Significance
Hepatitis B virus (HBV)-associated acute liver failure (ALF), also known as fulminant hepatitis B, is a rare but often fatal complication of acute HBV infection. The pathogenesis of ALF is still largely unknown due to the lack of experimental systems and the difficulties in obtaining liver samples. Our comprehensive study of both liver tissue and serum samples from ALF patients using cutting-edge technologies allowed us to identify viral and host factors uniquely associated with this disease. In contrast to classic acute hepatitis B where the liver damage appears to be T cell-mediated, this study demonstrates a major role of the humoral immunity in the pathogenesis of HBV-associated ALF, which may open new avenues for the diagnosis and treatment of this dramatic disease.
Keywords: hepatitis B virus, acute liver failure, hepatitis B core antigen, humoral immunity, pathogenesis
Abstract
Hepatitis B virus (HBV)-associated acute liver failure (ALF) is a dramatic clinical syndrome leading to death or liver transplantation in 80% of cases. Due to the extremely rapid clinical course, the difficulties in obtaining liver specimens, and the lack of an animal model, the pathogenesis of ALF remains largely unknown. Here, we performed a comprehensive genetic and functional characterization of the virus and the host in liver tissue from HBV-associated ALF and compared the results with those of classic acute hepatitis B in chimpanzees. In contrast with acute hepatitis B, HBV strains detected in ALF livers displayed highly mutated HBV core antigen (HBcAg), associated with increased HBcAg expression ex vivo, which was independent of viral replication levels. Combined gene and miRNA expression profiling revealed a dominant B cell disease signature, with extensive intrahepatic production of IgM and IgG in germline configuration exclusively targeting HBcAg with subnanomolar affinities, and complement deposition. Thus, HBV ALF appears to be an anomalous T cell-independent, HBV core-driven B cell disease, which results from the rare and unfortunate encounter between a host with an unusual B cell response and an infecting virus with a highly mutated core antigen.
Acute liver failure (ALF), previously known as fulminant hepatitis, is a rare but dramatic clinical syndrome characterized by the sudden loss of hepatocytes, leading to multiorgan failure in a person without preexisting liver disease (1). Hepatitis B virus (HBV) is a major cause of ALF worldwide (2). However, the difficulties in obtaining liver specimens and the extremely rapid clinical course of this disease, along with the lack of an animal model, have hampered pathogenesis studies. While in the classic form of acute hepatitis B liver damage and viral clearance are mediated by specific antiviral T cell responses (3–5) with a dominant intrahepatic T cell gene signature (6), little is known of the pathogenic mechanisms leading to ALF. On the viral side, variants of HBV containing precore or core promoter mutations that affect hepatitis B e antigen (HBeAg) expression have been associated with ALF (7–11), although the same mutations are also commonly detected in chronic hepatitis B surface antigen (HBsAg) carriers (12), suggesting that ALF is the result of a complex interplay between the virus and the host (3). On the host side, early studies conducted in the 1970s and 1980s demonstrated an unusually brisk antibody response against all HBV antigens, with significantly higher titers of IgM to hepatitis B core antigen (HBcAg) (13) and rapid clearance of HBsAg and HBeAg, compared with classic acute hepatitis B (13, 14), often accompanied by low levels of HBV replication (15–17). The question of whether an enhanced humoral immune response plays a role in the pathogenesis of HBV-associated ALF remains unanswered. More recently, high titers of IgM anticore were shown to differentiate ALF that follows primary HBV infection from ALF developing upon exacerbation of chronic HBV infection (18), suggesting that an exaggerated IgM response against HBcAg may be implicated in the pathogenesis of this disease. Consistent with these findings, we have previously shown that HBV-associated ALF is characterized by an overriding B cell gene signature centered in the liver, with extensive intrahepatic expression of IgG and IgM antibodies directed against the HBcAg (19).
Access to liver specimens from additional patients with HBV ALF provided us with the unique opportunity to confirm and extend our original observations (19) and to perform a genetic and functional characterization of the HBV strains associated with ALF. Here, we investigated the role of viral and host factors in the pathogenesis of HBV ALF in primary liver tissue, which is the anatomical site where the disease occurs, and compared the findings in ALF with those in classic acute hepatitis B in chimpanzees. By using cutting-edge molecular techniques, our study corroborates the pathogenic role of humoral immunity in HBV ALF.
Results
Clinical, Serologic, and Virologic Characteristics of the Patients.
Four healthy young adult individuals, two males and two females with a mean age ± SD of 42.2 ± 7.2 y, suddenly developed HBV ALF, with progressive coagulopathy and encephalopathy, and all underwent liver transplantation within 8 d of the onset of symptoms. Two patients had massive hepatic necrosis (patients 31 and 241) and two submassive hepatic necrosis (patients 32 and 219). Serologic HBV markers tested at admission and at liver transplantation showed rapid clearance of HBsAg and HBeAg and accelerated antibody seroconversion, with extraordinarily high titers of IgM anti-HBc (>1:500,000) at the time of liver transplantation in all patients, whereas the IgG anti-HBc titer was low (ranging from 1:40 to 1:400). The levels of HBV DNA ranged from 2.4 to 5.3 logs in serum and from 1.3 to 3.7 in liver; HBV RNA was also detected, demonstrating the persistence of HBV replication and viral gene expression at the time of liver transplantation (SI Appendix, Table S1). The titers of circulating IgM and IgG anti-HBc antibodies were also determined in patients with acute hepatitis B. We used archived serial serum samples derived from a prospective study conducted at the NIH on transfused heart surgery patients (20). Four patients were selected based on the availability of serial serum samples; two of them were infected with only HBV and two coinfected with HBV and hepatitis C virus. The IgM anti-HBc titer ranged from 1:4,096 to 1:41,943 (SI Appendix, Table S2), whereas the highest IgG anti-HBc titer was 1:128. There were no significant differences between monoinfected and coinfected patients, although these tests were performed on archived serum samples and there were no weekly samples throughout the course of the acute hepatitis. We also studied normal liver controls, including 10 liver donors and 7 subjects who underwent liver resection for hepatic hemangioma. None of them had evidence of active infection with hepatitis A, B, C, or D or other known viral infections. Their demographic, clinical, and liver pathology data are shown in SI Appendix, Table S3.
Genetic Characterization of HBV Strains in ALF by Next-Generation Sequencing.
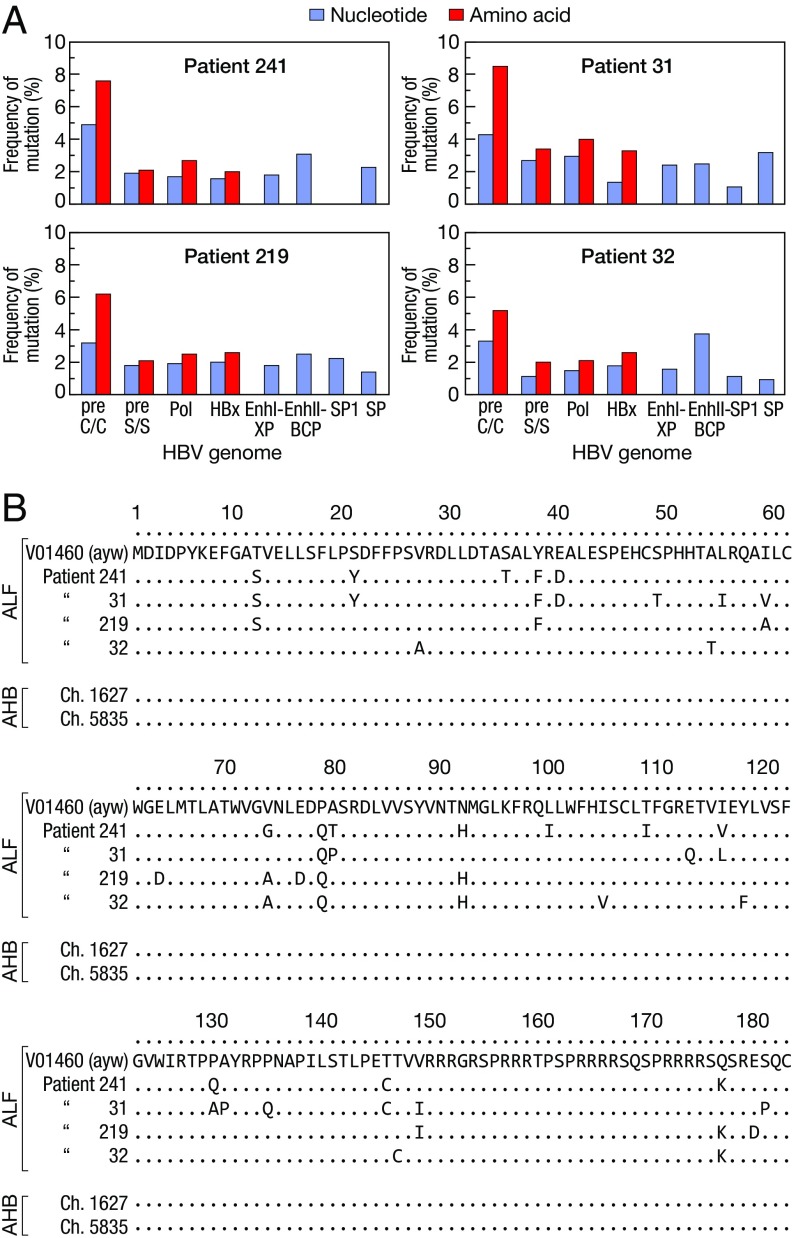
To characterize the HBV strains associated with ALF, we performed next-generation sequencing (NGS) of the entire HBV genome in serum and liver from each patient with ALF (SI Appendix, Fig. S1A). We found that all patients with ALF were infected with HBV genotype D, subgenotype D3, and harbored a highly homogeneous viral population (over 90% represented by a single dominant strain), but highly divergent from the reference HBV genotype D wild type (ayw) (21) (SI Appendix, Table S4). The precore stop codon mutation G1896A, which abrogates HBeAg production, was invariably present, whereas other mutations previously linked to ALF, namely A1762T and G1764A (9), were not found; no deletions, insertions, or nonsense mutations were detected (SI Appendix, Table S4). The precore/core was the most variable region of the entire HBV genome, with the highest rate of nonsynonymous mutations in all patients (Fig. 1A). Although the mutations were scattered throughout the HBcAg (Fig. 1B), we found two to three amino acid substitutions within the major B cell epitope (amino acids 73–85, positioned at the tip of the capsid spikes) (22) in each patient. Amino acid changes were also found at the level of the nuclear localization signal of HBcAg, at amino acid position 147–183 (Fig. 1B), whereas no mutations were detected in the major B cell epitopes of HBsAg (SI Appendix, Table S4). In contrast, no mutations were detected in HBcAg from the two chimpanzees with classic acute hepatitis B (Fig. 1B), as well as in HBcAg sequences from several patients with acute hepatitis B available in GenBank (SI Appendix, Table S5), indicating that ALF is specifically associated with a highly mutated HBcAg compared with the reference wild-type HBV.
Fig. 1.
Genetic heterogeneity of HBV in ALF. (A) Rate of mutation within the entire HBV genome in four patients with ALF compared with the wild-type reference HBV strain (ayw V01460) (21). (B) Sequence alignments of HBcAg in patients with ALF, patients with acute hepatitis B, and two chimpanzees with acute hepatitis B, compared with the wild-type HBV strain (ayw V01460). The sequence labels represent GenBank accession numbers. AHB denotes acute hepatitis B.
Functional Analysis of HBV Variants from Patients with ALF.
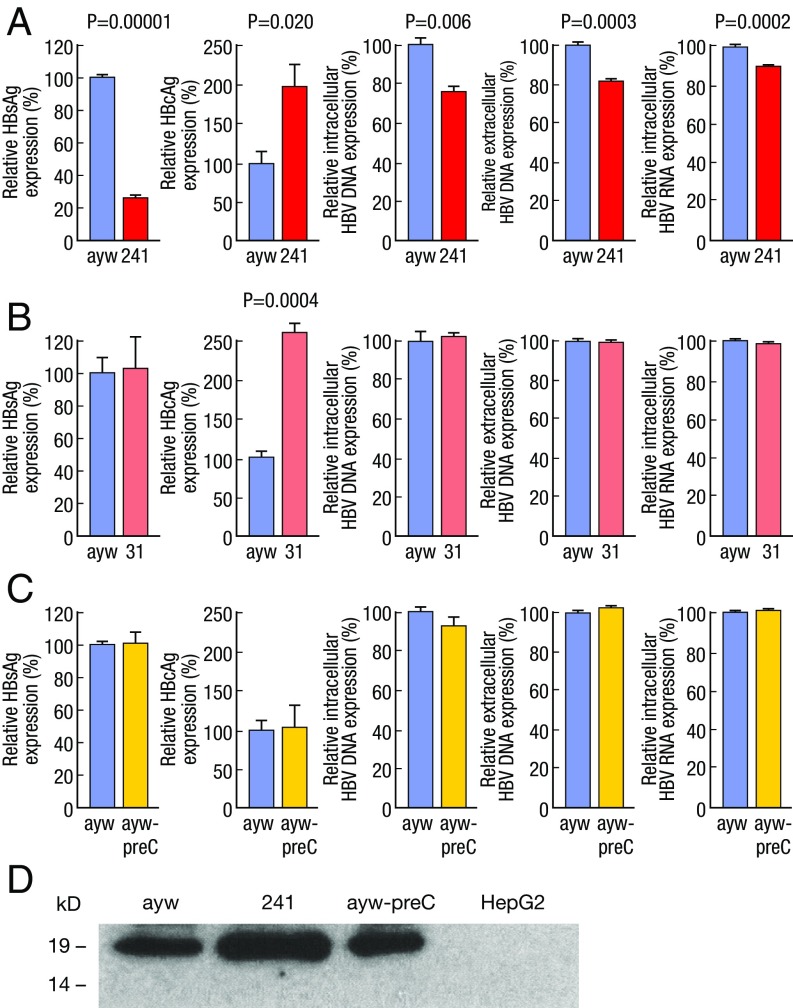
To get insights into the role of viral factors in the molecular pathogenesis of ALF, replication-competent HBV was cloned from the two patients with massive liver necrosis (patients 241 and 31) and expressed in HepG2 hepatoma cells (SI Appendix, Fig. S1B). In comparison with the wild-type HBV (ayw), HBV-241 showed significantly lower HBsAg production and viral replication, with significantly reduced levels of both extracellular and intracellular viral DNA and total intracellular viral RNA (Fig. 2A). However, HBV-241 showed increased HBcAg expression by approximately twofold in comparison with wild-type HBV (P = 0.020), as measured by ELISA and confirmed by immunoprecipitation and Western blot with antibodies that react equally well with core antigens from HBV-241 and wild-type HBV (Fig. 2 A and D and SI Appendix, Fig. S2A and Table S6). Similarly, HBV-31 showed increased HBcAg expression by more than twofold over wild-type HBV (P = 0.0004), while HBsAg expression and viral replication were similar to those of wild-type HBV (Fig. 2B and SI Appendix, Fig. S2B and Table S6). Kinetic analysis showed that the HBcAg level increased over time after transfection, with the highest level detected at day 5 posttransfection (SI Appendix, Fig. S2C).
Fig. 2.
Quantitative analysis of viral replication and antigen expression by patient-derived HBV variants versus wild-type HBV. HepG2 cells were transfected with monomeric linear full-length HBV genomes of wild type (ayw) (21), two HBV variants cloned from two patients with ALF (241 and 31), and the wild-type HBV mutant containing the single preC stop mutation (ayw-preC). The level of intracellular and extracellular HBV DNA and the level of total RNA were determined by real-time PCR and the level of HBsAg and HBcAg expression was measured by ELISA. Relative level of HBsAg in the culture supernatants, cellular HBcAg, intracellular and extracellular viral DNA, and total RNA of (A) wild-type HBV (ayw) versus 241 HBV, (B) wild-type HBV (ayw) versus 31 HBV, or (C) wild-type HBV (ayw) versus the preC mutant. Values were normalized to those obtained with the wild-type HBV and presented as percentage of expression. The data are the mean from three independent experiments with SE. The significance in difference was evaluated by unpaired two-tailed t test. The concentration of HBcAg in patient-derived ALF HBV variants (241 and 31) was significantly higher compared with that measured for the reference ayw strain, whereas no significant difference was detected between ayw and ayw-preC. (D) Western blot analysis of HBcAg expression in cells transfected with different HBV variants after immunoprecipitation with homologous anti-HBc antibodies. The patient-derived 241 HBV produced higher levels of HBcAg than the reference HBV strain or ayw-preC.
Since a prominent feature of HBV in ALF is the presence of the precore stop codon mutation, we generated an HBV ayw mutant containing only the precore stop codon mutation at nucleotide position 1896 (ayw-preC). The precore stop codon mutation did not affect the levels of viral antigen production (HBsAg and HBcAg) and viral replication (Fig. 2C), compared with those observed with the wild-type HBV control. Thus, these results suggest that other mutations, rather than the precore stop codon in patient-derived HBV strains, are responsible for the peculiar phenotype of HBcAg overexpression.
Cellular Localization of HBcAg.
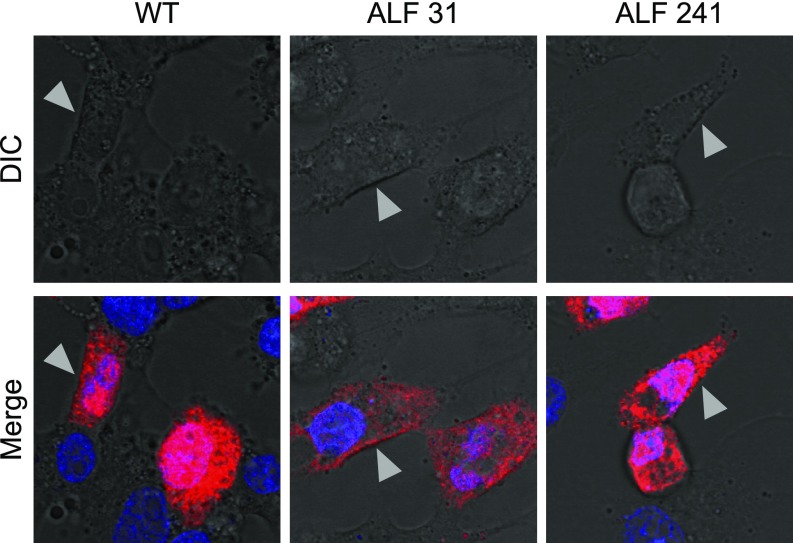
The expression of HBsAg and HBcAg was studied by confocal microscopy in HepG2 cells transfected with full-length, replication-competent linear DNA of wild-type HBV (ayw) or the HBV strains derived from two representative patients with massive liver necrosis (patients 31 and 241, with 95 and 100% of necrosis, respectively). The expression of HBcAg in cells fixed with only paraformaldehyde showed the presence of a strong cytoplasmic staining in cells transfected with all HBV strains, while the nuclear staining was negative (SI Appendix, Fig. S3). Instead, the expression of HBcAg in fixed and permeabilized cells was very intense both in the nucleus and in the cytoplasm for all HBV strains (SI Appendix, Figs. S4 and S5). The strong cytoplasmic HBcAg staining detected in the two strains derived from ALF patients (both HBeAg-negative at the time of liver transplantation; SI Appendix, Table S1) is consistent with the results of HBcAg localization studies using transfection of cloned HBcAg variants from anti-HBe–positive patients (23). Although in HBeAg-positive patients HBcAg tends to be nuclear (24), we detected a strong cytoplasmic staining also in cells transfected with the wild-type virus (SI Appendix, Figs. S3 and S4). Similar in vitro data were reported by Eyre et al. (25). Differential interference contrast (DIC) imaging was used to retrofit fluorescence images to show localization of the fluorescent signal on the plasma membrane and inside of the cells. By this technique we were able to visualize HBcAg on the cellular surface with all HBV strains, with an uneven, coarse granular distribution (Fig. 3). Only a minority of cells had isolated nuclear or cytoplasmic HBcAg localization. Whereas the vast majority of wild-type-transfected cells positive for HBcAg were also positive for HBsAg, in cells transfected with the two ALF strains not all cells positive for HBcAg were also positive for HBsAg (SI Appendix, Fig. S4).
Fig. 3.
Intracellular localization of hepatitis B core antigen (HBcAg) in HepG2 cells analyzed by confocal microscopy. HepG2 cells were transfected with full-length, replication-competent linear DNA of wild-type HBV (ayw) or two HBV strains derived from two representative patients with ALF (patients 31 and 241) and 48 h later were fixed, permeabilized, and immunostained by indirect immunofluorescence. High magnification (63×) of individual cells analyzed by DIC (Upper) shows an overlay of HBcAg with the cellular membrane (Lower). HBcAg appears in red (Alexa 594) and the nucleus in blue (DAPI). WT denotes wild-type HBV strain (ayw). The arrows indicate areas of overlay of HBcAg and the cellular membrane as shown in gray by DIC.
HBcAg Cell-Surface Binding and Complement Fixation.
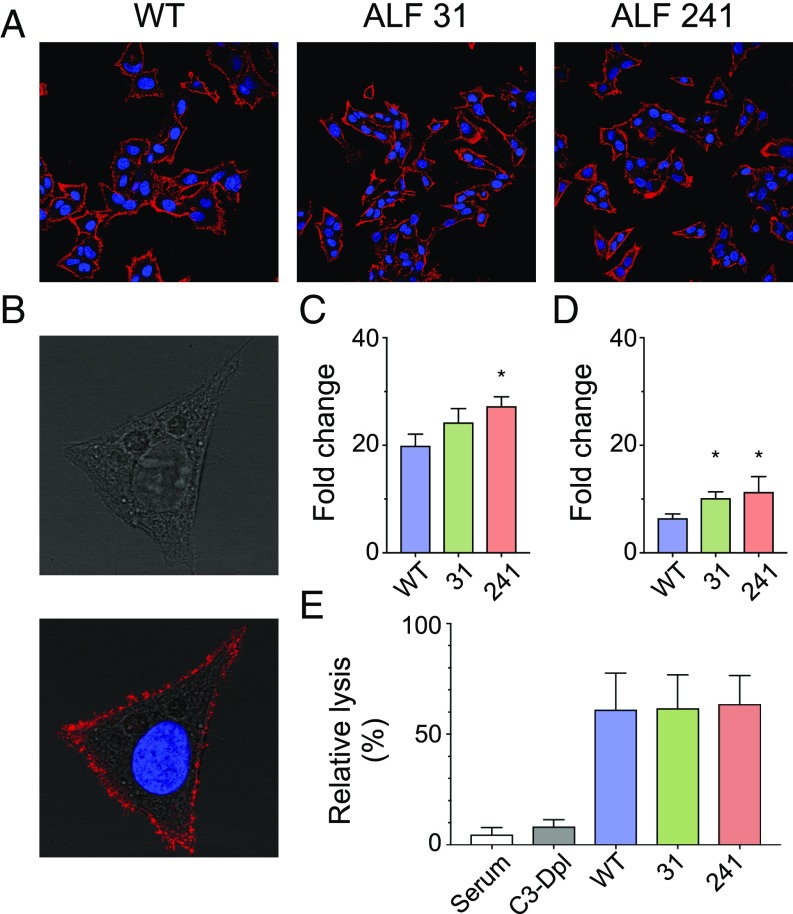
An alternative mechanism for the localization of HBcAg on the cellular membrane could be binding of HBcAg released from dying hepatocytes to the surface of hepatocytes, including bystander uninfected hepatocytes. To test this hypothesis, we performed an HBcAg cell surface-binding assay whereby untransfected HepG2 cells were incubated with HBcAg from wild-type HBV and from the ALF HBV strains 31 and 241; binding was revealed by immunostaining and flow cytometry using a specific anti-HBcAg Fab derived from an ALF patient (Fab E3). The staining unequivocally showed that HBcAg binds to the surface of HepG2 cells (Fig. 4 A and B), with a coarse pattern resembling that seen in cells transfected with full-length HBV strains (Fig. 3). Binding of HBcAg to the cellular membrane was confirmed by flow cytometry (Fig. 4C), which showed a stronger binding with the core from ALF strains, especially 241, than with the wild type.
Fig. 4.
Cell-surface binding of HBcAg to HepG2 cells, complement fixation, and cell lysis. Recombinant HBcAg synthesized from wild-type HBV and from ALF-derived HBV strains 31 and 241 was incubated with untransfected HepG2 cells. HBcAg binding was revealed by both immunostaining and flow cytometry using a specific anti-HBcAg FAb derived from an ALF patient (Fab E3) conjugated with a fluorophore (Alexa 647). (A) Immunostaining shows that HBcAg from all HBV strains binds to the entire liver cell membrane. (B) A higher magnification of a representative single cell bound to HBcAg using DIC shows a complete overlay of HBcAg staining with the cellular membrane, with a coarse granular pattern along the liver cell membrane. (C) Flow cytometry analysis confirmed the binding of HBcAg to the cellular membrane. The bars show the mean of fluorescence intensity (MFI) from three independent experiments expressed as fold change (mean + SD) with respect to the control without the addition of HBcAg. A stronger cell-surface binding was seen with HBcAg derived from ALF strains than with the wild type. The asterisk denotes P = 0.016 for the comparison between HBV 241 and ayw by unpaired two-tailed t test. (D) Flow cytometry analysis of C1q complement factor binding to HBcAg-human antibody complexes on the surface of HepG2 cells. The bars show the mean MFI (+SD) from three independent experiments expressed as fold change with respect to the control incubated with buffer only. (E) Complement-mediated cell lysis induced by HBcAg-containing immune complexes deposited on the surface of HepG2 cells. Cell lysis was measured by the uptake of propidium iodide and normalized to the uptake measured with the control (incubated with buffer only) and expressed as percent lysis. The data are the mean of three independent experiments (+ SD). The differences with cell lysis upon incubation with complete serum in the absence of immune complexes were evaluated by unpaired two-tailed t test.
Next, we investigated by flow cytometry if cell-surface immune complexes formed by HBcAg derived from all HBV strains in complex with a full-length ALF-derived anticore antibody (C7 IgG1) can bind to the C1q complement factor, trigger complement activation, and induce cell lysis. We found that C1q efficiently binds to the antigen–antibody complexes from all HBV strains (Fig. 4D), with a trend toward higher binding with the two ALF-derived HBV strains compared with the wild type. When complement-rich serum containing C1q was added to HepG2 cells bearing the immune complexes on their surface, cell lysis was detected in all samples with no significant differences between the wild type and the ALF HBV strains (Fig. 4E). Moreover, the cell lysis in cells incubated with C3-depleted serum was comparable to that in the cells incubated with complete serum in the absence of immune complexes. Altogether, these results confirmed that HBcAg can bind to the surface membrane of hepatocytes and form immune complexes, inducing complement deposition and activation of the classical pathway of the complement system.
Liver Gene and miRNA Expression Profiles in ALF.
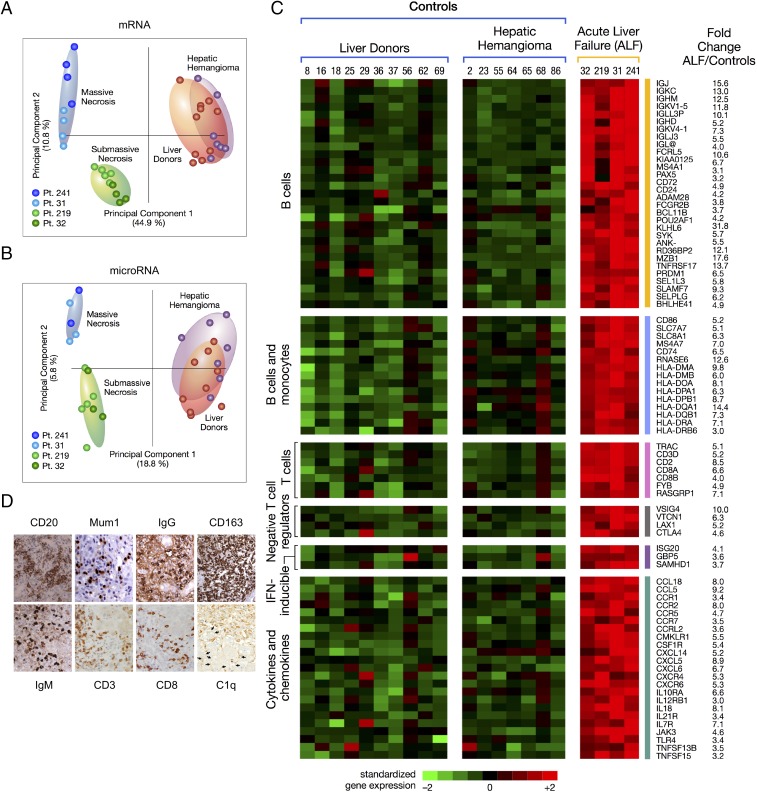
Next, we investigated the role of host factors in the pathogenesis of HBV-associated ALF (SI Appendix, Fig. S1A). Unsupervised principal component analysis of mRNA and miRNA showed a complete separation between ALF and controls (Fig. 5 A and B). Comparison between the liver of patients with ALF and controls showed a total of 1,351 mRNAs (SI Appendix, Fig. S6 A and B and Table S7) and 111 mature miRNAs (SI Appendix, Fig. S7 and Table S8) differentially expressed. Expression of both mRNA and miRNA was significantly associated with five major disease categories, with inflammatory and immunological diseases among the most prominent (SI Appendix, Fig. S8). Among mRNAs, humoral immune response genes were the most up-regulated, with an intrahepatic B cell gene signature (Fig. 5C) comprising a large number of Ig genes and genes involved in B cell development and maturation, including PRDM1/BLIMP1 and POU2AF1, two master regulators of germinal center formation and terminal B cell differentiation (26), MZB1 that is required for efficient humoral immune responses to T cell-independent and T cell-dependent antigens and promotes IgM assembly and secretion (27), FCRL5, which promotes B cell proliferation and the development of cells dyplaying multiple Ig (28), TNFRSF17/BCMA, which promote B cell maturation and survival (29), and BCL11B, an upstream gene regulator expressed by innate lymphoid cell types that respond exclusively to signaling via germline-encoded receptors (30). Another prominent gene signature was that shared by B cells and monocytes/macrophages (primarily HLA-II genes). Conversely, the number of T cell-associated genes was very limited (Fig. 5C), with no expression of the IFN-regulated chemokines CXCL9, CXCL10, and CXCL11. Of note, we detected four major negative regulators of T cell proliferation, activation, and T cell receptor signaling, namely, CTLA4, VSIG4, VTCN1, and LAX1 (Fig. 5C), which may provide an explanation for the limited T cell gene expression. In contrast with the findings in ALF, classic acute hepatitis B in chimpanzees was previously reported to be associated with a dominant intrahepatic T cell gene signature (6), including the IFN-regulated chemokines CXCL9, CXCL10, and CXCL11, that was temporally associated with viral clearance, whereas negative regulators of T cell activation were not detected, reflecting the key role of adaptive T cell responses in classic acute hepatitis B (6).
Fig. 5.
Principal component analysis of mRNA and miRNA, heat map of differentially expressed immune genes, and immunohistochemistry in the liver of patients with HBV-associated ALF. (A) Unsupervised principal component analyses of 6,996 mRNAs and (B) 1,105 mature miRNAs from four patients with ALF. The analysis also includes 17 samples from control livers, including 10 liver donors and 7 subjects who underwent liver resection for hepatic hemangioma. Each dot represents a single liver sample. Both plots show a complete separation between ALF and control livers. Within the ALF group, a separation was also observed between massive and submassive hepatic necrosis. (C) Heat map of immune genes differentially expressed in ALF. Each column represents a single patient. Data from the four ALF patients represent the average of multiple specimens analyzed (up to five liver specimens for each patient), whereas data from the 17 controls represent individual liver samples. Each cell represents the expression of a particular gene (rows) of a particular liver specimen (columns). Gene expression levels were log2-transformed and rowwise-standardized. Up-regulated genes are shown in shades of red and down-regulated genes in shades of green. (D) Immunohistochemical staining of liver tissue in ALF confirmed the prominent B cell signature with extensive infiltration of B cells (CD20), plasma blasts and plasma cells (Mum1), with strong cytoplasmic staining for IgM and IgG antibodies. The liver sections also showed the presence of T cells (CD3), mostly CD8-positive, extensive macrophage infiltration (CD163), and deposition of complement factor C1q. The arrows indicate weak linear intercellular staining of residual hepatocytes in a patient with submassive hepatic necrosis (magnification 40×).
Next, we investigated whether miRNAs expressed in ALF target B cell genes. We found that the vast majority of miRNAs (over 70%) target genes associated with B cell development, activation, and survival. The B cell-associated miRNAs expressed in ALF and their gene targets are shown in SI Appendix, Table S9. MiRNA expression in ALF was more similar to that of naïve B cells (up-regulation of let-7i, 342-3p, 342-5p, 361-3p, 200a, 221, 222, 200b, 10a, 150, 155, 99b-star; down-regulation of 455-3p, 106a, 17, 148a, 30a), than to that of centroblasts (up-regulation of 199b-3p, 15b, 138, 183 e 182) (31–35). We observed up-regulation of miRNAs involved in Ig somatic hypermutation and class-switch recombination (miR-155 and miR-181b, both of which target AID; miR-155 also targeting PU.1) (36–38),T cell-independent B cell responses (miR-182, the most up-regulated miRNA, with a fold change of 18.9) (39), and B cell differentiation and germinal centers (miR-150) (35, 40). In contrast, five members of the miR-30 family (30a-5p, 30b-5p, 30b-3p, 30c-1-3p, and 30e-5p) that target BCL6 were down-regulated, which may increase B cell survival (41). Thus, the integrated genome-wide analysis of mRNA and miRNA expression revealed a dominant B cell signature in HBV-associated ALF, consistent with a major role of humoral immunity in the pathogenesis of this disease.
Immunohistochemistry in ALF and Acute Hepatitis B.
In line with the prominent intrahepatic B cell lineage gene signature, we detected in all ALF patients an extensive liver infiltration of CD20+ mature B cells, plasma blasts and plasma cells strongly positive for cytoplasmic IgM and IgG, and mononuclear phagocytic cells expressing CD163 (Fig. 5D and SI Appendix, Fig. S9), whereas control normal liver tissues showed only rare B cells and plasma cells, a few CD3+ cells, and CD163+ Kupffer cells (SI Appendix, Fig. S9). Stainings with specific markers for naïve B cells (CD27 and IgD) showed that the vast majority of B cells were not classic naïve B cells, being negative for IgD and positive for CD27 (SI Appendix, Fig. S10). Despite the limited expression of T cell genes, the liver was infiltrated by CD3+ cells, predominantly CD8+ (Fig. 5D and SI Appendix, Fig. S9). In contrast, chimpanzees with acute hepatitis B (SI Appendix, Fig. S11A) showed low numbers of CD20+ B cells in portal spaces and rare plasma cells in the lobules, whereas the liver was mainly infiltrated by CD8+ T cells (SI Appendix, Fig. S11B). Thus, the extensive intrahepatic infiltration of CD20+ cells and plasma cells producing IgM and IgG was specifically associated with ALF. This intrahepatic expression of immunoglobulins was in all cases accompanied by complement deposition. We documented C1q deposition in liver tissue, predominantly on Kupffer cells, suggesting the presence of immune complexes phagocyted by these cells, along with a weak linear intercellular C1q staining in the two patients with submassive liver necrosis (Fig. 5D and SI Appendix, Fig. S12), consistent with membrane staining of residual hepatocytes; complement deposition was not detected in control liver tissues. In ALF, HBsAg and HBcAg were not detected in residual hepatocytes, whereas HBsAg was seen in the cytoplasm of both acutely HBV-infected chimpanzees; HBcAg was strongly positive in the nuclei of both animals with a diffuse cytoplasmic staining in one (SI Appendix, Fig. S13A).
Antigenic Target and Characteristics of Intrahepatic Antibodies in ALF.
Next, we investigated the reactivity of intrahepatic antibodies detected in ALF against autologous HBV antigens by phage display libraries (SI Appendix, Fig. S1D). Because of the extensive mutations we documented particularly in HBcAg, we reasoned that if specific anti-HBc antibodies are associated with the disease development they should react with the mutant antigens derived from these patients. Thus, viral genomic sequences encoding HBsAg and HBcAg from each patient were cloned, expressed, and used to screen Fab phage libraries constructed from the liver of the autologous patients (SI Appendix, Fig. S1C). The vast majority of the clones (92–99%; both IgG and IgM) were reactive with the autologous HBcAg, while none were reactive with HBsAg (SI Appendix, Table S10), indicating that HBcAg is the antigenic target of the intrahepatic B cell response in ALF. To validate these findings, we extracted IgM and IgG directly from frozen liver tissues of ALF patients, as well as from a normal liver donor, negative for HBV markers, as a control. Whereas IgM and IgG were recovered from all of the liver tissues tested, only IgM and IgG from patients with HBV ALF reacted specifically with three different HBV core antigens, while those from the control liver were negative (SI Appendix, Table S11). Thus, these results confirmed the presence of intrahepatic anticore IgM and IgG in ALF patients.
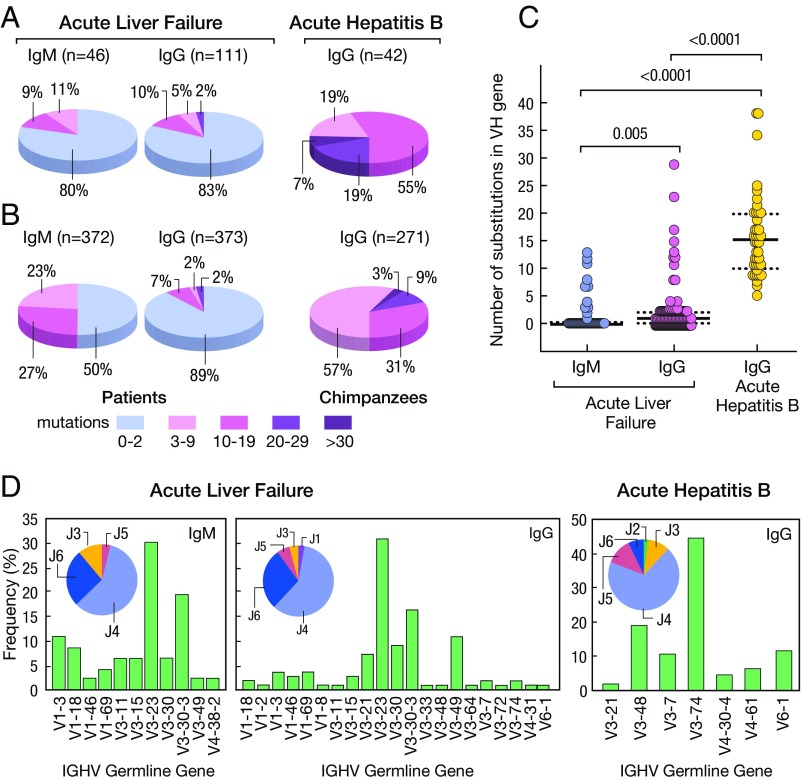
Sequence analysis of VH genes from the anti-HBc Fabs identified 46 unique IgM clones and 111 unique IgG clones (Fig. 6A). Strikingly, 80% of the IgM and 83% of the IgG anti-HBc antibodies were in germline configuration (Fig. 6A), suggesting that most of the anti-HBc are produced by naïve-like B cells (42). Analysis of the abundance of unmutated IgM and IgG clones showed that 89% of the IgG and 50% of the IgM were in germline configuration (Fig. 6B); furthermore, several pairs of IgM and IgG in each patient shared identical variable regions (SI Appendix, Tables S12 and S13), indicating that isotype switch from IgM to IgG had occurred without affinity maturation, a characteristic of T cell-independent B cell responses (43), and that unrelated patients share germline antibodies specific for HBcAg.
Fig. 6.
Characteristics of HBcAg-specific antibodies isolated from liver tissues of four patients with HBV-associated ALF and two chimpanzees with classic self-limited acute hepatitis B. (A) Proportion of variable genes from IgM (n = 46) and IgG (n = 111) detected in the livers of four patients with ALF or IgG (n = 46) from two chimpanzees with acute hepatitis B. The number of somatic mutations is identified by a different color. The percentage of antibodies in germline configuration as well as of those with different degrees of somatic hypermutation mutations are shown both in ALF and in classic acute hepatitis B in chimpanzees. (B) Abundance of antibodies with different degrees of mutations following three cycles of panning of the IgG and IgM libraries on HBcAg. (C) Average frequency of somatic mutations among IgM and IgG sequences of ALF patients or IgG from the two chimpanzees. Solid lines show the medians; dotted lines represent the interquartile range. P values refer to comparisons performed using the two-tailed Mann–Whitney test. (D) Frequency of IGHV and HJ genes used by HBcAg-specific IgM and IgG obtained from patients with ALF and IgG from chimpanzees with acute hepatitis B.
To investigate if the unusual anti-HBc antibodies in germline configuration are unique to ALF, we studied samples from the two chimpanzees with classic acute hepatitis B. PCR amplification of IgG and IgM Fab-encoding genes from the livers of the two chimpanzees, performed when high titer of serum IgM anticore antibodies were still detectable (SI Appendix, Fig. S13B), resulted in amplification of IgG, but not of IgM. Screening of IgG Fab phage display libraries from both chimpanzees against homologous HBsAg and HBcAg identified only antibodies specific for HBcAg, but not for HBsAg (SI Appendix, Table S10). A total of 42 distinct IgG clones were identified, 57% of which showed 3–9 somatic mutations and 43% 10–38 somatic mutations (Fig. 6 A and B). The number of somatic hypermutations in chimpanzees was significantly higher than in ALF (P < 0.0001) (Fig. 6C). In ALF, there was a strongly biased usage of the VH3 family, with about 50% of the clones using V3-23 and V3-30-3, a high prevalence of JH4 usage (Fig. 6D). A strong bias toward VH3 and JH4 was also seen in chimpanzees (Fig. 6D). Therefore, the overall pattern of gene usage for anticore heavy chain was not different from the normal gene usage distribution (44). However, it is interesting to note that genes V3-23 and V3-30-3, which were used at high frequency in the patients, were absent in chimpanzees since human and chimpanzees share a 99% homology at the genomic level (45) and show comparable antibody repertoires (www.vgenerepertoire.org). To ensure that phage libraries were unbiasied toward to certain V genes, the Fd amplicons which were used in the construction of IgM and IgG libraries of four ALF patients and one liver donor were subjected to NGS. As shown in SI Appendix, Fig. S14, almost all of the possible VH genes were amplified in both IgM and IgG libraries of each sample, indicating that full repertoires of IgM and IgG were unbiasedly represented in the libraries. Interestingly, the five high-affinity germline anticore antibody clones were identified almost exclusively from the ALF patients in whom the antibodies were originally recovered. To estimate the prevalence of a particular transcript obtained by NGS, we used two methods: (i) quantification of duplicate reads and (ii) quantification of unique duplicate sequences (SI Appendix, SI Materials and Methods). Both methods provided similar results. We found anti-HBc clones to be relatively rare, with frequencies between 1 × 10−3 and 2.4 × 10−6 (SI Appendix, Table S14).
To investigate the biological potency of intrahepatic antibodies in ALF, we measured the binding affinity of 11 representative Fab clones, six in germline configuration and five with 3–27 nucleotide substitutions, to both autologous and heterologous HBcAg by surface plasmon resonance. All of the germline IgM and IgG exhibited very high binding affinities, especially for autologous HBcAg, with Kd values in the nanomolar to subnanomolar range (Table 1 and SI Appendix, Fig. S15 and Table S15), as typically seen for affinity-matured hypermutated antibodies (46), as well as a broad range of reactivity against heterologous HBcAg. Surprisingly, the three Fabs with higher numbers of mutations (11–27) had overall lower binding affinities. For comparison, analysis of four representative chimpanzee Fab clones, all hypermutated with 6–19 nucleotide substitutions in the VH gene, showed high binding affinities to the homologous HBcAg, with Kd in the nanomolar to subnanomolar range, but limited cross-reactivity with core antigens derived from patients with ALF (only in two of four Fabs) (Table 1 and SI Appendix, Table S15).
Table 1.
Number of somatic mutations within the variable heavy chain (VH) gene and binding affinity to hepatitis B core antigen (HBcAg) measured by surface plasmon resonance of 10 fabs recovered from the livers of patients with ALF and chimpanzees with acute hepatitis B
| Disease outcome | Fab | Origin | Gene usage | Reads | No. of mutations in VH gene | Binding to HBcAg* Kd, nM | ||||
| 241 | 31 | 32 | 219 | ayw | ||||||
| ALF | F10 | 241-IgM/IgG | V3-15,D5-18,J6 | 14/70 | 0 | 7.14 | 269.0 | 15.5 | 46.2 | 207.0 |
| B7 | 31-IgM/IgG | V1-18,D3-16,J4 | 18/7 | 0 | 29.7 | 4.25 | 18.2 | 2.24 | 0.3 | |
| F4 | 31-IgG | V3-23,D1-1,J3 | 11 | 0 | 34.6 | 1.34 | 13.9 | 70.1 | 57.0 | |
| B8 | 31-IgG | V3-23,D1-1,J3 | 3 | 2 | 49.7 | 1.64 | 10.8 | 57.3 | 60.2 | |
| G3 | 32-IgM/IgG | V3-23,D3-16,J3 | 3/59 | 0 | 1.45 | 0.17 | 0.25 | 0.56 | 0.28 | |
| C7 | 219-IgG | V3-49,D2-8,J5 | 23 | 0 | 0.14 | 0.41 | 2.65 | 0.14 | 0.05 | |
| Acute hepatitis B | D10 | CH1627-IgG | V6-1,D6-19,J3 | 69 | 6 | ND | ND | 273.0 | ND | 9.62 |
| F8 | CH1627-IgG | V3-74,D2-2,J4 | 14 | 19 | 808.0 | 631.0 | 8.97 | 1.33 | 5.4 | |
| B9 | CH5835-IgG | V3-74,D5-12,J4 | 41 | 9 | ND | ND | 847.0 | 687.0 | 0.44 | |
| A1 | CH5835-IgG | V3-74,D4-11,J4 | 12 | 11 | ND | ND | 822.0 | 25.9 | 0.44 | |
ND denotes no detectable binding.
HBcAg was derived from patients 241, 31, 219, and 32 or wild-type HBV (ayw) (21).
Discussion
Due to its fulminant course and the lack of suitable animal models, HBV-associated ALF is a difficult disease to study and, therefore, its pathogenesis is still poorly understood. While in most previous studies serum was the sole clinical material available, access to multiple liver specimens and serum from well-characterized patients with HBV ALF provided us with the opportunity to investigate the molecular events occurring within the site of viral replication and tissue damage. Given the ethical barriers to obtaining liver tissue from patients with acute hepatitis B, we utilized archived liver specimens from two chimpanzees acutely infected with HBV for comparison.
Several distinctive features of the virus and the host common to all of our ALF patients were identified: (i) a remarkably high titer of IgM anti-HBc in serum despite low levels of HBV DNA replication; (ii) a highly homogeneous HBV population, yet highly divergent from wild-type HBV (ayw) (21), with precore/core being the most variable region and the G1896A precore stop codon mutation invariably present; (iii) a peculiar HBV phenotype characterized by a significant increase in HBcAg production regardless of the viral replication level; (iv) a prominent B cell gene signature centered in the liver, with limited T cell gene expression, along with an extensive infiltration of liver tissue by cells of the B-lymphoid lineage accompanied by complement deposition; (v) a predominance of miRNAs targeting B-lineage-associated genes; and (vi) intrahepatic IgM and IgG antibodies in germline configuration exclusively targeting HBcAg with nanomolar or even picomolar affinities. All these features are in sharp contrast with those observed in classic acute hepatitis B, indicating that they are specifically associated with ALF.
Although the current study stemmed from our initial findings on the distinctive B cell, rather than T cell, disease signature in ALF (19), it expanded its scope to shed light on the pathogenesis of HBV ALF. We increased the number of ALF cases from two to four, we performed a thorough genetic and functional characterization of the ALF HBV strains, we compared the disease features with those of classic acute HBV in chimpanzees, and we cloned and expressed autologous viral antigens to isolate and characterize the intrahepatic pathogenic antibodies. Thus, the current study overcame the limitations of the previous study which was based on wild-type unmutated viral antigens, presumably irrelevant to the pathogenesis of ALF.
NGS of HBV strains obtained from both liver and serum of each patient demonstrated that ALF is associated with a highly homogeneous viral population, but highly divergent from the wild-type, with a highly mutated HBcAg. This genetic divergence could have been present in the original transmitted virus or might have emerged in vivo as a consequence of the anomalous immune response against HBcAg. However, the fact that antibodies in germline configuration displayed an exceptionally high binding affinity for the autologous core proteins, along with an extraordinarily high titer of IgM anti-HBc and the hyperfulminant course of the disease in our patients, strongly suggest that the mutations were already present in the source of infection, in line with the results of a few studies showing that the source and the infected patient shared the same HBV genomic sequences (10, 47–49). Thus, our data suggest that mutations in HBcAg and their relationship with germline high-affinity anti-HBc antibodies play a central role in the development of this disease.
Mutations in the HBV precore/core genomic region and core promoter frequently associated with ALF were previously reported to induce an enhanced expression of HBcAg and viral replication in vitro (9, 50). Our functional analysis of HBV variants derived from two of our patients confirmed the enhanced production of HBcAg; however, this was not associated with increased levels of viral replication and was not due to the precore stop codon mutation. The data presented here and in other studies strongly suggest that HBV replication activity does not play a major role in the pathogenesis of ALF (15–18, 47, 51). The overproduction of HBcAg that we observed with the strains derived from two of our patients is of particular interest because a high level of cytoplasmic expression could potentially lead to the export of the core antigen to the cellular membrane, making it accessible to anti-HBc antibodies, as described in chronic infection (52, 53). Our data by DIC confocal microscopy using transfected HepG2 cells with wild-type and ALF-derived HBV strains provided evidence that HBcAg can be detected on the cellular membrane. Moreover, in vitro studies have shown that HBcAg expression may induce direct hepatocellular injury (54), leading to the extracellular release of HBcAg. Here, we showed that HBcAg strongly binds to the surface of untransfected HepG2 cells, in line with previous reports of HBcAg binding to the surface of B cells and other cell types (55). Of note, cell-surface binding was stronger with HBcAg derived from ALF HBV strains (especially the 241 strain) than with the wild-type HBcAg. Thus, our in vitro data support an alternative mechanism for HBcAg localization on the cellular membrane, whereby release of HBcAg from dying hepatocytes binds to the surface of hepatocytes, including bystander uninfected cells, leading to the formation of immune complexes and complement-mediated cell lysis.
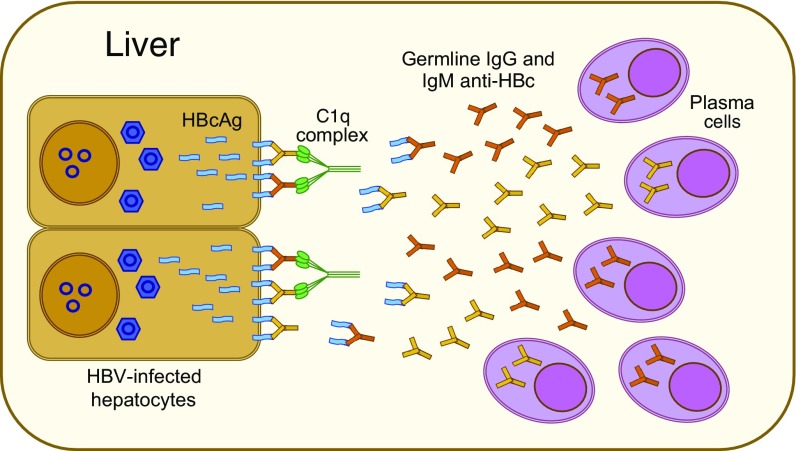
In contrast with classic hepatitis B, we found that HBV ALF is associated with an anomalous humoral immune response centered in the liver against the HBV core antigen, which is an extremely immunogenic protein that can act both as a T cell-independent and -dependent antigen (56). Although an early and enhanced antibody seroconversion against the core antigen had been recognized in ALF since the early 1980s (13), the pathogenic role of these antibodies has remained elusive for decades. Our study provides evidence that a large production of intrahepatic antibodies in germline configuration exclusively directed against HBcAg is unique to ALF, as none of the antibodies detected in the liver of classic acute hepatitis B in chimpanzees were in germline configuration. The extraordinary affinity of these anti-HBc antibodies is unusual for germline antibodies, and the fact that HBcAg is one of the few identified targets of human germline antibodies is peculiar and may be a key to the pathogenesis of this disease. These high-affinity antibodies appear to be produced in a T cell-independent manner upon direct high-affinity engagement by HBcAg without the need to undergo somatic hypermutation. Thus, altogether, our findings suggest a model of ALF pathogenesis (Fig. 7) whereby cell-surface HBcAg can be recognized by high-affinity intrahepatic germline antibodies, leading to widespread complement-dependent cytotoxicity and liver damage. This mechanism is consistent with the dramatic clinical course of ALF, where massive liver necrosis may occur within hours from the disease onset.
Fig. 7.
Potential model of HBV-associated ALF. Our data suggest a model for the pathogenesis of HBV-associated ALF, whereby liver damage is mediated by a T cell-independent B cell response centered in the liver with massive intrahepatic production of IgM and IgG antibodies in germline configuration with extraordinarily high binding affinity, exclusively directed against the HBcAg of HBV. This may result in the formation of antigen–antibody complexes on the surface of infected hepatocytes, leading to complement activation and massive liver necrosis.
While T cell immunity is central to the pathogenesis of classic acute hepatitis B (3–6), our data suggest that it does not play a prominent role in ALF. This concept is corroborated by an outbreak of fulminant hepatitis B in hemodialysis patients with a severely compromised cellular immunity (57). Despite the presence of infiltrating CD8-positive T cells in the livers of our patients, we found overexpression of negative regulators of T cell activation, including CTLA4, which inhibit T cell responses (58). These findings may explain the limited T cell gene signature detected in ALF along with the lack of markers of activated cytotoxic T lymphocytes, including IFN-regulated chemokines. These data raise the question of whether the lack of an early and robust T cell response may predispose to the development of ALF, since infected hepatocytes are not readily cleared by cytotoxic T cells, as occurs in classic acute hepatitis B (3–6).
Although the number of patients included in this study was limited, we emphasize that due to the rarity of ALF, its dramatic clinical course, and the difficulties in obtaining liver samples, there is not a single study published to date, except for ours in 2010 (19), in which the molecular pathogenesis of this disease has been investigated in the liver. Moreover, the liver specimens were obtained at a single time point, at the time of liver transplantation, but this occurred within 5 d from hospital admission. The finding that high titers of serum IgM anticore (>1:500,000) were still present in all ALF patients at the time of liver transplantation further suggests that the presence of antibodies in germline configuration of IgM and IgG class, with an unusual extraordinary high affinity against HBcAg and uniquely detected in the liver of ALF patients, does not reflect an epiphenomenon but rather suggests that these antibodies are pathogenic. This hypothesis is also supported by the lack of germline antibodies in the liver of chimpanzees with acute hepatitis B, obtained at the time of ALT peak, when the titer of IgM anticore was still high in both animals. In addition, the fact that our findings were highly consistent in all four patients studied provides strong evidence for the robustness of our data. Our patients were all infected with HBV genotype D. Although it would be interesting to study other genotypes, genotype D is one of the most frequently associated with ALF both in the United States (11, 18) and in other parts of the world (47, 49, 59).
In summary, our comprehensive study provides evidence for a major role of the humoral immunity in the pathogenesis of HBV-associated ALF, which appears to be the result of a rare and unfortunate encounter between a host with an unusual naïve B cell repertoire and an infecting virus with a highly mutated core antigen. This model of pathogenesis opens new perspectives for therapeutic and prophylactic strategies aimed at reducing the effects of the enhanced anti-HBc humoral immune response and may lead to the discovery of novel diagnostic markers for the early diagnosis of ALF.
Materials and Methods
Study Design.
We studied four patients with HBV-associated ALF. They were previously healthy individuals who suddenly developed ALF that was documented by clinical, biochemical, serologic, virologic, and histopathological criteria, as previously reported (60). The source of HBV infection was unknown for all four patients. None of them had a known risk factor or was an i.v. drug addict. The diagnosis of ALF was based on the occurrence of liver failure and hepatic encephalopathy in individuals with no prior liver disease according to previously established criteria (61). All four patients underwent liver transplantation at the Liver Transplantation Center in Cagliari, Italy, and the liver specimens were obtained from the explanted liver at the time of liver transplantation. The study design is illustrated in SI Appendix, Fig. S1. As a control group, we studied 10 liver donors and 7 subjects who underwent liver resection for hepatic hemangioma with normal liver.
Study Approval.
Written informed consent was obtained from each patient or the next of kin. Our study was approved by the Review Board of the Hospital Brotzu, Cagliari, Italy, and by the NIH Office of Human Subjects Research, granted on the condition that all samples were deidentified.
Chimpanzees.
To compare ALF with classic acute self-limited hepatitis B, we studied archival liver biopsies from two healthy, adult seronegative chimpanzees (Ch.5835 and Ch.1627) experimentally infected HBV (ayw) (5, 6), included in prior studies aimed at investigating the pathogenesis of acute hepatitis B. The animals were handled in compliance with guidelines and requirements specified by the Animal Research Committees at the National Institutes of Health (62). Both animals had been inoculated with 108 genome equivalents of HBV (serotype ayw) (63).
Full-Genome HBV NGS.
To investigate the genetic heterogeneity of HBV strains associated with ALF, HBV full-genome sequencing was performed both in serum and liver of each of the four patients with this disease using NGS (SI Appendix, SI Materials and Methods).
Bioinformatics Analysis of Sequences and Variants.
DNA sequences were analyzed against the HBV reference sequence of V01460 using VariantCaller 3.2 software on the Ion Torrent Server. The analysis pipeline was set at the high stringency somatic variant configuration. A nucleotide variant was called if the variant occurred >50 times with an average read depth ≥ 1,000× and a P value < 10−5 (quality score > 50), as previously described (64). The raw read data were also manually verified using a genome browser IVG (The Broad Institute).
HBV Genotype and Subgenotype Analysis.
The genotype and subgenotype of HBV sequences from our ALF patients were determined by phylogenetic analysis using HBV reference sequences for each HBV genotype: A (X02763), Aafr (AF297621), Ba (D00330), Bj (AB073858), C (AB033556), Caus (AB048704), D (X02496), E (X75657), F (X69798), G (AF160501), H (AY090454) and subgenotypes D1 (AF151735, AF280817), D2 (AB078033, X72702), D3 (X85254, V01460), D4 (FJ904442, AB048701), D5 (AB033558), and D7 (FJ904447, FJ904444), retrieved from GenBank.
Generation of Full-Length Replication-Competent HBV DNA Genomes.
The constructs were made using previously published methods (65, 66) with some modifications. Synthetic wild-type (genotype D, serotype ayw) or mutant (in accordance to the sequencing data from patients 241 and 31) linear monomeric HBV genomes with XhoI sites at the both terminal ends were synthesized (Biobasic) (SI Appendix, SI Materials and Methods).
Expression of HBsAg in Mammalian Cells.
Viral genomic sequences encoding HBsAg from each patient with ALF (patients 31, 32, 219, and 241) were cloned, expressed, and used to screen Fab phage libraries constructed from the liver of the autologous patients. Details are described in SI Appendix, SI Materials and Methods.
Expression and Purification of HBV Core Particles from Patients with HBV-Associated ALF.
Viral genomic sequences encoding HBcAg from each patient with ALF were cloned, expressed, and used to screen Fab phage libraries constructed from the liver of the autologous patients. Based on sequencing data, genes coding for HBcAg of patient 31, 32, 219, and 241 and the gene coding for wild-type ayw HBcAg (21) were synthesized by Gene Art (Invitrogen) (SI Appendix, SI Materials and Methods).
Gene Synthesis and Expression of HBcAg from Two Acutely Infected Chimpanzees.
Genes coding for HBcAg based on the sequence recovered from the two chimpanzees, which was identical to the reference wild-type HBV sequence (ayw) (21), were synthesized and expressed in Escherichia coli.
Kinetics Analysis of HBcAg Expression in Cell Culture.
Details are described in SI Appendix, SI Materials and Methods.
Immunofluorescence, Confocal Microscopy, HBcAg-Binding Assay, Flow Cytometry, and Complement Fixation.
Gene-Expression Profiling and Statistical Analysis.
Gene-expression and microRNA-expression profiling was performed in all 17 liver specimens obtained from the four patients with ALF and in all 17 controls, using Affymetrix Human U133 Plus 2 arrays and Affymetrix GeneChip miRNA 2.0 arrays (Affymetrix), respectively. Details on these methods and on the statistical analysis are reported in SI Appendix, SI Materials and Methods.
Generation of Fab-Display Phage Libraries.
Eight phage-display Fab libraries, one IgG1 and one IgM from each of the four patients with ALF, were constructed as previously reported (19, 60) using total RNA extracted from liver tissue. Each library had an average size of 1 × 108 individual clones. One IgG1 phage display Fab library was also generated for each of the two chimpanzees with an average size of 1 × 108 individual clones (SI Appendix, Fig. S1D). No IgM library was constructed from the chimpanzees because PCR amplification of Ig γ-chains was negative.
Analysis of Intrahepatic IgM and IgG Extracted from Liver Tissues of ALF Patients.
Details are described in SI Appendix, SI Materials and Methods.
Affinity Measurement by Surface Plasmon Resonance.
Surface plasmon resonance experiments were performed in a Biacore 3000 instrument (GE Healthcare). Details are described in SI Appendix, SI Materials and Methods.
NGS of Intrahepatic IgM and IgG Repertoire and Bioinformatics Analysis of Illumina Paired-End Sequencing.
Details are reported in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program of the NIH, National Institute of Allergy and Infectious Diseases, and National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
Data deposition: Next-generation sequencing data of HBV strains have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) (accession no. PRJNA422423). Microarray data (mRNA and miRNA) have been deposited in the NCBI Gene Expression Omnibus (GEO) database (accession nos. GSE96851 and GSE62037). Heavy-chain variable region sequences have been deposited in GenBank (accession nos. KX986360–KX986516 and KX986517–KX986558). Next-generation sequencing data of IgG and IgM antibodies have been deposited in the NCBI SRA (accession no. SRP126953).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809028115/-/DCSupplemental.
References
- 1.Lee WM. Acute liver failure. N Engl J Med. 1993;329:1862–1872. doi: 10.1056/NEJM199312163292508. [DOI] [PubMed] [Google Scholar]
- 2.Lee WM. Etiologies of acute liver failure. Semin Liver Dis. 2008;28:142–152. doi: 10.1055/s-2008-1073114. [DOI] [PubMed] [Google Scholar]
- 3.Chisari FV, Ferrari C. Hepatitis B virus immunopathogenesis. Annu Rev Immunol. 1995;13:29–60. doi: 10.1146/annurev.iy.13.040195.000333. [DOI] [PubMed] [Google Scholar]
- 4.Bertoletti A, Ferrari C. Adaptive immunity in HBV infection. J Hepatol. 2016;64(Suppl 1):S71–S83. doi: 10.1016/j.jhep.2016.01.026. [DOI] [PubMed] [Google Scholar]
- 5.Thimme R, et al. CD8(+) T cells mediate viral clearance and disease pathogenesis during acute hepatitis B virus infection. J Virol. 2003;77:68–76. doi: 10.1128/JVI.77.1.68-76.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wieland S, Thimme R, Purcell RH, Chisari FV. Genomic analysis of the host response to hepatitis B virus infection. Proc Natl Acad Sci USA. 2004;101:6669–6674. doi: 10.1073/pnas.0401771101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang TJ, et al. Hepatitis B virus precore mutation and fulminant hepatitis in the United States. A polymerase chain reaction-based assay for the detection of specific mutation. J Clin Invest. 1994;93:550–555. doi: 10.1172/JCI117006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carman WF, et al. Association of a precore genomic variant of hepatitis B virus with fulminant hepatitis. Hepatology. 1991;14:219–222. [PubMed] [Google Scholar]
- 9.Baumert TF, Rogers SA, Hasegawa K, Liang TJ. Two core promotor mutations identified in a hepatitis B virus strain associated with fulminant hepatitis result in enhanced viral replication. J Clin Invest. 1996;98:2268–2276. doi: 10.1172/JCI119037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ogata N, Miller RH, Ishak KG, Purcell RH. The complete nucleotide sequence of a pre-core mutant of hepatitis B virus implicated in fulminant hepatitis and its biological characterization in chimpanzees. Virology. 1993;194:263–276. doi: 10.1006/viro.1993.1257. [DOI] [PubMed] [Google Scholar]
- 11.Wai CT, et al. US Acute Liver Failure Study Group Clinical outcome and virological characteristics of hepatitis B-related acute liver failure in the United States. J Viral Hepat. 2005;12:192–198. doi: 10.1111/j.1365-2893.2005.00581.x. [DOI] [PubMed] [Google Scholar]
- 12.Bartholomeusz A, Locarnini S. Hepatitis B virus mutants and fulminant hepatitis B: Fitness plus phenotype. Hepatology. 2001;34:432–435. doi: 10.1053/jhep.2001.26764. [DOI] [PubMed] [Google Scholar]
- 13.Gimson AE, Tedder RS, White YS, Eddleston AL, Williams R. Serological markers in fulminant hepatitis B. Gut. 1983;24:615–617. doi: 10.1136/gut.24.7.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trepo CG, et al. Hepatitis B antigen (HBSAg) and/or antibodies (anti-HBS and anti-HBC) in fulminant hepatitis: Pathogenic and prognostic significance. Gut. 1976;17:10–13. doi: 10.1136/gut.17.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Cock KM, Govindarajan S, Valinluck B, Redeker AG. Hepatitis B virus DNA in fulminant hepatitis B. Ann Intern Med. 1986;105:546–547. doi: 10.7326/0003-4819-105-4-546. [DOI] [PubMed] [Google Scholar]
- 16.Brechot C, et al. Multiplication of hepatitis B virus in fulminant hepatitis B. Br Med J (Clin Res Ed) 1984;288:270–271. doi: 10.1136/bmj.288.6413.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liang TJ, Hasegawa K, Rimon N, Wands JR, Ben-Porath E. A hepatitis B virus mutant associated with an epidemic of fulminant hepatitis. N Engl J Med. 1991;324:1705–1709. doi: 10.1056/NEJM199106133242405. [DOI] [PubMed] [Google Scholar]
- 18.Dao DY, et al. Acute Liver Failure Study Group Two distinct subtypes of hepatitis B virus-related acute liver failure are separable by quantitative serum immunoglobulin M anti-hepatitis B core antibody and hepatitis B virus DNA levels. Hepatology. 2012;55:676–684. doi: 10.1002/hep.24732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farci P, et al. B cell gene signature with massive intrahepatic production of antibodies to hepatitis B core antigen in hepatitis B virus-associated acute liver failure. Proc Natl Acad Sci USA. 2010;107:8766–8771. doi: 10.1073/pnas.1003854107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Engle RE, et al. Transfusion-associated hepatitis before the screening of blood for hepatitis risk factors. Transfusion. 2014;54:2833–2841. doi: 10.1111/trf.12682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galibert F, Mandart E, Fitoussi F, Tiollais P, Charnay P. Nucleotide sequence of the hepatitis B virus genome (subtype ayw) cloned in E. coli. Nature. 1979;281:646–650. doi: 10.1038/281646a0. [DOI] [PubMed] [Google Scholar]
- 22.Colucci G, Beazer Y, Cantaluppi C, Tackney C. Identification of a major hepatitis B core antigen (HBcAg) determinant by using synthetic peptides and monoclonal antibodies. J Immunol. 1988;141:4376–4380. [PubMed] [Google Scholar]
- 23.Jazayeri SM, et al. Intracellular distribution of hepatitis B virus core protein expressed in vitro depends on the sequence of the isolate and the serologic pattern. J Infect Dis. 2004;189:1634–1645. doi: 10.1086/382190. [DOI] [PubMed] [Google Scholar]
- 24.Hsu HC, et al. Biologic and prognostic significance of hepatocyte hepatitis B core antigen expressions in the natural course of chronic hepatitis B virus infection. J Hepatol. 1987;5:45–50. doi: 10.1016/s0168-8278(87)80060-0. [DOI] [PubMed] [Google Scholar]
- 25.Eyre NS, et al. Hepatitis B virus and hepatitis C virus interaction in Huh-7 cells. J Hepatol. 2009;51:446–457. doi: 10.1016/j.jhep.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 26.Klein U, Dalla-Favera R. Germinal centres: Role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 27.Rosenbaum M, et al. MZB1 is a GRP94 cochaperone that enables proper immunoglobulin heavy chain biosynthesis upon ER stress. Genes Dev. 2014;28:1165–1178. doi: 10.1101/gad.240762.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dement-Brown J, et al. Fc receptor-like 5 promotes B cell proliferation and drives the development of cells displaying switched isotypes. J Leukoc Biol. 2012;91:59–67. doi: 10.1189/jlb.0211096. [DOI] [PubMed] [Google Scholar]
- 29.Coquery CM, Erickson LD. Regulatory roles of the tumor necrosis factor receptor BCMA. Crit Rev Immunol. 2012;32:287–305. doi: 10.1615/critrevimmunol.v32.i4.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. doi: 10.1146/annurev-immunol-032414-112032. [DOI] [PubMed] [Google Scholar]
- 31.Basso K, et al. Identification of the human mature B cell miRNome. Immunity. 2009;30:744–752. doi: 10.1016/j.immuni.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Di Lisio L, et al. The role of miRNAs in the pathogenesis and diagnosis of B-cell lymphomas. Blood. 2012;120:1782–1790. doi: 10.1182/blood-2012-05-402784. [DOI] [PubMed] [Google Scholar]
- 33.de Yébenes VG, Bartolomé-Izquierdo N, Ramiro AR. Regulation of B-cell development and function by microRNAs. Immunol Rev. 2013;253:25–39. doi: 10.1111/imr.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Musilova K, Mraz M. MicroRNAs in B-cell lymphomas: How a complex biology gets more complex. Leukemia. 2015;29:1004–1017. doi: 10.1038/leu.2014.351. [DOI] [PubMed] [Google Scholar]
- 35.Tan LP, et al. miRNA profiling of B-cell subsets: Specific miRNA profile for germinal center B cells with variation between centroblasts and centrocytes. Lab Invest. 2009;89:708–716. doi: 10.1038/labinvest.2009.26. [DOI] [PubMed] [Google Scholar]
- 36.de Yébenes VG, et al. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li YF, et al. Loss of miR-182 affects B-cell extrafollicular antibody response. Immunology. 2016;148:140–149. doi: 10.1111/imm.12592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 41.Lin J, et al. Follicular dendritic cell-induced microRNA-mediated upregulation of PRDM1 and downregulation of BCL-6 in non-Hodgkin’s B-cell lymphomas. Leukemia. 2011;25:145–152. doi: 10.1038/leu.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wrammert J, et al. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature. 2008;453:667–671. doi: 10.1038/nature06890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szomolanyi-Tsuda E, Welsh RM. T-cell-independent antiviral antibody responses. Curr Opin Immunol. 1998;10:431–435. doi: 10.1016/s0952-7915(98)80117-9. [DOI] [PubMed] [Google Scholar]
- 44.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 45.Chimpanzee Sequencing and Analysis Consortium Initial sequence of the chimpanzee genome and comparison with the human genome. Nature. 2005;437:69–87. doi: 10.1038/nature04072. [DOI] [PubMed] [Google Scholar]
- 46.Brezinschek HP, et al. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(-)/IgM+ B cells. J Clin Invest. 1997;99:2488–2501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Friedt M, et al. Mutations in the basic core promotor and the precore region of hepatitis B virus and their selection in children with fulminant and chronic hepatitis B. Hepatology. 1999;29:1252–1258. doi: 10.1002/hep.510290418. [DOI] [PubMed] [Google Scholar]
- 48.Karayiannis P, et al. Fulminant hepatitis associated with hepatitis B virus e antigen-negative infection: Importance of host factors. Hepatology. 1995;22:1628–1634. [PubMed] [Google Scholar]
- 49.Pollicino T, et al. Pre-S2 defective hepatitis B virus infection in patients with fulminant hepatitis. Hepatology. 1997;26:495–499. doi: 10.1002/hep.510260235. [DOI] [PubMed] [Google Scholar]
- 50.Baumert TF, Marrone A, Vergalla J, Liang TJ. Naturally occurring mutations define a novel function of the hepatitis B virus core promoter in core protein expression. J Virol. 1998;72:6785–6795. doi: 10.1128/jvi.72.8.6785-6795.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sterneck M, et al. Functional analysis of HBV genomes from patients with fulminant hepatitis. Hepatology. 1998;28:1390–1397. doi: 10.1002/hep.510280530. [DOI] [PubMed] [Google Scholar]
- 52.Chu CM, Liaw YF. Intrahepatic distribution of hepatitis B surface and core antigens in chronic hepatitis B virus infection. Hepatocyte with cytoplasmic/membranous hepatitis B core antigen as a possible target for immune hepatocytolysis. Gastroenterology. 1987;92:220–225. doi: 10.1016/0016-5085(87)90863-8. [DOI] [PubMed] [Google Scholar]
- 53.Trevisan A, et al. Core antigen-specific immunoglobulin G bound to the liver cell membrane in chronic hepatitis B. Gastroenterology. 1982;82:218–222. [PubMed] [Google Scholar]
- 54.Yoakum GH, et al. High-frequency transfection and cytopathology of the hepatitis B virus core antigen gene in human cells. Science. 1983;222:385–389. doi: 10.1126/science.6194563. [DOI] [PubMed] [Google Scholar]
- 55.Vanlandschoot P, Van Houtte F, Serruys B, Leroux-Roels G. The arginine-rich carboxy-terminal domain of the hepatitis B virus core protein mediates attachment of nucleocapsids to cell-surface-expressed heparan sulfate. J Gen Virol. 2005;86:75–84. doi: 10.1099/vir.0.80580-0. [DOI] [PubMed] [Google Scholar]
- 56.Milich DR, McLachlan A. The nucleocapsid of hepatitis B virus is both a T-cell-independent and a T-cell-dependent antigen. Science. 1986;234:1398–1401. doi: 10.1126/science.3491425. [DOI] [PubMed] [Google Scholar]
- 57.Igaki N, et al. An outbreak of fulminant hepatitis B in immunocompromised hemodialysis patients. J Gastroenterol. 2003;38:968–976. doi: 10.1007/s00535-003-1180-1. [DOI] [PubMed] [Google Scholar]
- 58.Vogt L, et al. VSIG4, a B7 family-related protein, is a negative regulator of T cell activation. J Clin Invest. 2006;116:2817–2826. doi: 10.1172/JCI25673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arankalle VA, et al. An outbreak of hepatitis B with high mortality in India: Association with precore, basal core promoter mutants and improperly sterilized syringes. J Viral Hepat. 2011;18:e20–e28. doi: 10.1111/j.1365-2893.2010.01391.x. [DOI] [PubMed] [Google Scholar]
- 60.Nissim O, et al. Liver regeneration signature in hepatitis B virus (HBV)-associated acute liver failure identified by gene expression profiling. PLoS One. 2012;7:e49611. doi: 10.1371/journal.pone.0049611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Trey C. The fulminant hepatic failure surveillance study. Brief review of the effects of presumed etiology and age of survival. Can Med Assoc J. 1972;106:525–528. [PMC free article] [PubMed] [Google Scholar]
- 62.ILARC . Guide for the Care and Use of Laboratory Animals. National Academies Press; Washington, DC: 1996. [PubMed] [Google Scholar]
- 63.Guidotti LG. Pathogenesis of viral hepatitis. J Biol Regul Homeost Agents. 2003;17:115–119. [PubMed] [Google Scholar]
- 64.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Günther S, et al. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol. 1995;69:5437–5444. doi: 10.1128/jvi.69.9.5437-5444.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pollicino T, et al. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology. 2006;130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.