenous RNF2 and Mdm2 by siRNA transfection and BMI1 inhibited for 48 h, transfected with plasmid DNA for 30 h, and labeled with CldU (20 min) and IdU (60 min) for fiber analysis. (C) Boxplot analysis of IdU-labeled tracks upon RNF2 depletion using two siRNAs with 10–90 percentile whiskers. (D) Representative images of labeled tracks after immunostaining of CldU (red) and IdU (green). (E) Immunoblot analysis of RNF2 after treatment described in B, confirming the depletion of RNF2 with two siRNAs. (F) H1299 cells were treated with 1 μM of the BMI1 inhibitor PTC-209 for 48 h, followed by fiber analysis. Fork progression analysis of the IdU label with representative images in G. (H) Immunoblot analysis detecting monoubiquitinated histone 2A (H2AK119ub1) as a readout of inhibitor activity after 48 h of treatment with BMI1 inhibitor. (I) Mdm2 and RNF2 were codepleted in H1299 cells for 48 h. Fiber analysis was carried out as described in B and displayed as fork rate in the IdU label. (J) Immunostained tracks of CldU (red) and IdU (green) in representative images. (K) Cells treated as described in I were subjected to immunoblotting and used to confirm depletion of Mdm2 and RNF2 as well as H2AK119ub1. (L) Immortalized retinal pigment epithelial (RPE) cells with a targeted deletion of TP53 were transfected with siRNA to deplete Mdm2 and/or RNF2 for 48 h. Subsequently, cells were labeled with CldU and IdU (25 μM; 20 min and 60 min, respectively) for fiber analysis, and fork rates were determined from the IdU label. (M) Labeled tracks were immunostained in red (CldU) and green (IdU). (N) Immunoblot analysis to confirm knockdown efficiency as well as the monoubiquitinated form of histone H2A at Lys119. (O) H1299 cells were first transfected with siRNAs, followed by a plasmid transfection after 24 h. After another 30 h, samples were subjected to fiber assay labeling with 25 μM CldU (20 min) and 25 μM IdU (60 min). Fork progression in the IdU label after RNF2 depletion and plasmid transfection to overexpress Mdm2 is displayed in boxplots with 10–90 percentile whiskers. (P) Fluorescently labeled tracks of CldU (red) and IdU (green). (Q) Immunoblot staining for Mdm2 and RNF2 confirms knockdown and overexpression described in O. (R) Fork rate of the IdU label after Mdm2 depletion and ectopic expression of RNF2. (S) Labeled tracks of CldU and IdU stained in red and green, respectively. (T) Total protein levels of Mdm2 and RNF2 were assessed to confirm transfection efficiencies. In addition to the DNA fiber assays shown in this figure, replicates are shown in SI Appendix, Fig. S2.
Mdm2 Supports DNA Replication Processivity as Well as Fork Restart.
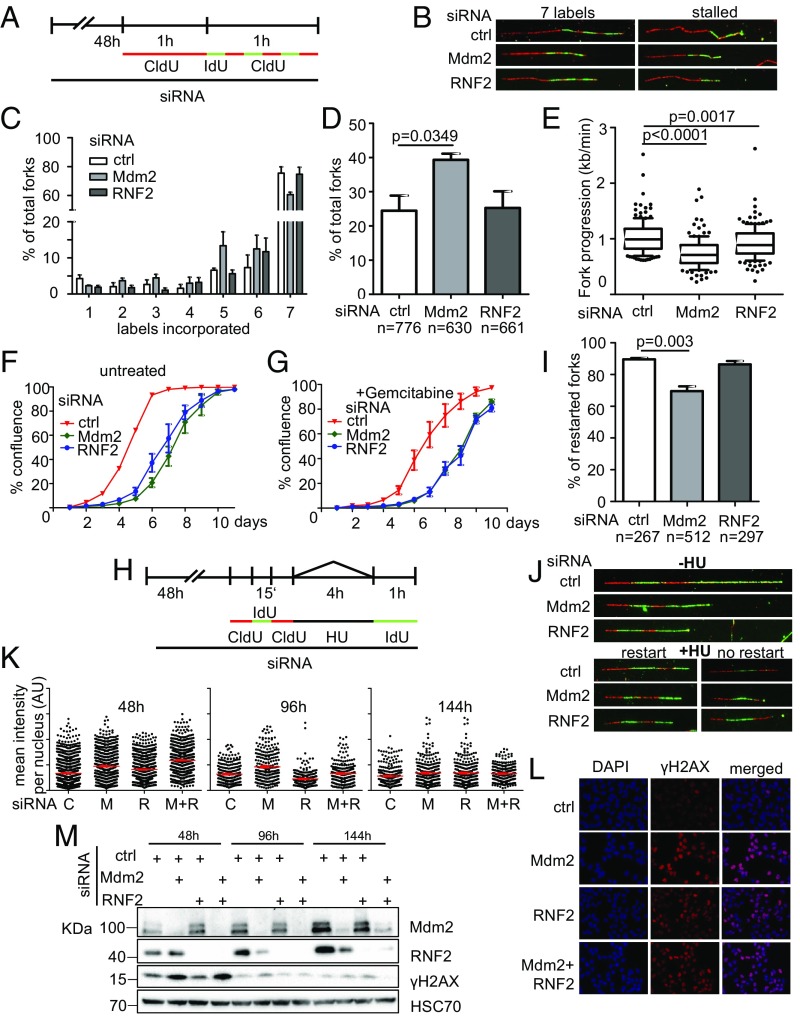
To assess whether the differences seen in fork progression rates are a result of increased fork stalling or decreased velocity of the replication fork, we applied a modified labeling protocol for DNA fiber assays as described in ref. 6, changing the label of newly synthesized DNA for multiple brief periods of time, allowing the exact determination when a replication fork had stalled. Mdm2 depletion led to an increase in fork stalling indicated by fibers with less than seven labels incorporated (Fig. 3 A–D). Fork velocity in continuously replicating stretches (labels 2–5 only) was affected by either depleting Mdm2 or RNF2 from the cells (Fig. 3E). In conclusion, Mdm2 is required for proper DNA replication fork processivity and also velocity, whereas RNF2 primarily contributes to fork velocity. In addition to direct effects on replication forks, cells depleted of Mdm2 and RNF2 were also less able to give rise to progeny (Fig. 3F and SI Appendix, Fig. S3 A–D). This effect was also seen when exogenous replicative stress was induced by a 24-h gemcitabine treatment (Fig. 3G).
Fig. 3.
Mdm2 depletion impairs genomic stability. (A) Fork processivity was analyzed as described previously (6). First, cells were transfected with scrambled siRNA (control) or siRNAs targeting Mdm2 and RNF2 for 48 h and then labeled with CldU for 1 h, followed by short (10 min) pulses of IdU and CldU for a total of seven labels. From this, the length of labels 2–5 was used for fork progression analysis and the number of labels incorporated for fork stalling analysis. (B) Representative images of replicated stretches that have incorporated all seven labels as well as stalled ones that contain fewer than seven labels. (C) The number of forks that proceeded through n labels is displayed for control, Mdm2, and RNF2 knockdown. Seven labels reflect full progression of the fork throughout the entire labeling time. Numbers lower than 7 indicate premature termination during the labeling time. (D) The percentages of forks with fewer than seven labels indicate that Mdm2 and RNF2 knockdowns cause replication to run in a less processive manner than cells transfected with scrambled (control) siRNA. (E) Fork velocity was determined through the length of labels 2–5 (kilobases per minute) which ensures that fork stalling (as seen in C and D) is not affecting velocity measurements. (F and G) To assay for cell proliferation, H1299 cells were transfected with siRNAs against Mdm2 and RNF2. Twenty-four hours posttransfection, cells were treated with 10 nM gemcitabine (in H2O) for another 24 h. Confluence of transfected cells (F) and with additional gemcitabine (10 nM) treatment (G) was analyzed every 24 h. (H) Fork restart was assessed by subjecting siRNA-transfected cells to three short pulses of CldU, IdU, and CldU (50 μM; 15 min each) followed by treatment with 2 mM hydroxyurea for 4 h and a subsequent long pulse of IdU of 60 min (50 μM). Controls were labeled with IdU immediately after the short pulses, omitting hydroxyurea treatment. (I) The percentage of forks that restart DNA synthesis after hydroxyurea treatment from three independent experiments is displayed for Mdm2- and RNF2-depleted cells as well as for control transfections. (J) Representative images of labeled tracks in the absence of HU treatment as well as restarted forks and those that have impaired fork restart and contain only the first three labels. (K) Immunofluorescent staining for the DNA damage marker γH2AX of cells codepleted of Mdm2 and RNF2, at 2, 4, and 6 d post knockdown. Fluorescence intensity was quantified by Fiji software and normalized to the median intensity of the control siRNA experiment at each time point. (L) Representative images of immunostaining 48 h after Mdm2 and RNF2 depletion stained for γH2AX. (M) Immunoblot analysis of protein lysates harvested in parallel to immunofluorescent samples in K. Stainings for Mdm2 and RNF2 to determine knockdown efficiency as well as for γH2AX to reflect the DNA damage response. A biological replicate of the immunofluorescence data shown in this figure is displayed in SI Appendix, Fig. S3.
To assess whether Mdm2 or RNF2 might also support the integrity of stalled replication forks and prevent their collapse, we first treated Mdm2- or RNF2-depleted cells with hydroxyurea (HU) to stop DNA replication. Subsequent release from this block revealed that most control-transfected and also RNF2-depleted cells retained the capability to restart such replication forks. In contrast, Mdm2-depleted cells contained a substantial fraction of replication forks that failed to restart (Fig. 3 H–J). This argues that Mdm2 is also essential for the maintenance of replication forks upon stalling. Finally, we observed that the DNA damage response, as revealed by H2AX phosphorylation, was induced upon depletion of Mdm2 and/or RNF2, consistent with replication stress (Fig. 3 K and L). After 4 or 6 d, however, this response was no longer detectable, despite continued knockdown (Fig. 3M), suggesting that the cells can adapt to the depletion in the long run.
The Ring Finger Domain of Mdm2 Is Necessary to Sustain DNA Replication.
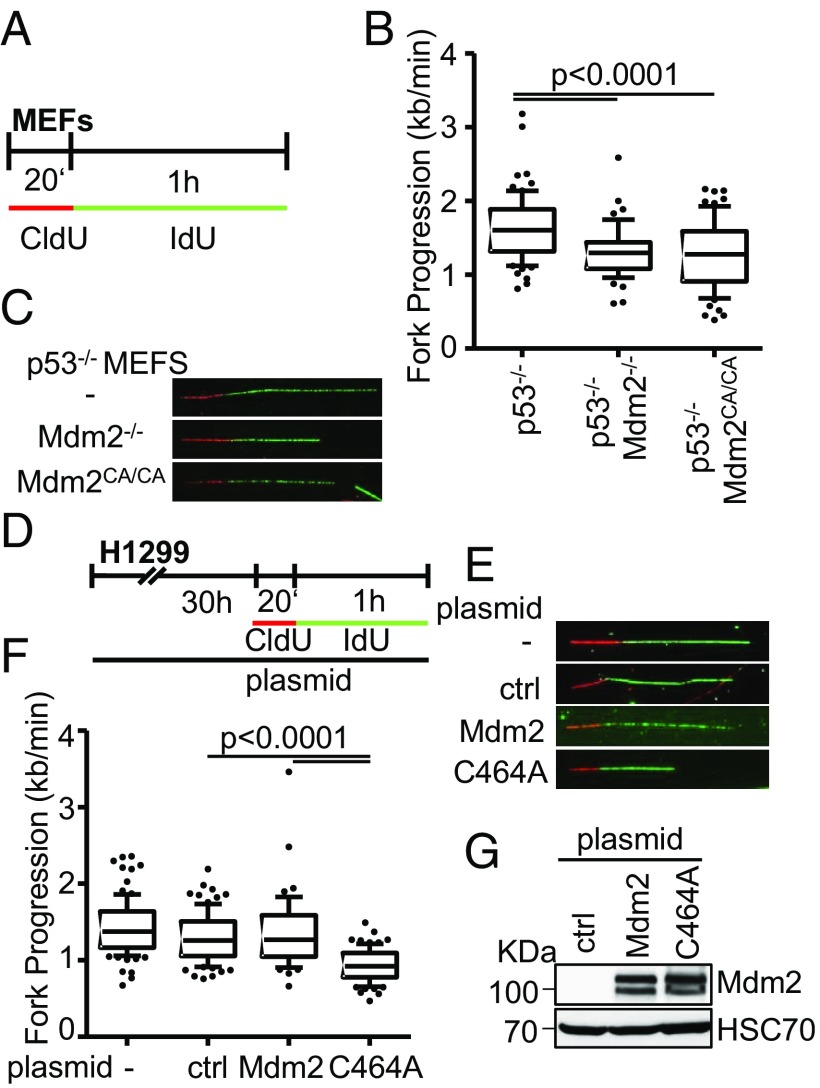
The chromatin modifier function of Mdm2 requires its Ring finger domain, suggesting that the E3 ubiquitin ligase activity of this domain is involved (1). We therefore sought to test whether the same is true for the support of DNA replication by Mdm2. We first employed murine embryonic fibroblasts (MEFs) from animals that either had wild-type (WT) Mdm2 on a p53−/− background, or otherwise lacked both Mdm2 and p53, or had the p53−/− background with a biallelic point mutation in the Mdm2 gene that gave rise to the mutation C464A, disrupting the Ring finger structure (18) (Fig. 4A). Strikingly, the knockout of Mdm2, as well as the Ring finger knockin, each led to reduced DNA replication fork progression compared with p53 single knockout cells (Fig. 4 B and C and SI Appendix, Fig. S4 A and B). Similar results were obtained with H1299 cells. While Mdm2 overexpression had little effect on DNA replication, the overexpression of Mdm2 with a Ring finger mutation markedly reduced fork progression, presumably as a result of a dominant negative effect (Fig. 4 D–G and SI Appendix, Fig. S4 C and D). Taken together, these results strongly suggest that Mdm2 requires the Ring finger domain and its ubiquitin ligase activity to sustain DNA replication independent of p53.
Fig. 4.
The Ring finger domain of Mdm2 supports replication fork progression. (A) Mouse embryonic fibroblasts lacking p53, alone or in combination with a deletion of Mdm2, or a biallelic point mutation in the RING domain of Mdm2 (C462A), were subjected to fiber assay labeling with CldU (20 min) and IdU (60 min). (B) Boxplot analysis of fork progression in the IdU label when either Mdm2 or just its RING domain activity was depleted with 10–90 percentile whiskers. (C) Representative images of the labeled tracks of CldU (red) and IdU (green). (D) H1299 cells were subjected to plasmid transfection for 30 h, followed by DNA fiber assay labeling with CldU for 20 min and IdU for 60 min. (E) Fluorescently labeled tracks of CldU (red) and IdU (green) in untransfected samples as well as upon transfection with the control plasmid pcDNA3 or the expression plasmids pCMV-Mdm2 and pCMV-Mdm2C464A (note that human Mdm2 carries the corresponding cysteine residue that forms part of the Ring finger structure at position 464). (F) Analysis of fork progression in the IdU label shows a significant reduction upon overexpression of RING-mutant Mdm2 but not wild-type Mdm2. (G) Immunoblot analysis upon transfection as in D confirms overexpression of Mdm2 with both a wild-type and a mutant plasmid carrying a point mutation in the RING finger. Two further replicates to the fiber assays in this figure are shown in SI Appendix, Fig. S4.
H2A Ubiquitination Is Essential for DNA Replication Fork Progression.
For mechanistic understanding, we first determined the levels of helicases and other factors involved in RNA folding (senataxin, aquarius, DHX9, and hnRNP K) by immunoblot analysis upon depletion of Mdm2 and RNF2, based on the possibility that ubiquitin ligases might destabilize them. However, we did not observe obvious changes in the corresponding protein levels (SI Appendix, Fig. S2J). Thus, we considered the hypothesis that histone ubiquitination might be key to DNA replication. Previous work revealed that Mdm2, like RNF2, governs the ubiquitination of histone 2A (H2A) at K119 (1, 17). To find out whether this ubiquitination is required to sustain DNA replication, we overexpressed H2A from plasmids, either as WT or with a mutation at the ubiquitination sites K118/K119 (Fig. 5A) (19), followed by fiber assays. In the presence of the mutation, fork progression was considerably impaired compared with WT H2A (Fig. 5 B–D and SI Appendix, Fig. S5 A–C). Furthermore, while the ubiquitination-competent H2A rescued the defect of replication in Mdm2- or RNF2-depleted cells, the H2A mutant largely failed to do so (Fig. 5 E–G and SI Appendix, Fig. S5 D and E). We conclude that the ubiquitinated residues near the carboxy terminus of H2A are key to DNA replication. Next, we investigated the impact of BAP1, a bona fide deubiquitinating enzyme for H2A (20). Depletion of BAP1 impaired the progression of replication forks (Fig. 5 H–J and SI Appendix, Fig. S5 F and G), as previously reported in different cell systems (21), while overexpression of BAP1, but not a defective mutant of it (22) considerably enhanced the progression of replication forks (Fig. 5 K–M and SI Appendix, Fig. S5 H and I). This argues that not only the ubiquitination of H2A, but also its timely deubiquitination support DNA replication. In accordance, overexpressing BAP1 failed to rescue the defect imposed by Mdm2 depletion (Fig. 5 N–P and SI Appendix, Fig. S5 J and K), further suggesting that the dynamics of ubiquitination as well as deubiquitination are determining fork progression.
Fig. 5.
Dynamic changes in H2A ubiquitination are required for DNA replication. (A) H1299 cells were transfected with siRNA for a total of 48 h, and with plasmid DNA for 30 h before DNA fiber or immunoblot analysis. (B) Cells transfected with plasmids to express wild-type H2A (pcDNA3.1-Flag-H2A) or a mutant lacking the lysine residue that is ubiquitinated by Mdm2 and RNF2 (K119; pcDNA3.1-Flag-H2A-K118-119R) were assessed regarding their fork progression in the IdU label. (C) Representative images of fluorescently labeled tracks corresponding to experiment in B. (D) Immunoblot staining of cells treated as in B for Flag-tagged H2A expressed from plasmids. Note that the Upper band represents ubiquitinated H2A and is observed only with wild-type H2A. (E) Cells were transfected with siRNA targeting Mdm2 followed by ectopic expression of wild-type or mutant (K118-119R) H2A or an empty control plasmid. Fiber analysis was carried out 30 h post plasmid transfection using 50 μM CldU and IdU (20 and 60 min). (F) Representative labeled tracks of CldU (red) and IdU (green) of Mdm2-depleted cells with ectoptic H2A expression. (G) Immunoblot analysis to confirm Mdm2 depletion and the expression of Flag-tagged H2A plasmids. (H) H1299 cells depleted of BAP1 using three different siRNAs. Transfections were carried out for 48 h, followed by DNA fiber analysis to monitor replication fork progression. The track lengths of the IdU label are displayed in a boxplot. (I) Fluorescently labeled tracks of control and BAP1-siRNA transfected H1299 cells corresponding to H. (J) Immunoblot analysis of samples treated as in H to confirm depletion of BAP1 with siRNAs. (K) H1299 cells were transfected with plasmids expressing the deubiquitinating enzyme BAP1 and a catalytically inactive point mutant of BAP1 (C91A) for 30 h. Fiber analysis was carried out using 50 μM CldU and IdU for 20 and 60 min, respectively. (L) Fluorescent staining of CldU (red) and IdU (green)-labeled tracks corresponding to K. (M) Ectopic expression of BAP1 from plasmids was assessed by immunoblotting and staining with an antibody to BAP1 after 30 h of transfection. (N and O) Cells depleted of Mdm2 and RNF2 were transfected with plasmids encoding BAP1 as wild-type or a catalytically inactive mutant and subjected to fork progression analysis. Representative images of labeled tracks in N and fork progression rates in O. (P) Lysates of cells treated as in O were assessed for levels of Mdm2, RNF2, BAP1, as well as the substrate of the three enzymes, H2AK119ub1. Replicates to the fiber assays in this figure are shown in SI Appendix, Fig. S5.
Mdm2 Depletion Enhances R-Loop Formation.
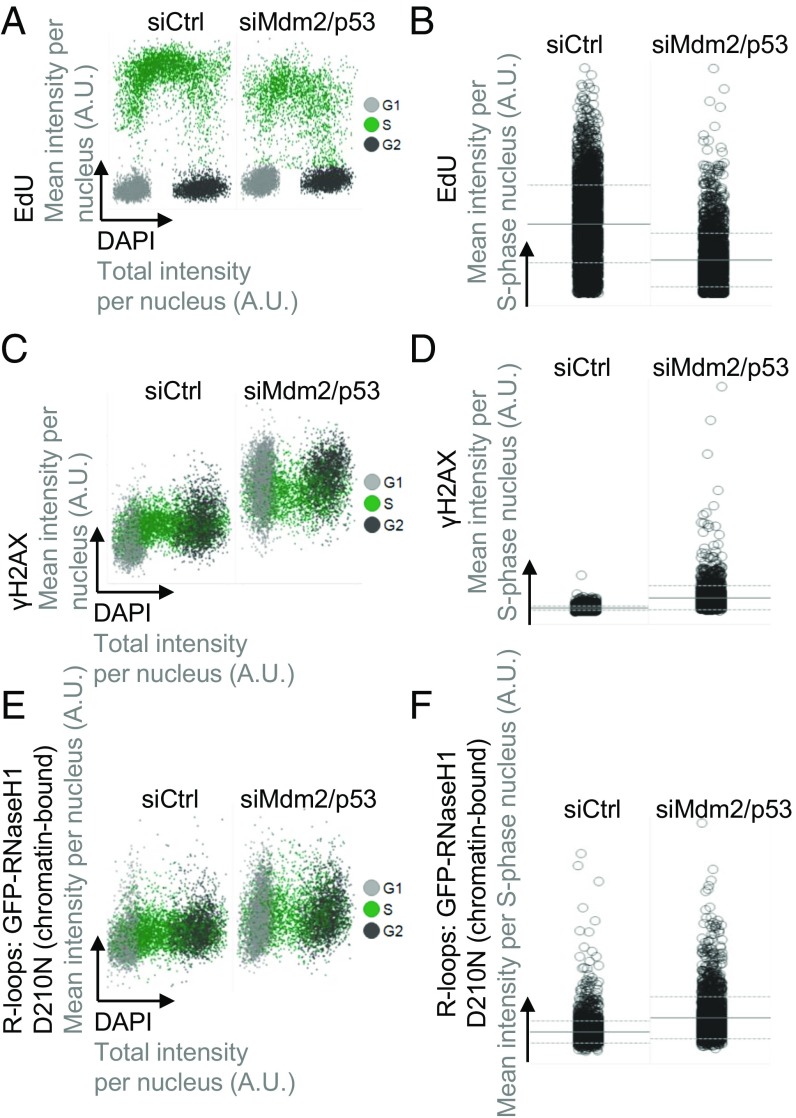
In search of a mechanism that might enhance DNA replication through histone ubiquitination, we next tested the role of R loops, i.e., DNA:RNA hybrids that typically form during dysregulated transcription and can represent obstacles to DNA polymerases during DNA replication (23). PRC-mediated histone modifications often repress transcription (4). In parallel, we had previously observed similar repression patterns and histone modifications as a function of Mdm2 (1). Therefore, we reasoned that R-loop formation might occur in response to the depletion of PRC components or Mdm2, providing a plausible explanation for impaired DNA replication fork progression. RNaseH1 is an enzyme that recognizes RNA:DNA hybrid structures and cleaves the RNA moiety. Mutagenic modifications of the catalytic triad in its C terminus render RNaseH1 catalytically inactive while retaining its RNA:DNA hybrid binding ability (24). We made use of an inducible cell system carrying the D210N mutation in GFP-tagged RNaseH1 to specifically detect R loops by quantitative image-based cytometry (25, 26) (SI Appendix, Fig. S6 A and B). In parallel to R loops, we also detected DNA synthesis and DNA damage signaling in the same system. In line with our fiber studies, EdU incorporation was reduced after depleting Mdm2 and p53 (Fig. 6 A and B and SI Appendix, Fig. S6 A and B), reflecting impaired DNA replication. Furthermore, we observed increased levels of the DNA damage marker γH2AX (Fig. 6 C and D and SI Appendix, Fig. S6 C and D), again reflecting replication stress. Strikingly, the amounts of chromatin-bound RNase-H1-D210N were elevated in the absence of Mdm2, indicating that depletion of Mdm2 increases R-loop formation (Fig. 6 E and F and SI Appendix, Fig. S6 E and F). In conclusion, loss of Mdm2 increases the persistence of nonphysiological levels of R loops.
Fig. 6.
Mdm2 depletion increases replicative stress and R-loop formation. U2OS cells with stably integrated, inducible noncatalytic GFP-RNaseH1-D210N mutant were transfected with siRNA against p53 and Mdm2 for 48 h, before mutant RNaseH1 induction by doxycycline treatment for another 24 h. Next, the cells were pulsed with EdU to detect DNA synthesis for 20 min, preextracted, fixed, and stained. The signals from DAPI, EdU, γH2AX, and chromatin-bound RNaseH1-D210N were quantified by high-content microscopy. The D210N mutation abolishes RNase activity but retains binding to RNA:DNA hybrids and can thus be used to detect them. For this analysis, the total number of cells analyzed was n = 7,545 in control knockdown and 7,412 in p53/Mdm2-depleted cells (A, C, and E). S phase-specific data were collected from n = 3,457 in controls and n = 2,225 in p53/Mdm2-depleted samples (B, D, and F). (A) Two-dimensional cell cycle staging was performed based on the EdU and DAPI signals. (B) The S phase-specific EdU signals were plotted for Mdm2/p53-depleted cells versus a control siRNA transfection. Horizontal lines represent averages and SDs. (C) Levels of the DNA damage marker γH2AX increase upon depletion of Mdm2 and p53 throughout the cell cycle. (D) The S phase-specific γH2AX signals were plotted for Mdm2/p53-depleted cells versus a control siRNA transfection. Horizontal lines represent averages and SDs. (E) Chromatin-bound RNaseH1-D210 marks RNA:DNA hybrids, and this signal increased when Mdm2 and p53 were depleted. (F) The S phase-specific signals of chromatin-bound RNaseH1-D210N were plotted for Mdm2/p53-depleted cells versus a control siRNA transfection. Horizontal lines represent averages and SDs. A schematic diagram and replicate of the experiment in this figure is shown in SI Appendix, Fig. S6.
CDK9 Inhibition and RNaseH1 Overexpression Each Allow Processive DNA Replication Despite the Removal of Mdm2 or RNF2.
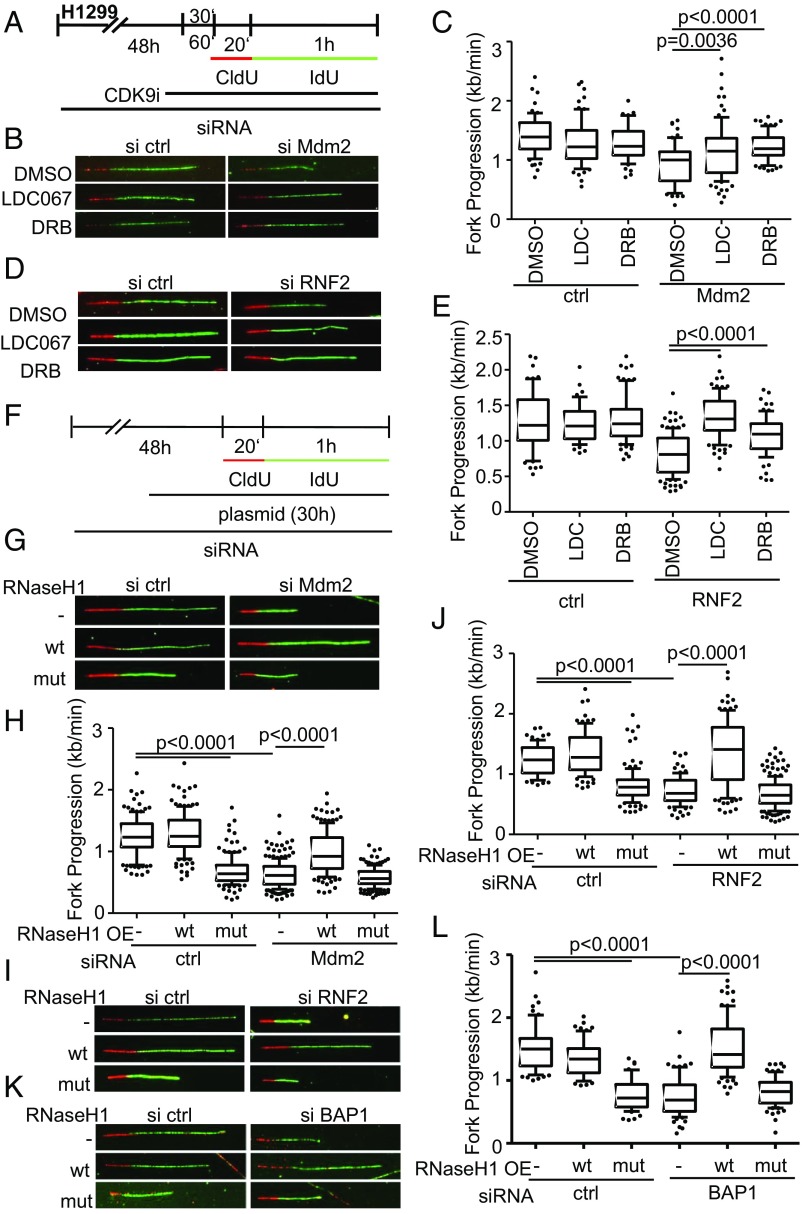
We next sought to determine whether conflicts with transcription cause replication stress when Mdm2 is depleted. One way of addressing these conflicts is by inhibiting CDK9, a kinase essential for transcriptional elongation (27). And indeed, CDK9 inhibition was previously found to restore DNA replication upon p53 knockdown (7). Importantly, we observed that two different CDK9 inhibitors restored DNA replication upon depletion of Mdm2 (Fig. 7 A–C and SI Appendix, Fig. S7 A–C) and RNF2 (Fig. 7 D and E and SI Appendix, Fig. S7 D and E). Thus, ongoing transcription and perhaps the resulting R-loop formation represent a prerequisite for impaired DNA replication upon Mdm2 depletion.
Fig. 7.
CDK9 inhibition and RNaseH1 overexpression each allow processive DNA replication despite the removal of Mdm2 or RNF2. (A) H1299 cells were transfected with siRNA against Mdm2 for 48 h. After 47 h, cells were additionally treated with 10 μM LDC067 and 25 μM 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB), two CDK9 inhibitors, and a solvent control (DMSO). After a 1-h preincubation with the inhibitors, the cells were incubated with nucleoside analogs CldU (20 min) and IdU (60 min) containing inhibitors and solvent control. (B) Representative images of labeled tracks of CldU (red) and IdU (green). (C) Fork progression analysis was carried out from IdU-labeled tracks and displayed in a boxplot with 10–90 percentile whiskers. (D) H1299 cells transfected with RNF2 siRNA and pretreated with 10 μM LDC067 and 25 μM DRB for 30 min. Representative images of labeled tracks of CldU (red) and IdU (green) corresponding to E. (E) Fiber assay was carried out as described in A. (F) H1299 cells were first transfected with siRNA targeting Mdm2/RNF2 and with a plasmid containing wild-type RNAseH1 (pICE-RNAseH1-NLS-mCherry) as well as a catalytically inactive mutant (pICE-RNASeH1-D10-E48R-NLS-mCherry) and a control plasmid (pcDNA3) 24 h after the first transfection. After another 30 h, samples were used for fiber assay labeling by incubation with 25 μM CldU (20 min) and 25 μM IdU (60 min). (G) Representative images of immunostained tracks of CldU (red) and IdU (green) after Mdm2 depletion and RNAseH1 overexpression. (H) Boxplot analysis of fork progression in the IdU label of the fiber assay indicates that active RNase H restores DNA replication. (I) Fluorescently labeled tracks of CldU (red) and IdU (green). Fork progression rates of samples depleted of RNF2 and with overexpressed RNAseH1 are shown in J, again indicating a rescue of DNA replication. (K) H1299 cells were transfected with BAP1-targeting siRNA for 48 h in total and plasmids (pICE-NLS-mCherry, pICE-RNAseH1-NLS-mCherry, and pICE-RNAseH1-NLS-mCherry) for 30 h followed by fiber analysis. Representative images of the experiment display CldU in red and IdU-labeled tracks in green. (L) Boxplot analysis of fork progression in the IdU label of the fiber assay indicates that active RNase H restores DNA replication. In addition to replication assays shown here, two biological replicates can be found in SI Appendix, Fig. S7.
Finally, we tested whether DNA:RNA hybrids in Mdm2/RNF2-depleted cells represent an obstacle to DNA replication. We overexpressed RNase H1 to resolve hybrids that form while depleting Mdm2 or RNF2. Wild-type RNaseH1 but not a catalytically inactive mutant of it gave rise to a robust rescue of DNA replication fork progression upon Mdm2 depletion (Fig. 7 F–H and SI Appendix, Fig. S7 F, H, and J), and also upon RNF2 knockdown (Fig. 7 I and J and SI Appendix, Fig. S7 G, I, and J). RNaseH1 also rescued DNA replication upon depletion of BAP1 (Fig. 7 K and L and SI Appendix, Fig. S7K). Taken together, these results strongly suggest that Mdm2 prevents the persistence of R loops and replicative stress.
Discussion
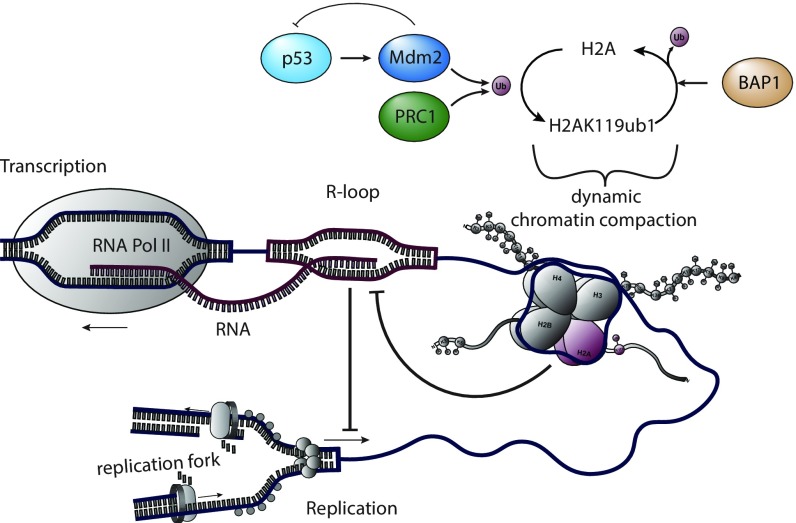
Mdm2 and PRCs not only display similar patterns in chromatin modification and gene regulation (1, 2) but also enable DNA replication with high processivity, independent of p53. Our results strongly suggest that these two activities are carried out through overlapping mechanisms. Both Mdm2 and PRC1/RNF2 have an E3 ubiquitin ligase function, and this sustains DNA replication. Moreover, Mdm2 and RNF2 can substitute for each other in this regard. Accordingly, H2A ubiquitination and deubiquitination are each required for unperturbed fork progression. When R loops are removed by RNase H1, replication is restored despite the depletion of Mdm2 or RNF2. Thus, we propose that Mdm2 and PRC1 each support DNA replication through their ability to ubiquitinate histones and prevent supraphysiological R-loop formation (Fig. 8).
Fig. 8.
The interdependence of histone ubiquitination and DNA replication. We propose that the ubiquitination of histone 2A by Mdm2 and RNF2, as well as its deubiquitination by BAP1, provide a dynamic system to prevent the formation of R loops and thereby the occurrence of replication stress.
Interestingly, members of PRC2 were found in association with DNA replication forks (28) and necessary for their symmetric progression (29). This raises the possibility that, on top of dampening unscheduled transcription, PRC2 might directly support DNA replication, perhaps enabling immediate histone modifications simultaneously with DNA synthesis. Of note, EZH2 activity recruits Mus81 to destabilize stalled replication forks (16). This might disrupt such forks on the one hand but might enable faster resumption of replication on the other hand. It is thus conceivable that PRC2 not only dampens transcription but also alters the dynamics of DNA replication by recruiting DNA processing factors.
Mdm2 and RNF2 each enhance the monoubiquitination of histone 2A at K119 (1, 17), and BAP1 mediates the removal of this modification (20). All three factors are required for proper DNA replication fork progression, while H2A overexpression only supports replication when the ubiquitination sites K118/119 are present. Thus, both the ubiquitination and the deubiquitination of H2A determine DNA polymerization. RNF2 and BAP1 as well as ubiquitinated H2A were also found associated with replication forks by isolation of proteins on nascent DNA (iPOND) (21). Interestingly, the ubiquitination of H2A by RNF168 (at K13/15) was recently reported to govern DNA replication as well (30). It is thus tempting to speculate that both the addition and the removal of ubiquitin to and from H2A, each at specific stages of DNA synthesis and histone incorporation, coordinate the synthesis of DNA with the concomitant duplication of chromatin structures.
Curiously, the overexpression of Mdm2 has previously been found to enhance rather than diminish replication stress (31). Of note, however, Mdm2 has long been known to impair cell proliferation when overexpressed (32), whereas our study is mainly based on the depletion of endogenous Mdm2. We therefore propose that Mdm2, when present at physiological levels, actually supports DNA replication.
Depletion of Mdm2 enhances detectable DNA:RNA hybrids, and this notion might explain previously reported observations. Both p53 (7) and Mdm2 (33) contribute to the resistance of cells to topoisomerase inhibitors, a class of drugs that induces R loops (34). Moreover, depleting p53 sensitizes cells toward codepletion of DHX9 (35), a helicase capable of removing R loops (36).
In a broader sense, RNA metabolism is becoming a major focus in current research on DNA replication stress. RNA processing factors constitute a major fraction of kinase substrates upon DNA damage (37, 38), making it plausible that such factors might affect DNA integrity, perhaps through R-loop formation. Recently, the oncogenic EWS–FLI1 fusion protein was found to cause R loops and thereby DNA damage (39). Our results expand this concept, implicating Mdm2 and polycomb repressors not only in the formation of a closed chromatin state, but also in the suppression of R loops, thereby enabling proper DNA replication. A number of mechanisms were suggested for R loops forming obstacles to DNA replication (40, 41). However, the exact nature of such conflicts between RNA:DNA hybrids and DNA replication still remains to be determined.
Besides its chromatin modifier function, Mdm2 is also capable of interfering with DNA repair through binding Nbs1, one of the MRN complex members (42, 43). However, this association negatively regulates MRN-mediated repair of double-stranded DNA breaks. Since Mdm2, according to our data, is a positive factor in DNA replication fork progression, we propose that this function is independent of Nbs1 binding. Of note, however, Mdm2 might also prevent the degradation of stalled replication forks, as has been reported for BRCA2 (44). Our observations suggest that Mdm2 is required for successful fork restart after transient, hyroxyurea-mediated fork stalling.
Since a minimum Mdm2 level is necessary for proper DNA replication, events that lower the amounts of intracellular Mdm2 might confer replication stress. For instance, Mdm2 levels drop in response to DNA damage, through phosphorylation and subsequent autoubiquitination of Mdm2 (45, 46). This suggests that impaired DNA replication might occur in response to DNA damage, not only as a direct result of DNA lesions, but also as a consequence of Mdm2 degradation. Furthermore, most cancers show impaired p53 activity, e.g., due to p53 mutations, resulting in lowered expression of the p53-responsive gene encoding Mdm2. Thus, the generally enhanced DNA damage observed in many tumors (47) might at least in part result from insufficient Mdm2 levels and the resulting DNA replication stress. Taken together, Mdm2 may not only serve as a p53 antagonist, but also as an effector of p53-mediated tumor suppression, modifying chromatin, avoiding R loops, and supporting replication.
On the other hand, Mdm2 represents an oncogene, and its overexpression can result from gene amplification, most notably in sarcomas. Our results suggest that not only the inactivation of p53 but also the modification of chromatin with subsequently enhanced DNA replication processivity may contribute to this oncogenicity. Importantly, Mdm2 has frequently been targeted by drug candidates, but with limited success in the clinics. To our knowledge, all Mdm2-targeting drugs that are currently evaluated in clinical studies interfere with the interaction of p53 and Mdm2, but not with the ubiquitin ligase activity of the Mdm2 Ring finger domain (48). Furthermore, our previous work indicates that the prototype drug of this kind, nutlin-3a, does not interfere with the chromatin modifier function of Mdm2 (1). Therefore, targeting the Ring finger domain rather than the p53 binding domain of Mdm2 can be expected to have a more profound impact on cancer cell proliferation, by interfering with chromatin modification and DNA replication. This encourages the continued evaluation of Mdm2 as a drug target, regarding its various activities on p53 as well as on chromatin and on DNA replication.
Materials and Methods
For more complete descriptions, see SI Appendix, Supplemental Materials and Methods.
H1299 (nonsmall cell lung carcinoma, p53 null), U2OS (osteosarcoma, p53 proficient), and mouse embryonic fibroblasts were cultured in DMEM, HCT116 p53−/− (colon carcinoma) cells in RPMI-1640. U2OS-derived clones for the inducible expression of catalytically inactive GFP-RNAseH1 D210N were kindly provided by Pavel Janscak, University of Zurich, Zurich. Lipofectamine 3000 was used for transfections.
DNA fiber assays to analyze replication fork progression and processivity were carried out essentially as in ref. 6.
Quantitative image-based cytometry was performed essentially as described before (26) using doxycycline-inducible GFP-RNaseH1 D210N cells.
Supplementary Material
Acknowledgments
We thank Yanping Zhang for MEFs with p53/Mdm2 deletions/mutation and Pavel Janscak for providing GFP-RNAseH1 D210N cells. pCMV-MDM2 was a gift from Bert Vogelstein (Addgene plasmid no. 16441), pCMV-MDM2(C464A) was a gift from Tyler Jacks (Addgene plasmid no. 12086), pLenti6/V5-DEST-RNF2 was a gift from Lynda Chin (Addgene plasmid no. 31216), and pICE-RNaseHI-WT-NLS-mCherry (Addgene plasmid no. 60365) as well as pICE-RNaseHI-D10R-E48R-NLS-mCherry (Addgene plasmid no. 60367) were gifts from Patrick Calsou. BAP1 expression plasmids were from Anjana Rao (Addgene plasmid nos. 81024 and 81025) and H2A expression plasmids from Titia Sixma (Addgene plasmid nos. 63561 and 63564). This work was supported by the Else Kröner Fresenius Stiftung, the Wilhelm Sander Stiftung, the Deutsche José Carreras Leukämie Stiftung, the Deutsche Krebshilfe (M.D. and K.W.), the Deutsche Forschungsgemeinschaft and the Boehringer Ingelheim Fonds (I.K.). Further support came from the Swiss National Science Foundation Grant (PP00P3_150690 to M.A.), the European Research Council Grant (ERC-2016-STG 714326 DiVineGenoMe to M.A.), and the Forschungskredit Candoc Program of the University of Zurich Grant (FK-16-053 to F.T.). I.K. was a member of the Göttingen Graduate School for Neurosciences, Biophysics, and Molecular Biosciences (GGNB) and of the International Max Planck Research School (IMPRS)/MSc./PhD program Molecular Biology at Göttingen during this work. F.T. is a member of the Molecular Life Science Program of the Life Science Zurich Graduate School.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809592115/-/DCSupplemental.
References
- 1.Wienken M, et al. MDM2 associates with polycomb repressor complex 2 and enhances stemness-promoting chromatin modifications independent of p53. Mol Cell. 2016;61:68–83. doi: 10.1016/j.molcel.2015.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wienken M, Moll UM, Dobbelstein M. Mdm2 as a chromatin modifier. J Mol Cell Biol. 2017;9:74–80. doi: 10.1093/jmcb/mjw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wen W, et al. Knockdown of RNF2 induces apoptosis by regulating MDM2 and p53 stability. Oncogene. 2014;33:421–428. doi: 10.1038/onc.2012.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blackledge NP, Rose NR, Klose RJ. Targeting polycomb systems to regulate gene expression: Modifications to a complex story. Nat Rev Mol Cell Biol. 2015;16:643–649. doi: 10.1038/nrm4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwartz YB, Pirrotta V. A new world of polycombs: Unexpected partnerships and emerging functions. Nat Rev Genet. 2013;14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 6.Klusmann I, et al. p53 activity results in DNA replication fork processivity. Cell Rep. 2016;17:1845–1857. doi: 10.1016/j.celrep.2016.10.036. [DOI] [PubMed] [Google Scholar]
- 7.Yeo CQX, et al. p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 2016;15:132–146. doi: 10.1016/j.celrep.2016.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Lerner LK, et al. Predominant role of DNA polymerase eta and p53-dependent translesion synthesis in the survival of ultraviolet-irradiated human cells. Nucleic Acids Res. 2017;45:1270–1280. doi: 10.1093/nar/gkw1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roy S, et al. p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. eLife. 2018;7:e31723. doi: 10.7554/eLife.31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mansilla SF, et al. Cyclin Kinase-independent role of p21CDKN1A in the promotion of nascent DNA elongation in unstressed cells. eLife. 2016;5:e18020. doi: 10.7554/eLife.18020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hampp S, et al. DNA damage tolerance pathway involving DNA polymerase ι and the tumor suppressor p53 regulates DNA replication fork progression. Proc Natl Acad Sci USA. 2016;113:E4311–E4319. doi: 10.1073/pnas.1605828113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mansilla SF, et al. UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic Acids Res. 2013;41:6942–6951. doi: 10.1093/nar/gkt475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bermejo R, Lai MS, Foiani M. Preventing replication stress to maintain genome stability: Resolving conflicts between replication and transcription. Mol Cell. 2012;45:710–718. doi: 10.1016/j.molcel.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Aguilera A, García-Muse T. R Loops: From transcription byproducts to threats to genome stability. Mol Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 15.Gazdar AF, Girard L, Lockwood WW, Lam WL, Minna JD. Lung cancer cell lines as tools for biomedical discovery and research. J Natl Cancer Inst. 2010;102:1310–1321. doi: 10.1093/jnci/djq279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rondinelli B, et al. EZH2 promotes degradation of stalled replication forks by recruiting MUS81 through histone H3 trimethylation. Nat Cell Biol. 2017;19:1371–1378. doi: 10.1038/ncb3626. [DOI] [PubMed] [Google Scholar]
- 17.Minsky N, Oren M. The RING domain of Mdm2 mediates histone ubiquitylation and transcriptional repression. Mol Cell. 2004;16:631–639. doi: 10.1016/j.molcel.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 18.Itahana K, et al. Targeted inactivation of Mdm2 RING finger E3 ubiquitin ligase activity in the mouse reveals mechanistic insights into p53 regulation. Cancer Cell. 2007;12:355–366. doi: 10.1016/j.ccr.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 19.Mattiroli F, et al. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell. 2012;150:1182–1195. doi: 10.1016/j.cell.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 20.Sahtoe DD, van Dijk WJ, Ekkebus R, Ovaa H, Sixma TK. BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat Commun. 2016;7:10292. doi: 10.1038/ncomms10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee HS, Lee SA, Hur SK, Seo JW, Kwon J. Stabilization and targeting of INO80 to replication forks by BAP1 during normal DNA synthesis. Nat Commun. 2014;5:5128. doi: 10.1038/ncomms6128. [DOI] [PubMed] [Google Scholar]
- 22.Balasubramani A, et al. Cancer-associated ASXL1 mutations may act as gain-of-function mutations of the ASXL1-BAP1 complex. Nat Commun. 2015;6:7307. doi: 10.1038/ncomms8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santos-Pereira JM, Aguilera A. R loops: New modulators of genome dynamics and function. Nat Rev Genet. 2015;16:583–597. doi: 10.1038/nrg3961. [DOI] [PubMed] [Google Scholar]
- 24.Wu H, Lima WF, Crooke ST. Investigating the structure of human RNase H1 by site-directed mutagenesis. J Biol Chem. 2001;276:23547–23553. doi: 10.1074/jbc.M009676200. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen HD, et al. Functions of replication protein a as a sensor of R loops and a regulator of RNaseH1. Mol Cell. 2017;65:832–847.e4. doi: 10.1016/j.molcel.2017.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pellegrino S, Michelena J, Teloni F, Imhof R, Altmeyer M. Replication-coupled dilution of H4K20me2 guides 53BP1 to pre-replicative chromatin. Cell Rep. 2017;19:1819–1831. doi: 10.1016/j.celrep.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morales F, Giordano A. Overview of CDK9 as a target in cancer research. Cell Cycle. 2016;15:519–527. doi: 10.1080/15384101.2016.1138186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leung KH, Abou El Hassan M, Bremner R. A rapid and efficient method to purify proteins at replication forks under native conditions. Biotechniques. 2013;55:204–206. doi: 10.2144/000114089. [DOI] [PubMed] [Google Scholar]
- 29.Piunti A, et al. Polycomb proteins control proliferation and transformation independently of cell cycle checkpoints by regulating DNA replication. Nat Commun. 2014;5:3649. doi: 10.1038/ncomms4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmid JA, et al. Histone ubiquitination by the DNA damage response is required for efficient DNA replication in unperturbed S phase. Mol Cell. 2018;71:897–910.e8. doi: 10.1016/j.molcel.2018.07.011. [DOI] [PubMed] [Google Scholar]
- 31.Frum RA, et al. The human oncoprotein MDM2 induces replication stress eliciting early intra-S-phase checkpoint response and inhibition of DNA replication origin firing. Nucleic Acids Res. 2014;42:926–940. doi: 10.1093/nar/gkt944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown DR, Thomas CA, Deb SP. The human oncoprotein MDM2 arrests the cell cycle: Elimination of its cell-cycle-inhibitory function induces tumorigenesis. EMBO J. 1998;17:2513–2525. doi: 10.1093/emboj/17.9.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conradt L, et al. Mdm2 inhibitors synergize with topoisomerase II inhibitors to induce p53-independent pancreatic cancer cell death. Int J Cancer. 2013;132:2248–2257. doi: 10.1002/ijc.27916. [DOI] [PubMed] [Google Scholar]
- 34.Marinello J, Chillemi G, Bueno S, Manzo SG, Capranico G. Antisense transcripts enhanced by camptothecin at divergent CpG-island promoters associated with bursts of topoisomerase I-DNA cleavage complex and R-loop formation. Nucleic Acids Res. 2013;41:10110–10123. doi: 10.1093/nar/gkt778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee T, Pelletier J. Dependence of p53-deficient cells on the DHX9 DExH-box helicase. Oncotarget. 2017;8:30908–30921. doi: 10.18632/oncotarget.15889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cristini A, Groh M, Kristiansen MS, Gromak N. RNA/DNA hybrid interactome identifies DXH9 as a molecular player in transcriptional termination and R-loop-associated DNA damage. Cell Rep. 2018;23:1891–1905. doi: 10.1016/j.celrep.2018.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuoka S, et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160–1166. doi: 10.1126/science.1140321. [DOI] [PubMed] [Google Scholar]
- 38.Beli P, et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol Cell. 2012;46:212–225. doi: 10.1016/j.molcel.2012.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gorthi A, et al. EWS-FLI1 increases transcription to cause R-loops and block BRCA1 repair in Ewing sarcoma. Nature. 2018;555:387–391. doi: 10.1038/nature25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Muse T, Aguilera A. Transcription-replication conflicts: How they occur and how they are resolved. Nat Rev Mol Cell Biol. 2016;17:553–563. doi: 10.1038/nrm.2016.88. [DOI] [PubMed] [Google Scholar]
- 41.Aguilera A, Gómez-González B. DNA-RNA hybrids: The risks of DNA breakage during transcription. Nat Struct Mol Biol. 2017;24:439–443. doi: 10.1038/nsmb.3395. [DOI] [PubMed] [Google Scholar]
- 42.Alt JR, et al. Mdm2 binds to Nbs1 at sites of DNA damage and regulates double strand break repair. J Biol Chem. 2005;280:18771–18781. doi: 10.1074/jbc.M413387200. [DOI] [PubMed] [Google Scholar]
- 43.Bouska A, Lushnikova T, Plaza S, Eischen CM. Mdm2 promotes genetic instability and transformation independent of p53. Mol Cell Biol. 2008;28:4862–4874. doi: 10.1128/MCB.01584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlacher K, et al. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li J, Kurokawa M. Regulation of MDM2 stability after DNA damage. J Cell Physiol. 2015;230:2318–2327. doi: 10.1002/jcp.24994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Carr MI, Roderick JE, Gannon HS, Kelliher MA, Jones SN. Mdm2 phosphorylation regulates its stability and has contrasting effects on oncogene and radiation-induced tumorigenesis. Cell Rep. 2016;16:2618–2629. doi: 10.1016/j.celrep.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 48.Burgess A, et al. Clinical overview of MDM2/X-targeted therapies. Front Oncol. 2016;6:7. doi: 10.3389/fonc.2016.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.