Significance
The “pygmy” phenotype is a classic example of convergent adaptation in humans, having evolved independently in both African and Asian rainforest hunter-gatherers. By focusing on indications of subtle allele-frequency changes occurring in aggregate across variants in many genes (polygenic adaptation), we observed signatures of positive natural selection on the same growth-related pathways in rainforest hunter-gatherer populations from both continents. Unexpectedly, we also observed signatures of convergent positive selection on heart development pathway genes. We hypothesize that the heart pathway result may reflect compensatory changes following height-related adaptation in the GH/IGF1 pathway, which in addition to general growth processes also affects heart development. Our results exemplify the insights that can be gained from comparative studies of diverse human populations.
Keywords: rainforest hunter-gatherers, convergent evolution, population genomics, stature
Abstract
Different human populations facing similar environmental challenges have sometimes evolved convergent biological adaptations, for example, hypoxia resistance at high altitudes and depigmented skin in northern latitudes on separate continents. The “pygmy” phenotype (small adult body size), characteristic of hunter-gatherer populations inhabiting both African and Asian tropical rainforests, is often highlighted as another case of convergent adaptation in humans. However, the degree to which phenotypic convergence in this polygenic trait is due to convergent versus population-specific genetic changes is unknown. To address this question, we analyzed high-coverage sequence data from the protein-coding portion of the genomes of two pairs of populations: Batwa rainforest hunter-gatherers and neighboring Bakiga agriculturalists from Uganda and Andamanese rainforest hunter-gatherers and Brahmin agriculturalists from India. We observed signatures of convergent positive selection between the rainforest hunter-gatherers across the set of genes with “growth factor binding” functions (). Unexpectedly, for the rainforest groups, we also observed convergent and population-specific signatures of positive selection in pathways related to cardiac development (e.g., “cardiac muscle tissue development”; ). We hypothesize that the growth hormone subresponsiveness likely underlying the adult small body-size phenotype may have led to compensatory changes in cardiac pathways, in which this hormone also plays an essential role. Importantly, in the agriculturalist populations, we did not observe similar patterns of positive selection on sets of genes associated with growth or cardiac development, indicating our results most likely reflect a history of convergent adaptation to the similar ecology of rainforests rather than a more general evolutionary pattern.
Similar ecological challenges may repeatedly result in similar evolutionary outcomes, and many instances of phenotypic convergence arising from parallel changes in the same genetic loci have been uncovered (reviewed in refs. 1–3). Many examples of convergent genetic evolution reported to date are for simple monogenic traits, for example depigmentation in independent populations of Mexican cave fish living in lightless habitats (4, 5) and persistence of the ability to digest lactose in adulthood in both European and African agriculturalist/pastoralist humans (6). Most biological traits, however, are highly polygenic. Since the reliable detection of positive selection in aggregate on multiple loci of individually small effect (i.e., polygenic adaptation) is relatively difficult (7–11), the extent to which either convergent genetic changes (at the same loci and functional pathways) or changes affecting distinct genetic pathways may underlie these complex traits is less clear.
Human height is a classic example of a polygenic trait with approximately 800 known loci significantly associated with stature in Europeans collectively accounting for 27.4% of the heritable portion of height variation in this population (12). A stature phenotype also represents one of the most striking examples of convergent evolution in humans. Small body size (or the “pygmy” phenotype, e.g., average adult male stature 155 cm) appears to have evolved independently in rainforest hunter-gatherer populations from Africa, Asia, and South America (13), as groups on different continents do not share common ancestry to the exclusion of nearby agriculturalists (14, 15). Positive correlations between stature and the degree of admixture with neighboring agriculturalists have confirmed that the adult small body-size phenotype is, at least in part, genetically mediated and therefore potentially subject to natural selection (16–20).
Indeed, previous population genetic studies have identified signatures of strong positive natural selection across the genomes of various worldwide rainforest hunter-gatherer groups (15, 19, 21, 22). In some cases, the candidate positive selection regions were significantly enriched for genes involved in growth processes and pathways (15, 19). However, in one rainforest hunter-gatherer population, the Batwa from Uganda, an admixture mapping approach was used to identify 16 genetic loci specifically associated with the adult small body-size phenotype (17). While these genomic regions were enriched for genes involved in the growth hormone pathway and for variants associated with stature variation in Europeans, there was no significant overlap between the small body-size phenotype-associated regions and the strongest signals of positive selection in the Batwa genome. Rather, subtle shifts in allele frequencies were observed across these regions in aggregate, consistent with a history of polygenic adaptation for the Batwa small body-size phenotype (17) and underscoring the importance of using different types of population genetic approaches to study the evolutionary history of this trait. In addition to growth (15) and various growth-factor signaling pathways (19), studies focused on other rainforest hunter-gatherer groups have also reported enrichments for signatures of selection on genes involved in immunity (19, 21, 22), metabolism (19, 21, 22), development (15, 22), and reproduction (19, 21, 22).
Here, we investigate population-specific and convergent patterns of positive selection in African and Asian hunter-gatherer populations, using genome-wide sequence data from two sets of populations: the Batwa rainforest hunter-gatherers and the nearby Bakiga agriculturalists of Uganda in East Africa (23) and the Jarawa and Onge rainforest hunter-gatherers of the Andaman Islands in South Asia and the Uttar Pradesh Brahmin agriculturalists from mainland India (24, 25). We specifically test whether convergent or population-specific signatures of positive selection, detected both with “outlier” tests designed to identify strong signatures of positive selection and with tests designed to identify signatures of polygenic adaptation, are enriched for genes with growth-related functions. After studying patterns of convergent- and population-specific evolution in the Batwa and Andamanese hunter-gatherers, we then repeat these analyses in the paired Bakiga and Brahmin agriculturalists to evaluate whether the evolutionary patterns most likely relate to adaptation to hunter-gatherer subsistence in rainforest habitats, rather than being more generalized evolutionary patterns for human populations.
Results
We sequenced the protein-coding portions of the genomes (exomes) of 50 Batwa rainforest hunter-gatherers and 50 Bakiga agriculturalists (dataset originally reported in ref. 23), identified single-nucleotide polymorphisms (SNPs), and analyzed the resultant data alongside those derived from published whole-genome sequence data for 10 Andamanese rainforest hunter-gatherers and 10 Brahmin agriculturalists (dataset from ref. 25). We restricted our analysis to exonic SNPs, for comparable analysis of the Asian whole-genome sequence data with the African exome sequence data. To polarize allele-frequency differences observed between each pair of hunter-gatherer and agriculturalist populations, we merged these data with those from outgroup comparison populations from the 1,000 Genomes Project (26): exome sequences of 30 unrelated British individuals from England and Scotland (GBR) for comparison with the Batwa/Bakiga data and exome sequences of 30 Luhya individuals from Webuye, Kenya (LWK) for comparison with the Andamanese/Brahmin data. Outgroup populations were selected for genetic equidistance from the test populations. While minor levels of introgression from a population with European ancestry have been observed for the Batwa and Bakiga (23, 27), the statistic we used is relatively robust to low levels of admixture (28).
To identify regions of the genome that may have been affected by positive selection in each of our test populations, we computed the population branch statistic (PBS) (29) for each exonic SNP identified among or between the Batwa and Bakiga and the Andamanese and Brahmin populations (SI Appendix, Figs. S1 and S2 and Dataset S1a). The PBS is an estimate of the magnitude of allele-frequency change that occurred along each population lineage following divergence of the most closely related populations, with the allele-frequency information from the outgroup population used to polarize frequency changes to one or both branches. Larger PBS values for a population reflect greater allele-frequency change on that branch, which in some cases could reflect a history of positive selection (29).
For each analyzed population, we computed a PBS selection index for each gene by comparing the mean PBS for all SNPs located within that gene to a distribution of values estimated by shuffling SNP–gene associations (without replacement) and recomputing the mean PBS value for that gene 100,000 times (SI Appendix, Table S15). The PBS selection index is the percentage of permuted values that are higher than the actual (observed) mean PBS value for that gene, such that a very low PBS selection index gene is one with a higher mean PBS value than would be expected by chance. Per-gene PBS selection index values were not significantly correlated with gene size (linear regression of log-adjusted selection indexes against gene length: adjusted , -statistic ; SI Appendix, Fig. S3), suggesting that this metric is not overtly biased by gene size.
Convergent evolution can operate at different scales, including on the same DNA or amino acid change, on different genetic variants within the same gene, or across different genes involved in the same biomolecular pathway or function. Given that our motivating phenotype is a complex trait and signatures of polygenic adaptation are expected to be relatively subtle and especially difficult to detect at the individual mutation and gene levels, in this study we principally consider patterns of convergence versus population specificity at the functional pathway/annotation level. We do note that when we applied the same approaches described in this study to individual SNPs, we identified several individual alleles with patterns of convergent allele-frequency evolution between the Batwa and Andamanese that may warrant further study (Dataset S1b), including a nonsynonymous SNP in the gene FIG4, which when disrupted in mice results in a phenotype of small but proportional body size (30). However, likely related to the above-discussed challenges of identifying signatures of polygenic adaptation at the locus-specific level, the results of our individual SNP and gene analyses were otherwise largely unremarkable, and thus the remainder of our report and discussion focuses on pathway-level analyses.
Outlier Signatures of Strong Convergent and Population-Specific Selection.
The set of genes with the lowest (outlier) PBS index values for each population may be enriched for genes with histories of relatively strong positive natural selection. We used a permutation-based analysis to test whether curated sets of genome-wide growth-associated genes (four lists tested separately ranging from 266 to 3,996 genes; 4,888 total genes; SI Appendix, Text S1) or individual Gene Ontology (GO) annotated functional categories of genes (GO categories with fewer than 50 genes were excluded) have significant convergent excesses of genes with low PBS selection index values () in both of two cross-continental populations, for example the Batwa and Andamanese. Specifically, for each functional category set of genes and population, we first used Fisher’s exact tests to estimate the probability that the number of genes with PBS selection index values was greater than that expected by chance. We then reshuffled the PBS selection indexes across all genes 1,000 different times for each population to generate distributions of permuted enrichment P values for each functional category set of genes. We compared our observed Batwa and Andamanese Fisher’s exact test P values to those from the randomly generated distributions as follows. We computed the joint probability of the null hypotheses for both the Andamanese and Batwa being false as , where and are the P values of the Fisher’s exact test, and we compared this joint probability estimate to the same statistic computed for the P values from the random iterations. We then defined the P value of our empirical test for convergent evolution as the probability that this statistic was more extreme (lower) for the observed values than for the randomly generated values. The resultant P value summarizes the test of the null hypothesis that both results could have been jointly generated under random chance. While each individual population’s outlier-based test results are not significant after multiple-test correction, this joint approach provides increased power to identify potential signatures of convergent selection by assessing the probability of obtaining two false positives in these independent samples.
Several GO biological processes were significantly overrepresented—even when accounting for the number of tests performed—among the sets of genes with outlier signatures of positive selection in both the Batwa and Andamanese hunter-gatherer populations (empirical test for convergence ; SI Appendix, Table S1 and Fig. 1A). These GO categories include “limb morphogenesis” (GO:0035108; empirical test for convergence ; ; Batwa, genes observed = 5, expected = 1.69, Fisher’s exact ; Andamanese, observed = 6, expected = 2.27, Fisher’s exact ).
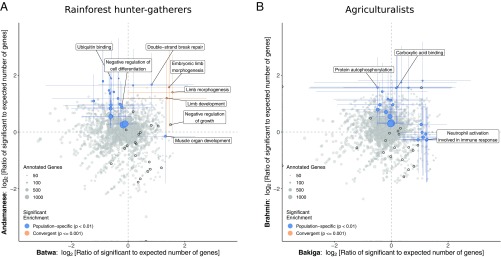
Fig. 1.
GO categories with significant enrichment for signatures of strong positive selection. (A and B) GO functional categories’ ratios of expected to observed counts of outlier genes (with PBS selection index 0.01) in the Batwa and Andamanese rainforest hunter-gatherers (A) and Bakiga and Brahmin agriculturalist control comparison (B). Results are shown for GO biological processes and molecular functions. Point size is scaled to number of annotated genes in each category. Terms that are significantly overrepresented for genes with signatures of potential positive selection (Fisher’s ) in either population are shown in blue and for both populations convergently (empirical permutation-based ) are shown in orange. Colored lines represent 95% CI for significant categories estimated by bootstrapping genes within pathways. Black outlines indicate growth-associated terms: the “growth” biological process (GO:0040007) and its descendant terms or the molecular functions “growth factor binding,” “growth factor receptor binding,” “growth hormone receptor activity,” and “growth factor activity” and their subcategories.
Other functional categories of genes were overrepresented in the sets of outlier loci for one of these hunter-gatherer populations but not the other (Fig. 1A; SI Appendix, Table S2; and Dataset S1j). The top population-specific enrichments for genes with outlier PBS selection index values for the Batwa were associated with growth and development: “muscle organ development” (GO:0007517; observed genes, 10; expected genes, 4.02; ) and “negative regulation of growth” (GO:0045926; observed genes, 7; expected genes, 2.48; ). Significantly overrepresented GO biological processes for the Andamanese included “negative regulation of cell differentiation” (GO:0045596; observed genes, 18; expected genes, 9.79; ). However, these population-specific enrichments were not significant following multiple-test correction (false discovery rate for both Batwa terms and for the Andamanese result).
In contrast, no GO functional categories were observed to have similarly significant convergent excesses of “outlier” genes with signatures of positive selection across the two agriculturalist populations to those observed for the rainforest hunter-gatherer populations (Fig. 1B and Dataset S1e), and the top-ranked GO categories from both the convergent evolution analysis and the population-specific analyses were absent any obvious connections to skeletal growth. The top-ranked functional categories with enrichments for genes with outlier PBS selection index values for the individual agriculturalist populations included “neutrophil activation involved in immune response” for the Bakiga (GO:0002283; observed = 13; expected = 5.43; ; ) and “protein autophosphorylation” for the Brahmin (GO:0046777; observed = 11; expected = 3.71; ; ; Dataset S1j).
Signatures of Convergent and Population-Specific Polygenic Adaptation.
Outlier-based approaches such as that presented above are expected to have limited power to identify signatures of polygenic adaptation (7–11), which is our expectation for the small adult body-size phenotype (17). Unlike the previous analyses in which we identified functional categories with an enriched number of genes with outlier PBS selection index values, for our polygenic evolution analysis we computed a “distribution shift-based” statistic to instead identify functionally grouped sets of loci with relative shifts in their distributions of PBS selection indexes. Specifically, we used the Kolmogorov–Smirnov (KS) test to quantify the distance between the distribution of PBS selection indexes for the genes within a functional category to that of the genome-wide distribution. Significantly positive shifts in the PBS selection index distribution for a particular functional category may reflect individually subtle but consistent allele-frequency shifts across genes within the category, which could result from either a relaxation of functional constraint or a history of polygenic adaptation. Our approach is similar to another recent method that was used to detect polygenic signatures of pathogen-mediated adaptation in humans (31). As above, we identified functional categories with convergently high KS values between cross-continental groups by repeating these tests 1,000 times on permuted gene–PBS values and computing the joint probability of both null hypotheses being false for the two populations. We then compared this value from the random iterations to the same statistic computed with the observed KS P values for each functional category. For example, for the Batwa and Andamanese, we tallied the number of random iterations for which the joint probability of both null hypotheses being false was more extreme (lower) than those of the random iterations. In this way we tested the null hypothesis that both of our observed P values could have been jointly generated by random chance.
The GO molecular function with the strongest signature of a convergent polygenic shift in PBS selection indexes across the Batwa and Andamanese populations was growth factor binding (SI Appendix, Table S3 and Fig. 2A; GO:0019838; Batwa ; Andamanese ; Fisher’s combined ; empirical test for convergence ; ), and the top GO biological process was “organ growth” (GO:0035265; Batwa ; Andamanese ; Fisher’s combined ; empirical test for convergence ; ). The other top Batwa–Andamanese convergent GO biological processes are not as obviously related to growth, but instead involve muscles, particularly heart muscles. A significant convergent shift in PBS selection indexes across both hunter-gatherer populations was observed for cardiac muscle tissue development (GO:0048738; Batwa ; Andamanese ; Fisher’s combined ; empirical test for convergence ; ).
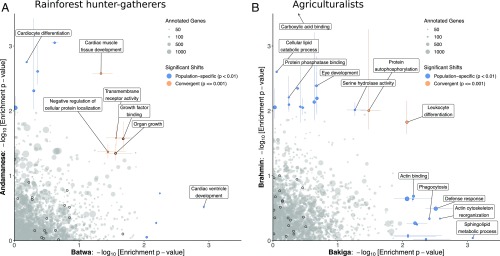
Fig. 2.
GO categories with significant enrichment for signatures of polygenic selection. (A and B) GO functional categories’ distribution shift test P values, indicating a shift in the PBS selection index values for these genes, in the Batwa and Andamanese rainforest hunter-gatherers (A) and Bakiga and Brahmin agriculturalist control comparison (B). Results are shown for GO biological processes and molecular functions. Point size is scaled to number of annotated genes in each category. Terms that are significantly enriched for genes with signatures of potential positive selection (KS ) in either population are shown in blue and for both populations convergently (empirical permutation-based ) are shown in orange. Colored lines represent 95% CI for significant categories estimated by bootstrapping genes within pathways. Black outlines indicate growth-associated terms: the growth biological process (GO:0040007) and its descendant terms or the molecular functions growth factor binding, growth factor receptor binding, growth hormone receptor activity, and growth factor activity and their subcategories. One GO molecular function, “carboxylic acid binding” (GO:0031406; Brahmin 7.3 10−5; ) is not shown, but indicated with an arrow.
In contrast, when this analysis was repeated on the agriculturalist populations, no growth- or muscle-related functional annotations were observed with significantly convergent shifts in both populations (Fig. 2B and Dataset S1l). The GO categories with evidence of potential convergent evolution between the agriculturalists were the biological processes “leukocyte differentiation” (GO:0002521; Bakiga ; Brahmin ; Fisher’s combined ; convergence empirical ; ) and protein autophosphorylation (GO:0046777; Bakiga ; Brahmin ; Fisher’s combined ; convergence empirical ; ).
We also used Bayenv, a Bayesian linear modeling method for identifying loci with allele frequencies that covary with an ecological variable (9, 32), to assess the level of consistency with our convergent polygenic PBS shift results. Specifically, we used Bayenv to test whether the inclusion of a binary variable indicating subsistence strategy would increase the power to explain patterns of genetic diversity for a given functional category of loci over a model that considered only population history (as inferred from the covariance of genome-wide allele frequencies in the dataset). We converted Bayes factors into per-gene index values via permutation of SNP–gene associations (Dataset S1g) and identified GO terms with significant shifts in the Bayenv Bayes factor index distribution (9, 32) (Dataset S1m). The top results from this analysis included growth factor activity (GO:0008083; ; ), categories related to enzyme regulation (e.g., “enzyme regulator activity”; GO:0030234; ; ), and categories related to muscle cell function (e.g., “microtubule binding”; GO:0008017; ; ). There were more GO terms that were highly ranked () in both the hunter-gatherer PBS shift-based empirical test of convergence and the Bayenv analysis than expected by chance (for biological processes GO terms, observed categories in common = 13, expected = 8.03, Fisher’s exact test ; for molecular function GO terms, observed categories in common = 4, expected = 1.45, Fisher’s exact test ).
While we did not observe any significant population-specific shifts in PBS selection index values for growth-associated GO functional categories in any of our studied populations (SI Appendix, Table S4 and Text S1), for each individual rainforest hunter-gatherer population we did observe nominal shifts in separate biological process categories involving the heart (Fig. 2A). For the Batwa, “cardiac ventricle development” (GO:0003231) was the top population-specific result (median PBS index = 0.272 vs. genome-wide median PBS index = 0.528; ; ). For the Andamanese, “cardiocyte differentiation” (GO:0035051) was also ranked highly (median PBS index = 0.353 vs. genome-wide median PBS index = 0.552; ; ). We note that while these are separate population-specific signatures, 17 genes are shared between the above two cardiac-related pathways (of 61 total cardiocyte differentiation genes, 28%; of 71 total cardiac ventricle development genes, 24%; Dataset S1n).
In contrast, cardiac development-related GO categories were not observed among those with highly ranked population-specific polygenic shifts in selection index values for either the Bakiga or the Brahmin agriculturalists (Fig. 2B and Dataset S1o). The only GO term with a significant population-specific shift in the agriculturalists after multiple-test correction was molecular function carboxylic acid binding in the Brahmins (GO:0031406; ; ).
To ensure that our results were robust to several possible biases, we repeated the above analyses with several modifications. First, to control for potential biases related to variation in gene length and SNP minor allele frequency (MAF), we repeated all analyses after computing the PBS selection index with binning of genes by length and SNPs by MAF, respectively. Our results were not materially different (SI Appendix, Tables S5, S6, S9, S10, S13, S14, S17, and S18; Figs. S4–S6, S9, and S10; and Text S3). Second, to account for the effect of linkage disequilibrium among SNPs within a gene, we recomputed the empirical test for convergence P values by permuting gene–GO relationships when generating the random null distributions for the PBS selection index values instead of gene–PBS relationships as in our original analysis. Again, downstream results were largely unchanged (SI Appendix, Tables S21 and S22 and Text S4). Finally, to confirm that allele-frequency uncertainty due to sample size differences among the populations in our study did not materially affect our major results, we performed several rounds of analyses after randomly subsampling sets of individuals from some populations. Our main growth- and heart-related results did not change substantially (SI Appendix, Text S5; Figs. S7, S8, S11, and S12; and Tables S7, S8, S11, S12, S15, S16, S19, and S20).
Discussion
The independent evolution of adult small body size in multiple different tropical rainforest environments worldwide presents a natural human model for comparative study of the genetic and evolutionary bases of growth and body size. Through an evolutionary genomic comparison of African and Asian rainforest hunter-gatherer populations to one another and with nearby agriculturalists, we have gained additional, indirect insight into the genetic structure of body size, a fundamental biological trait. Specifically, we identified a signature of potential convergent positive selection on the growth factor binding pathway that could partially underlie the independent evolution of small body size in African and Asian rainforest hunter-gatherers.
Unexpectedly, we also observed signatures of potential polygenic selection across functional categories of genes related to heart development in the rainforest hunter-gatherer populations, both convergently and on a population-specific basis. To a minor extent, the growth factor- and heart-related functional categories highlighted in our study do overlap: Of the 123 total genes annotated across the three heart-related categories (cardiac muscle tissue development, GO:0048738; cardiac ventricle development, GO:0003231; and cardiocyte differentiation, GO:0035051), 9 (7.3%) are also included among the 66 annotated genes in the growth factor binding category (GO:0019838). However, even after excluding these 9 genes from our dataset, we still observed similar polygenic PBS shifts in the Batwa and Andamanese for both growth factor- and heart-related functional categories (SI Appendix, Text S2), demonstrating that our observations are not driven solely by cross-annotated genes.
We hypothesize that the evolution of growth hormone subresponsiveness, which appears to at least partly underlie short stature in some rainforest hunter-gatherer populations (33–37), may in turn have also resulted in strong selection pressure for compensatory adaptations in cardiac pathways. The important roles of growth hormone (GH) in the heart are evident from studies of patients deficient in the hormone. For example, patients with GH deficiency are known to be at an increased risk of atherosclerosis and mortality from cardiovascular disease (38) and have worse cardiac function (39). More broadly, shorter people have elevated risk of coronary artery disease (40), likely due to the pleiotropic effects of variants affecting height and atherosclerosis development (41). Such health outcomes may relate to the important roles that GH plays in the development and function in the myocardium (42, 43), which contains a relatively high concentration of receptors for GH (44).
Given their short stature and hypothesized altered GH response, rainforest hunter-gatherer individuals would theoretically be expected to have higher rates of cardiovascular disease symptoms, including elevated systolic and pulse blood pressure [associated with short stature (45) and GH deficiency (46)] and arterial stiffening [associated with GH deficiency (47) but not height (48)], compared with taller populations. Yet several studies of rainforest hunter-gatherer cardiovascular health instead reported relatively lower or comparable blood pressure or rates of hypertension [Republic of Congo (49), Cameroon (50–52), Brazil (53), and Democratic Republic of the Congo (54)], reduced age-associated rise in blood pressure [Cameroon (51) and Democratic Republic of the Congo (54)], and lower or comparable aortic stiffness with less arterial distensibility due to age and blood pressure [Cameroon (51, 52)] in rainforest hunter-gatherer populations relative to agricultural populations compared in the same studies. We hypothesize that the signatures of positive selection on genes in cardiac pathways in rainforest hunter-gatherer populations that we observed in this study may reflect compensatory adaptation in response to the evolution of the small body-size phenotype, thereby potentially explaining the otherwise surprisingly low levels of cardiovascular risk factors that have been reported for rainforest hunter-gatherer populations.
Alternative explanations for our finding of potential convergent positive selection on cardiac-related pathways could include selection pressures related to the ecological stresses of full-time tropical rainforest occupation by human hunter-gatherers that have acted on cardiac function directly, rather than indirectly through height-related adaptation. Especially before the ability to trade forest products for cultivated goods with agriculturalists, the diets of full-time rainforest hunter-gatherers may have been calorically and nutritionally restricted on at least a seasonal basis (13). Caloric restriction has a direct functional impact on cardiac metabolism and function, with modest fasting in mice leading to the depletion of myocardial phospholipids, which potentially act as a metabolic reserve to ensure energy to essential heart functions (55). In human rainforest hunter-gatherers, selection may have favored variants conferring cardiac phenotypes optimized to maintain myocardial homeostasis during the nutritional stress that these populations may have experienced in the past. Other aspects of the shared rainforest ecology may have instead or also exerted selective pressure on heart phenotypes, including increased exposure to the many tropical protozoan and helminth parasites known to directly or indirectly impact cardiac health (56), providing a logical starting point for follow-up study of this possibility.
An important caveat to our study is the lack of statistical significance for our population-specific analyses after controlling for the multiplicity of tests resulting from hierarchically nested GO terms. The absence of strong signals of positive selection that are robust to the multiple-testing burden likely reflects both the expected subtlety of evolutionary signals of selection on polygenic traits and the restriction of our dataset to gene coding-region sequences. However, our comparative approach to identify signatures of convergent evolution is more robust. Therefore, while we cannot yet accurately estimate the extent to which signatures of positive selection that potentially underlie the evolution of the adult small body-size phenotype occurred in the same versus distinct genetic pathways between the Batwa and Andamanese, we do feel confident in our findings of convergent growth-related and cardiac-related pathways evolution. The concurrent signatures of convergent evolution across these two pathways in both African and Asian rainforest hunter-gatherers is an example of the insight into a biomedically relevant phenotype that can be gained from the comparative study of human populations with nonpathological natural variation.
Materials and Methods
Sample Collection and Dataset Generation.
Sample collection, processing, and sequencing have been previously described (17, 23). Sampling of biomaterials (blood or saliva) from Batwa rainforest hunter-gatherers and Bakiga agriculturalists of southwestern Uganda took place in 2010 (17). The study was approved by the Institutional Review Boards (IRBs) of both the University of Chicago (16986A) and Makerere University, Kampala, Uganda (2009-137), and local community approval and individual informed consent were obtained before collection. DNA samples of 50 Batwa and 50 Bakiga adults were included in the present study. Exome capture, sequencing, and variant calling were described previously (23). Briefly, sequence reads were aligned to the hg19/GRCh37 genome with BWA v.0.7.7 mem with default settings (57), PCR duplicates were detected with Picard Tools v.1.94 (broadinstitute.github.io/picard), and realignment around indels and base quality recalibration was done with GATK v3.5 (58) using the known indel sites from the 1,000 Genomes Project (26). Variants were called individually with GATK HaplotypeCaller (58), and variants were pooled together with GATK GenotypeGVCF and filtered using variant quality score recalibration (VQSR). Only biallelic SNPs with a minimum depth of 5× and less than 85% missingness that were polymorphic in the entire dataset were retained for analyses.
Variant data for the Andamanese individuals (Jarawa and Onge) and an outgroup mainland Indian population (Uttar Pradesh Brahmins) from ref. 25 were downloaded in variant call format (VCF) from a public website. To ensure the exome capture-derived African and whole-genome shotgun sequencing-derived Asian datasets were comparable, we restricted our analyses of these data to exonic SNPs only.
Merging with 1,000 Genomes Data.
We chose outgroup comparison populations from the 1,000 Genomes Project (26) to be equally distantly related to the ingroup populations: Reads from a random sample of 30 unrelated individuals from British in England and Scotland (GBR) and Luhya in Webuye, Kenya (LWK) were chosen for the Batwa/Bakiga and Andamanese/Brahmin datasets, respectively. We recalled variants in each 1,000 Genomes comparison population at loci that were variable in the ingroup populations, using GATK UnifiedGenotyper (58). Variants were filtered to exclude those with QualByDepth 2.0, RMSMappingQuality (MQ) 40.0, FisherStrand 60.0, HaplotypeScore 13.0, MQRankSum −12.5, or ReadPosRankSum −8.0. We removed SNPs for which fewer than 10 of the 30 individuals from the 1,000 Genomes datasets had genotypes.
Computation of the PBS and the Per-Gene PBS Index.
Using these merged datasets, we computed between population pairs using the unbiased estimator of Weir and Cockerham (59), transformed it to a measure of population divergence [], and then calculated the PBS, as in ref. 29. The PBS was computed on a per-SNP basis. We computed an empirical P value for each SNP, simply the proportion of coding SNPs with a PBS greater than the value for this SNP, which we adjusted for false discovery rate (FDR).
SNPs were annotated with gene-based information using ANNOVAR (60) with refGene (Release 76) (61) and PolyPhen (62) data. As the Andamanese/Brahmin dataset spanned the genome and the Batwa/Bakiga exome dataset included off-target intronic sequences as well as untranslated regions (UTRs) and microRNAs, we restricted our analysis to only exonic SNPs. For both the Batwa/Bakiga and Andamanese/Brahmin datasets, we computed a “PBS selection index” for each gene as follows. We compared the mean PBS for all SNPs located within that gene to a distribution of values estimated by shuffling SNP–gene associations (without replacement) and recomputing the mean PBS value for that gene 10,000 times. We defined the PBS selection index of the gene as the percentage of these empirical mean values that is higher than its observed mean PBS value. When identifying outlier genes, gene-based indexes were adjusted for FDR.
To assess potential biases related to variation in gene length and SNP MAFs, we repeated all analyses after computing the PBS selection index with binning of genes by length or SNPs by MAF. Complete details of these methods are included in SI Appendix, Text S3.
To identify SNPs with allele frequencies correlated with subsistence strategy (hunter-gatherer, Andamanese and Batwa; agriculturalists, Bakiga and Brahmin), we used Bayenv2.0 (32) to assess whether the addition of a binary variable denoting subsistence strategy improved the Bayesian model that already took into account covariance between samples due to ancestry. As with the PBS results, we computed an index for each gene by sampling new values for each SNP from the distribution of all Bayes factors and comparing the actual average for this gene to those of the bootstrapped replicates.
Creation of a Priori Lists of Growth-Related Genes.
To test the hypothesis that genes with known influence on growth would show increased positive selection in rainforest hunter-gatherer populations, we curated a priori lists of growth-related genes as described fully in SI Appendix, Text S1. Briefly, we obtained the following gene lists: (i) 3,996 genes that affect growth or size in mice (MP:0005378) from the Mouse/Human Orthology with Phenotype Annotations database (63); (ii) 266 genes associated with abnormal skeletal growth syndromes in the Online Mendelian Inheritance in Man (OMIM) database (https://omim.org), as assembled by ref. 64; (iii) 427 genes expressed substantially more highly in the mouse growth plate, the cartilaginous region on the end of long bones where bone elongation occurs, than in soft tissues [lung, kidney, heart; 2.0-fold change (65)]; and (iv) 955 genes annotated with the GO growth biological process (GO:0040007). As the GH/IGF1 pathway is a major regulator of growth and disruptions to the pathway have been implicated in the adult small body-size phenotype, we also collected lists of genes associated with GH and IGF1, respectively, from the Online Predicted Human Interaction Database (OPHID) of protein–protein interaction (PPI) networks (66). Separately, we also used a list of genes found to be associated with the adult small body-size phenotype in the Batwa (17).
Statistical Overrepresentation and Distribution Shift Tests.
Using the PBS and Bayenv indexes, we next tested for a statistical overrepresentation of extreme values () for the above a priori gene lists as well as all GO terms, using the topGO package of Bioconductor (67), gene-to-GO mapping from the “org.Hs.eg.db” package (68), and Fisher’s exact test in “classic” mode (i.e., without adjustment for GO hierarchy). We similarly performed a statistical enrichment test using the KS test again in classic mode, which tested for a shift in the distribution of the PBS or Bayenv statistic, rather than an excess of extreme values. In all cases, we pruned the GO hierarchy to exclude GO terms with fewer than 50 annotated genes to reduce the number of tests, leaving 1,742 and 1,816 GO biological processes and 266 and 285 GO molecular functions tested for the African and Asian datasets, respectively. To further reduce the number of redundant tests, we also computed the semantic similarity between GO terms to remove very similar terms. We computed the similarity metric of ref. 69 as implemented in the GoSemSim R package (70), a measure of the overlapping information content in each term using the annotation statistics of their common ancestor terms, and then clustered based on these pairwise distances between GO terms using Ward hierarchical clustering. We then pruned GO terms by cutting the tree at a height of 0.5 and retaining the term in each cluster with the lowest P value. With this reduced set of GO overrepresentation and distribution shift results, we adjusted the P value for FDR.
Identification of Signatures of Convergent Evolution.
We used two methods to identify convergent evolution: (i) computation of simple combined P values for SNPs, genes, and GO overrepresentation and distribution shift tests using Fisher’s and Edgington’s methods and (ii) a permutation-based approach to identify GO pathways for which both the Batwa and Andamanese overrepresentation or distribution shift test results are more extreme than is to be expected by chance (the “empirical test for convergence”). These two approaches are summarized below.
We searched for convergence between Batwa and Andamanese individuals by computing the joint P value for the PBS on a per-SNP, per-gene, and per-GO term basis. We calculated all joint P values using Fisher’s method [as the sum of the natural logarithms of the uncorrected P values for the Batwa and Andamanese tests (71) as well as via Edgington’s method [based on the sum of all P values (72)]. Metaanalysis of P values was done via custom script and the metap R package (73).
We also assessed the probability of getting two false positives in the Batwa and Andamanese selection results by shuffling the genes’ PBS indexes 1,000 times and performing GO overrepresentation and distribution shift tests on these permuted values. We compared the observed Batwa and Andamanese P values to this generated distribution of P values, as described above. We computed the joint probability of both null hypotheses being false for the Andamanese and Batwa as ()(), where and are the P values of Fisher’s exact test or of the KS test for the outlier- and shift-based tests, respectively, and we compared the joint probability to the same statistic computed for the P values from the random iterations. The empirical test for convergence P value was simply the number of iterations for which this statistic was more extreme (lower) for the observed values than for the randomly generated values.
We also performed a variation of this analysis, but to preserve patterns of linkage disequilibrium among SNPs within a gene in the null distribution, instead of permuting gene–PBS relationships to generate the random null distributions for the PBS selection index values of the two populations considered jointly, we instead permuted the gene–GO relationships. That is, to compute the PBS selection index, the one-to-many relationships between genes and GO terms were shuffled when generating the null distribution, maintaining the groupings of GO terms that were assigned together to an original gene. Full details of this analysis are available in SI Appendix, Text S4.
Script and Data Availability.
All scripts used in the analysis are available at https://github.com/bergeycm/rhg-convergence-analysis and released under the GNU General Public License v3. Exome data for the Batwa and Bakiga populations have previously been deposited in the European Genome–phenome Archive available at https://ega-archive.org/ under accession code EGAS00001002457.
Supplementary Material
Acknowledgments
We thank the Batwa and Bakiga communities and all individuals who participated in this study and J. A. Hodgson and E. C. Reeves for helpful discussions. This work was supported by NIH Grant R01-GM115656 (to G.H.P. and L.B.B.), NIH Grant 1 F32 GM125228-01A1 (to C.M.B), and Agence Nationale de la Recherche AGRHUM ANR-14-CE02-0003-01 (to L.Q.-M.). M.L. was supported by the Fondation pour la Recherche Médicale (FDT20170436932). This research was conducted with Advanced CyberInfrastructure computational resources provided by The Institute for CyberScience at The Pennsylvania State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Exome data for the Batwa and Bakiga populations have been deposited in the European Genome–Phenome Archive (EGA), https://ega-archive.org/ (accession no. EGAS00001002457). All scripts used in the analysis are available on GitHub (https://github.com/bergeycm/rhg-convergence-analysis).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812135115/-/DCSupplemental.
References
- 1.Stern DL. The genetic causes of convergent evolution. Nat Rev Genet. 2013;14:751–764. doi: 10.1038/nrg3483. [DOI] [PubMed] [Google Scholar]
- 2.Elmer KR, Meyer A. Adaptation in the age of ecological genomics: Insights from parallelism and convergence. Trends Ecol Evol. 2011;26:298–306. doi: 10.1016/j.tree.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Christin PA, Weinreich DM, Besnard G. Causes and evolutionary significance of genetic convergence. Trends Genet. 2010;26:400–405. doi: 10.1016/j.tig.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Protas ME, et al. Genetic analysis of cavefish reveals molecular convergence in the evolution of albinism. Nat Genet. 2006;38:107–111. doi: 10.1038/ng1700. [DOI] [PubMed] [Google Scholar]
- 5.Gross JB, Borowsky R, Tabin CJ. A novel role for Mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax mexicanus. PLoS Genet. 2009;5:e1000326. doi: 10.1371/journal.pgen.1000326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tishkoff SA, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40. doi: 10.1038/ng1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pritchard JK, Di Rienzo A. Adaptation–Not by sweeps alone. Nat Rev Genet. 2010;11:665–667. doi: 10.1038/nrg2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: Hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–R215. doi: 10.1016/j.cub.2009.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–1423. doi: 10.1534/genetics.110.114819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stephan W. Signatures of positive selection: From selective sweeps at individual loci to subtle allele frequency changes in polygenic adaptation. Mol Ecol. 2016;25:79–88. doi: 10.1111/mec.13288. [DOI] [PubMed] [Google Scholar]
- 11.Wellenreuther M, Hansson B. Detecting polygenic evolution: Problems, pitfalls, and promises. Trends Genet. 2016;32:155–164. doi: 10.1016/j.tig.2015.12.004. [DOI] [PubMed] [Google Scholar]
- 12.Marouli E, et al. Rare and low-frequency coding variants alter human adult height. Nature. 2017;542:186–190. doi: 10.1038/nature21039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perry GH, Dominy NJ. Evolution of the human pygmy phenotype. Trends Ecol Evol. 2009;24:218–225. doi: 10.1016/j.tree.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen M, Guo X, Wang Y. An Aboriginal Australian genome reveals separate human dispersals into Asia. Science. 2011;334:94–98. doi: 10.1126/science.1211177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Migliano AB, et al. Evolution of the pygmy phenotype: Evidence of positive selection from genome-wide scans in African, Asian, and Melanesian Pygmies. Hum Biol. 2013;85:251–284. doi: 10.3378/027.085.0313. [DOI] [PubMed] [Google Scholar]
- 16.Perry GH, Verdu P. Genomic perspectives on the history and evolutionary ecology of tropical rainforest occupation by humans. Quat Int. 2016;448:150–157. [Google Scholar]
- 17.Perry GH, et al. Adaptive, convergent origins of the pygmy phenotype in African rainforest hunter-gatherers. Proc Natl Acad Sci USA. 2014;111:E3596–E3603. doi: 10.1073/pnas.1402875111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker NSA, et al. Indirect evidence for the genetic determination of short stature in African Pygmies. Am J Phys Anthropol. 2011;145:390–401. doi: 10.1002/ajpa.21512. [DOI] [PubMed] [Google Scholar]
- 19.Jarvis JP, et al. Patterns of ancestry, signatures of natural selection, and genetic association with stature in Western African Pygmies. PLoS Genet. 2012;8:e1002641. doi: 10.1371/journal.pgen.1002641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pemberton TJ, et al. A genome scan for genes underlying adult body size differences between Central African hunter-gatherers and farmers. Hum Genet. 2018;137:487–509. doi: 10.1007/s00439-018-1902-3. [DOI] [PubMed] [Google Scholar]
- 21.Lachance J, et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsieh P, et al. Whole genome sequence analyses of Western Central African Pygmy hunter-gatherers reveal a complex demographic history and identify candidate genes under positive natural selection. Genome Res. 2015;26:279–290. doi: 10.1101/gr.192971.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lopez M, et al. The demographic history and mutational load of African hunter-gatherers and farmers. Nat Ecol Evol. 2018;2:721–730. doi: 10.1038/s41559-018-0496-4. [DOI] [PubMed] [Google Scholar]
- 24.Mondal M, et al. Genomic analysis of Andamanese provides insights into ancient human migration into Asia and adaptation. Nat Genet. 2016;48:1066–1070. doi: 10.1038/ng.3621. [DOI] [PubMed] [Google Scholar]
- 25.Mondal M, Casals F, Majumder PP, Bertranpetit J. 2016. Further confirmation for unknown archaic ancestry in Andaman and South Asia. bioRxiv:10.1101/071175. Preprint, posted August 23, 2016.
- 26.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patin E, et al. The impact of agricultural emergence on the genetic history of African rainforest hunter-gatherers and agriculturalists. Nat Commun. 2014;5:3163. doi: 10.1038/ncomms4163. [DOI] [PubMed] [Google Scholar]
- 28.Huerta-Sánchez E, et al. Genetic signatures reveal high-altitude adaptation in a set of Ethiopian populations. Mol Biol Evol. 2013;30:1877–1888. doi: 10.1093/molbev/mst089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yi X, et al. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Campeau PM, et al. Yunis-Varón syndrome is caused by mutations in FIG4, encoding a phosphoinositide phosphatase. Am J Hum Genet. 2013;92:781–791. doi: 10.1016/j.ajhg.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Daub JT, et al. Evidence for polygenic adaptation to pathogens in the human genome. Mol Biol Evol. 2013;30:1544–1558. doi: 10.1093/molbev/mst080. [DOI] [PubMed] [Google Scholar]
- 32.Günther T, Coop G. Robust identification of local adaptation from allele frequencies. Genetics. 2013;195:205–220. doi: 10.1534/genetics.113.152462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rimoin DL, Merimee TJ, Rabinowitz D, Cavalli-Sforza LL, McKusick VA. Peripheral subresponsiveness to human growth hormone in the African Pygmies. N Engl J Med. 1969;281:1383–1388. doi: 10.1056/NEJM196912182812502. [DOI] [PubMed] [Google Scholar]
- 34.Merimee TJ, Rimoin DL, Cavalli-Sforza LC, Rabinowitz D, McKusick VA. Metabolic effects of human growth hormone in the African pygmy. Lancet. 1968;292:194–195. doi: 10.1016/s0140-6736(68)92624-x. [DOI] [PubMed] [Google Scholar]
- 35.Merimee TJ, Rimoin DL, Cavalli-Sforza LL. Metabolic studies in the African pygmy. J Clin Invest. 1972;51:395–401. doi: 10.1172/JCI106825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geffner ME, Bailey RC, Bersch N, Vera JC, Golde DW. Insulin-like growth factor-I unresponsiveness in an Efe Pygmy. Biochem Biophysical Res Commun. 1993;193:1216–1223. doi: 10.1006/bbrc.1993.1755. [DOI] [PubMed] [Google Scholar]
- 37.Geffner ME, Bersch N, Bailey RC, Golde DW. Insulin-like growth factor I resistance in immortalized T cell lines from African Efe Pygmies. J Clin Endocrinol Metab. 1995;80:3732–3738. doi: 10.1210/jcem.80.12.8530626. [DOI] [PubMed] [Google Scholar]
- 38.Carroll PV, et al. Growth hormone deficiency in adulthood and the effects of growth hormone replacement: A review. J Clin Endocrinol Metab. 1998;83:382–395. doi: 10.1210/jcem.83.2.4594. [DOI] [PubMed] [Google Scholar]
- 39.Arcopinto M, et al. Growth hormone deficiency is associated with worse cardiac function, physical performance, and outcome in chronic heart failure: Insights from the T.O.S.CA. GHD study. PLoS One. 2017;12:e0170058. doi: 10.1371/journal.pone.0170058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paajanen TA, Oksala NK, Kuukasjärvi P, Karhunen PJ. Short stature is associated with coronary heart disease: A systematic review of the literature and a meta-analysis. Eur Heart J. 2010;31:1802–1809. doi: 10.1093/eurheartj/ehq155. [DOI] [PubMed] [Google Scholar]
- 41.Nelson CP, et al. Genetically determined height and coronary artery disease. N Engl J Med. 2015;372:1608–1618. doi: 10.1056/NEJMoa1404881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devesa J, Almengló C, Devesa P. Multiple effects of growth hormone in the body: Is it really the hormone for growth? Clin Med Insights Endocrinol Diabetes. 2016;9:47–71. doi: 10.4137/CMED.S38201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Meyers DE, Cuneo RC. Controversies regarding the effects of growth hormone on the heart. Mayo Clin Proc. 2003;78:1521–1526. doi: 10.4065/78.12.1521. [DOI] [PubMed] [Google Scholar]
- 44.Mathews LS, Enberg B, Norstedt G. Regulation of rat growth hormone receptor gene expression. J Biol Chem. 1989;264:9905–9910. [PubMed] [Google Scholar]
- 45.Langenberg C, Hardy R, Kuh D, Wadsworth ME. Influence of height, leg and trunk length on pulse pressure, systolic and diastolic blood pressure. J Hypertens. 2003;21:537–543. doi: 10.1097/00004872-200303000-00019. [DOI] [PubMed] [Google Scholar]
- 46.Barreto-Filho JAS, et al. Familial isolated growth hormone deficiency is associated with increased systolic blood pressure, central obesity, and dyslipidemia. J Clin Endocrinol Metab. 2002;87:2018–2023. doi: 10.1210/jcem.87.5.8474. [DOI] [PubMed] [Google Scholar]
- 47.Markussis V, et al. Abnormal carotid arterial wall dynamics in symptom-free hypopituitary adults. Eur J Endocrinol. 1997;136:157–164. doi: 10.1530/eje.0.1360157. [DOI] [PubMed] [Google Scholar]
- 48.Reeve JC, Abhayaratna WP, Davies JE, Sharman JE. Central hemodynamics could explain the inverse association between height and cardiovascular mortality. Am J Hypertens. 2014;27:392–400. doi: 10.1093/ajh/hpt222. [DOI] [PubMed] [Google Scholar]
- 49.Moussouami SI, et al. Prevalence and risk factors of cardiovascular diseases in the Congo-Brazzaville Pygmies. World J Cardiovasc Dis. 2016;6:211–217. [Google Scholar]
- 50.Kesteloot H, Ndam N, Sasaki S, Kowo M, Seghers V. A survey of blood pressure distribution in pygmy and Bantu populations in Cameroon. Hypertension. 1996;27:108–113. doi: 10.1161/01.hyp.27.1.108. [DOI] [PubMed] [Google Scholar]
- 51.Lemogoum D, et al. Effects of hunter-gatherer subsistence mode on arterial distensibility in Cameroonian Pygmies. Hypertension. 2012;60:123–128. doi: 10.1161/HYPERTENSIONAHA.111.187757. [DOI] [PubMed] [Google Scholar]
- 52.Ngatchou W, et al. Arterial stiffness and cardiometabolic phenotype of Cameroonian Pygmies and Bantus. J Hypertens. 2018;36:520–527. doi: 10.1097/HJH.0000000000001577. [DOI] [PubMed] [Google Scholar]
- 53.Mancilha-Carvalho JJ, Esposito R. Blood pressure and electrolyte excretion in the Yanomamo Indians, an isolated population. J Hum Hypertens. 1989;3:309–314. [PubMed] [Google Scholar]
- 54.Mann GV, Roels OA, Price DL, Merrill JM. Cardiovascular disease in African Pygmies: A survey of the health status, serum lipids and diet of Pygmies in Congo. J Chronic Dis. 1961;15:341–371. doi: 10.1016/0021-9681(62)90082-6. [DOI] [PubMed] [Google Scholar]
- 55.Han X, Cheng H, Mancuso DJ, Gross RW. Caloric restriction results in phospholipid depletion, membrane remodeling, and triacylglycerol accumulation in murine myocardium. Biochemistry. 2004;43:15584–15594. doi: 10.1021/bi048307o. [DOI] [PubMed] [Google Scholar]
- 56.Hidron A, et al. Cardiac involvement with parasitic infections. Clin Microbiol Rev. 2010;23:324–349. doi: 10.1128/CMR.00054-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- 60.Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Leary NA, et al. Reference sequence (RefSeq) database at NCBI: Current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44:D733–D745. doi: 10.1093/nar/gkv1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blake JA, et al. Mouse Genome Database (MGD)-2017: Community knowledge resource for the laboratory mouse. Nucleic Acids Res. 2017;45:D723–D729. doi: 10.1093/nar/gkw1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lui JC, et al. Synthesizing genome-wide association studies and expression microarray reveals novel genes that act in the human growth plate to modulate height. Hum Mol Genet. 2012;21:5193–5201. doi: 10.1093/hmg/dds347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brown KR, Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 67.Alexa A, Rahnenfuhrer J. 2016 topGO: Enrichment Analysis for Gene Ontology, R Package Version 2(26). Availavle at https://bioconductor.org/packages/release/bioc/html/topGO.html. Accessed October 26, 2018.
- 68.Carlson M. 2017 org.Hs.eg.db: Genomewide Annotation for Human. Available at https://bioconductor.org/packages/release/data/annotation/html/org.Hs.eg.db.html. Accessed October 26, 2018.
- 69.Jiang JJ, Conrath DW. 1997. Semantic similarity based on corpus statistics and lexical taxonomy. Proceedings of International Conference Research on Computational Linguistics (ROCLING X), eds K-J Chen, C-R Huang, R Sproat (The Association for Computational Linguistics and Chinese Language Processing (ACLCLP), Taipei, Taiwan), pp 19–33.
- 70.Yu G, et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics. 2010;26:976–978. doi: 10.1093/bioinformatics/btq064. [DOI] [PubMed] [Google Scholar]
- 71.Mosteller F, Fisher R. Questions and answers. Am Stat. 1948;2:30–31. [Google Scholar]
- 72.Edgington ES. An additive method for combining probability values from independent experiments. J Psychol. 1972;80:351–363. [Google Scholar]
- 73.Dewey M. metap: Meta-Analysis of Significance Values. 2017 Available at https://cran.r-project.org/web/packages/metap/index.html. Accessed October 26, 2018.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.