Significance
Heat stress is deleterious to living organisms and is being exacerbated by climate change. Although heat is known thermodynamically to unfold RNA in the test tube, the effect of heat stress on the global transcriptome within the complex environment of the living cell has not been investigated in any organism. We harnessed innovative methods for genome-wide probing of RNA structure in vivo to quantify the effect of heat shock on the RNA structurome in the eukaryote rice, a vitally important crop that is vulnerable to temperature stress. By coupling these assays with measurements of the temperature-regulated transcriptome and translatome, we reveal previously unknown relationships between temperature modulation of mRNA structure melting and mRNA abundance loss, with implications for crop improvement.
Keywords: heat shock, rice, RNA-seq, RNA thermometer, Structure-seq
Abstract
The heat shock response is crucial for organism survival in natural environments. RNA structure is known to influence numerous processes related to gene expression, but there have been few studies on the global RNA structurome as it prevails in vivo. Moreover, how heat shock rapidly affects RNA structure genome-wide in living systems remains unknown. We report here in vivo heat-regulated RNA structuromes. We applied Structure-seq chemical [dimethyl sulfate (DMS)] structure probing to rice (Oryza sativa L.) seedlings with and without 10 min of 42 °C heat shock and obtained structural data on >14,000 mRNAs. We show that RNA secondary structure broadly regulates gene expression in response to heat shock in this essential crop species. Our results indicate significant heat-induced elevation of DMS reactivity in the global transcriptome, revealing RNA unfolding over this biological temperature range. Our parallel Ribo-seq analysis provides no evidence for a correlation between RNA unfolding and heat-induced changes in translation, in contrast to the paradigm established in prokaryotes, wherein melting of RNA thermometers promotes translation. Instead, we find that heat-induced DMS reactivity increases correlate with significant decreases in transcript abundance, as quantified from an RNA-seq time course, indicating that mRNA unfolding promotes transcript degradation. The mechanistic basis for this outcome appears to be mRNA unfolding at both 5′ and 3′-UTRs that facilitates access to the RNA degradation machinery. Our results thus reveal unexpected paradigms governing RNA structural changes and the eukaryotic RNA life cycle.
Heat stress can have dramatic effects on organisms. After exposure to high temperatures, severe cellular damage occurs in many living systems, including in crop species such as rice (Oryza sativa L.), the staple food for almost half the human population (1). Increasing temperatures and climate variability seriously threaten crop production levels and food security (2), and vulnerability to heat stress results in direct negative effects on yield (3, 4).
A variety of regulatory reprogramming mechanisms occur in organisms in response to high temperature stress, including changes in the transcriptome, proteome, and metabolome (5–7). RNA secondary and tertiary structure are known to influence numerous processes related to gene expression (8), including transcription (9), RNA maturation (10), translation initiation (11), and transcript degradation (12). However, how heat stress affects RNA structure on a genome-wide scale in vivo is an important yet missing piece of the puzzle concerning temperature-based gene regulation.
The combination of RNA structure probing methods and high-throughput sequencing has made it possible to obtain genome-wide RNA structural information at nucleotide resolution in one assay, essentially overcoming many of the limitations of length and abundance of RNA molecules that arise in gel probing of individual RNA species. In yeast, melting temperatures have been obtained for RNA structures genome-wide in vitro by probing with V1 nuclease, which cleaves at double-stranded regions (13). In the bacterium Yersinia pseudotuberculosis, in vitro RNA structuromes were mapped at different temperatures using both V1 and the single-stranded nuclease S1 (14). In several other bacterial species, temperature-induced changes in the structures of individual RNA thermometers, as assessed in vitro, have been documented to modulate mRNA translation efficiency (15).
However, in contrast to the above in vitro data, the extent to which temperature stress functionally alters the RNA structurome in living cells is not understood, despite the advent of methods to probe RNA structure genome-wide in vivo (16–20) and extensive evidence that in vivo structure of an RNA molecule can differ dramatically from its in vitro or in silico structures (16, 18). Moreover, in vivo, RNA structures can be altered by numerous endogenous factors that are not present in the test tube, including cellular solutes, proteins, and endogenous crowding agents (21), leading to significant biological consequences. Here, we performed genome-wide investigation of how elevated temperatures regulate the in vivo structurome, applying our Structure-seq2 methodology (19) to profile in vivo RNA structure in the crop plant rice (O. sativa L.). We obtained structural data on 14,292 transcripts and assessed this dataset with respect to possible RNA thermometers of the type described in prokaryotes. We combined our RNA structurome data with Ribo-seq analyses to identify mRNAs undergoing translation, as well as RNA-seq time courses to quantify post-heat-shock transcriptomes. Our analysis of relationships among the structure, translation, and abundance of thousands of individual mRNAs identifies a heretofore unappreciated structural basis for the dynamic regulation of mRNA abundance after heat shock.
Results
Structure-Seq Reveals Heat-Induced Unfolding of the in Vivo Eukaryotic Transcriptome.
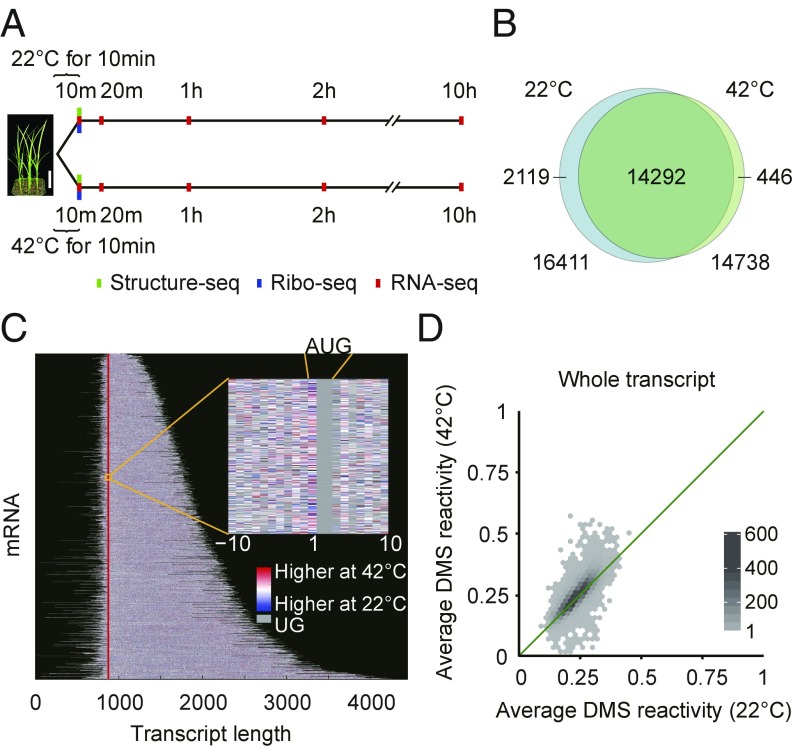
Our optimized Structure-seq2 methodology (19) employs structure probing with dimethyl sulfate (DMS), which methylates adenines and cytosines on their Watson–Crick face (N1 of A and N3 of C) when they are not base-paired or otherwise protected. This methylation results in termination of reverse transcription, thus providing a read-out of the position of the modified, non-base-paired nucleotide (SI Appendix, Fig. S1). Structure-seq libraries (SI Appendix, Table S1) were generated from 14-d-old rice shoot tissue after a brief (10 min) treatment at 22 °C (control) or 42 °C (heat shock) with or without DMS (Fig. 1A). Our data show high reproducibility between biological replicates (SI Appendix, Fig. S2) and the majority of the reads map to mRNAs (SI Appendix, Fig. S3 A–D). The data demonstrate the expected specificity for modification of A and C in DMS-treated samples (SI Appendix, Fig. S3 E–I).
Fig. 1.
Experimental design and Structure-seq library statistics. (A) Timeline of Structure-seq (green), Ribo-seq (blue), and RNA-seq (red) experiments. [Scale bar (white) for rice seedlings, 4 cm.] (B) Overlap of mRNAs with sufficient structure-probing coverage between 22 °C (blue) and 42 °C (green). (C) Heat stress-induced structural reactivity changes across the rice mRNA structurome. Each horizontal line represents a different mRNA. Reactivity information is obtained at single nucleotide resolution (Inset). Vertical line (red) marks start codon. (D) Average DMS reactivity is significantly greater at 42 °C than 22 °C (whole transcripts; P = 5.27 × 10−77; r = 0.82). [Scale bar (gradient), numbers of RNAs.] In the analyses of C and D, only transcripts with sufficient Structure-seq coverage under both temperature conditions are shown and used.
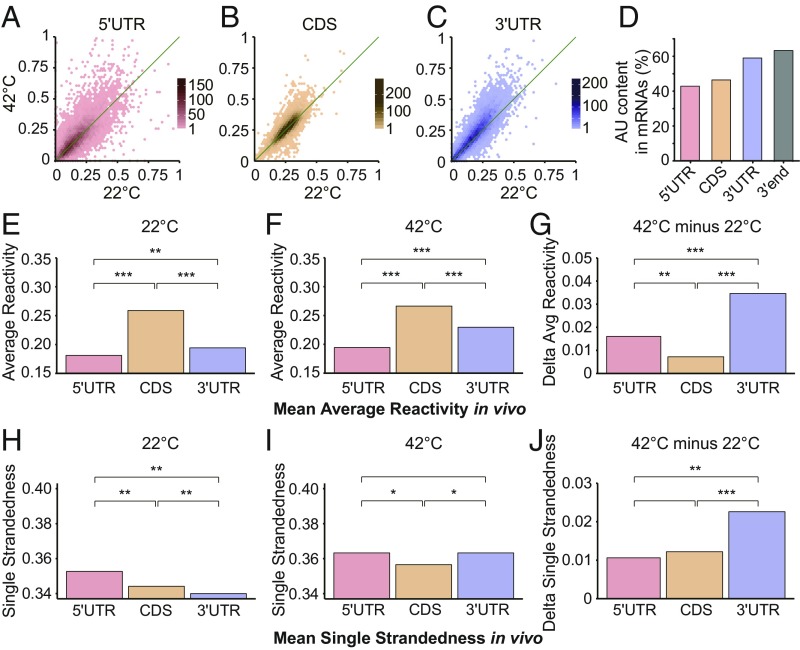
We chose a short, 10-min heat shock both to optimize study of direct temperature effects on the RNA structurome, which should be rapid, and because such acute events are commonplace in crop and forest canopies because of transient heating from sunflecks (22). We obtained sufficient structural coverage at both temperatures for 14,292 mRNAs (Fig. 1 B and C and SI Appendix, Fig. S4 A and B). After normalization for chemical reactivity differences between temperatures (SI Appendix, Materials and Methods), a global trend of significantly elevated average DMS reactivity at 42 °C compared with 22 °C was observed for entire transcripts (Fig. 1D), as well as for subregions (Fig. 2 A–C). Because RNA secondary structures can melt anywhere between 1 °C and 99 °C (23), these results suggest that secondary structures of many mRNAs in rice have evolved to melt in vivo over this biologically relevant temperature range. The 3′UTRs showed the most significant increase in average DMS reactivity under heat (P = 2.24 × 10−89; Fig. 2C and SI Appendix, Fig. S4 C and D). Interestingly, rice 3′UTRs harbor a higher AU content (Fig. 2D); given the weaker base-pairing of AU versus GC, this provides a mechanistic basis for melting of this region of the mRNA (SI Appendix, Fig. S5). After folding of whole transcripts using our in vivo restraints, we found that 3′UTRs are actually predicted to be more structured than 5′UTRs or coding sequences (CDSs) at 22 °C, as has also been reported for mammalian 3′UTRs (24), yet show the greatest gain in predicted single-strandedness at 42 °C, consistent with the marked increase in reactivity of 3′UTRs at 42 °C (Fig. 2 E–J). These results suggest that rice 3′UTRs have increased susceptibility versus other regions of the transcript to melting out on acute heat shock (Fig. 2 H–J).
Fig. 2.
Average DMS reactivity is higher on all mRNA regions at elevated temperature. Average DMS reactivity is significantly greater at 42 °C for all mRNA subregions. (A) 5′UTR (P = 4.00 × 10−18; r = 0.74). (B) CDS (P = 8.08 × 10−12; r = 0.83). (C) 3′UTR (P = 2.24 × 10−89; r = 0.87). DMS reactivities on whole transcripts were cross-normalized between temperatures to correct for the higher chemical reactivity of DMS at higher temperature (SI Appendix, Materials and Methods). [Scale bars (gradient) in A–C, numbers of mRNAs.] (D) Average AU content is significantly greater in 3′UTRs than in 5′UTRs or CDS, especially at the 3′ end (last 100 nt). (E and F) Mean of the average DMS reactivity at 22 °C (E) and 42 °C (F) in the 5′UTR, CDS, 3′UTR regions. (G) Change in average DMS reactivity (42 °C − 22 °C) in the 5′UTR, CDS, and 3′UTR regions. (H and I) Mean of single strandedness at 22 °C (H) and 42 °C (I) in the 5′UTR, CDS, 3′UTR regions. Here, single-strandedness is the percentage of single-stranded nucleotides in the RNA structure predicted with in vivo restraints. (J) Change in average single strandedness (42 °C − 22 °C) in the 5′UTR, CDS, 3′UTR regions. In the analyses of A–J, only transcripts with sufficient Structure-seq coverage under both temperature conditions were used. In E–J, *P < 0.01; **P < 10−10; ***P < 10−50, respectively. Specific P values for each comparison are provided in SI Appendix, Table S2.
Heat-Induced RNA Structural Changes in Rice Differ from Known Prokaryotic RNA Temperature-Sensing Mechanisms.
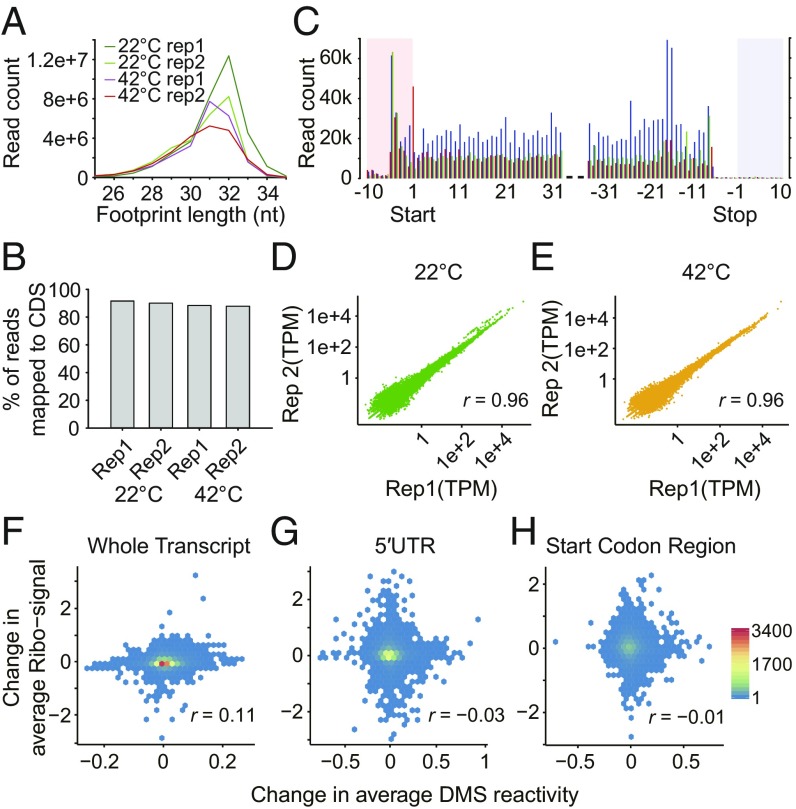
In bacteria, temperature-induced changes in 5′UTR structures of individual RNAs, referred to as RNA thermometers, modulate translation efficiency (15). In our rice RNA structuromes, variation in heat-induced structural reactivity change was greater in 5′UTRs (Fig. 2A) than in other transcript regions (Fig. 2 B and C). We investigated a possible relationship between mRNA structure and translation: Ribo-seq translatome profiles were determined after 10 min of the same temperature treatments as for Structure-seq libraries (Figs. 1A and 3 A–E and SI Appendix, Table S3). However, we found no correlation between the average temperature-induced change in DMS reactivity in the whole transcript, around the start codon, or in the entire 5′UTR, and change in ribosome association between temperatures (Fig. 3 F–H). Nor did we find evidence in the rice transcriptome or structurome itself for several specific known bacterial RNA thermometers (25) (SI Appendix, Supporting File). Thus, heat-induced RNA structural changes in rice identified here appear to differ from those described to date in prokaryotes. In future research, it would be interesting to discern whether application of global in vivo structure probing methods to prokaryotes would reveal temperature-dependent relationships between mRNA structure and mRNA abundance such as those described here.
Fig. 3.
Ribo-seq data statistics and absence of correlations between temperature-induced changes in DMS reactivity and in the translatome. (A) Distribution of sequence read length of Ribo-seq data, peaking at 30–32 nucleotides, as expected for ribosome footprinting. (B) Percentage of mRNA-mapped Ribo-seq reads that map to the CDS. (C) Distribution of sequence read count around start codon and stop codon. Shown are 32-nt reads as the example; reading frames are shown in red (first position), blue (second position), and green (third position), and UTRs are highlighted in pink and gray. (D and E) High correlation of transcript abundance between replicates of Ribo-seq libraries. Transcript abundance was calculated as TPM (transcripts per million). (D) 22 °C. (E) 42 °C. (F–H) No correlation detected between the change in average DMS reactivity (42 °C − 22 °C) and change in Ribo-seq signal (42 °C − 22 °C) for (F) whole transcripts (n = 14,197). (G) 5′UTR (n = 9,895), (H) start codon region (−50 nt to +50 nt; n = 8,726). n, number of candidates with both sufficient coverage in Structure-seq and presence in Ribo-seq datasets.
Heat-Induced Unfolding Promotes Transcript Degradation.
Rapid changes in plant mRNA transcriptomes in response to stimuli have been documented (26). We accordingly anticipated that acute heat shock might result in mRNA abundance changes, and we hypothesized that RNA structure could be regulatory of such changes.
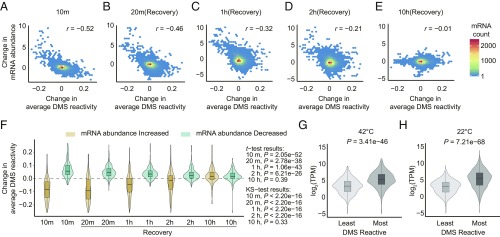
Indeed, we found that of the 14,292 transcripts for which we had Structure-seq data at both temperatures, 1,052 (7.4%) showed a statistically significant change in abundance between 42 °C heat shock and 22 °C control samples. We observed a strong inverse correlation between temperature-induced change in DMS reactivity and temperature-induced change in transcript abundance as quantified from –DMS libraries (note that reads from −DMS libraries are analogous to RNA-seq library reads; SI Appendix, Fig. S6 A and B). To further evaluate the relationship between RNA structure change and transcript abundance change, we next performed classical RNA-seq experiments that quantified transcript abundance change over a longer time course post-heat shock (Fig. 1A) after the same 10 min of 42 °C or 22 °C conditions as we employed in our RNA structurome experiments (SI Appendix, Figs. S7 and S8 and Table S4). RNA-seq data at 10 min were highly consistent with our mRNA abundance measurements from –DMS Structure-seq libraries (SI Appendix, Fig. S6 C and D). Our RNA-seq experiments confirmed a significant negative correlation between change in DMS reactivity and change in transcript abundance at 10–20 min after heat shock, and even out to 1 h (Fig. 4 A–C). After 2 h, and especially after 10 h, the correlation weakened and was eventually lost (Fig. 4 D and E), presumably reflecting a mechanism in which the structurome and transcriptome are rapidly affected by heat shock and then slowly recover (SI Appendix, Fig. S7). A converse analysis, in which mRNAs with the greatest increase or decrease in abundance between temperatures were first identified and then analyzed for DMS reactivity, confirmed this inverse relationship as well as its time dependence (Fig. 4F).
Fig. 4.
Strong negative correlation between heat-shock-induced DMS reactivity change and heat-shock-induced mRNA abundance (TPM) change that gradually dissipates after heat shock. (A–E) Change of average DMS reactivity (42 °C − 22 °C) from Structure-seq (all 10 min) vs. fold change (log2) in mRNA abundance (42 °C − 22 °C) from RNA-seq (see Fig. 1A for time course), calculated on all mRNAs with sufficient Structure-seq coverage. (A) 10 min (= end of 42 °C treatment), (B) 20 min, (C) 1 h, (D) 2 h, (E) 10 h. (F) Distribution of change in average DMS reactivity of all transcripts with sufficient Structure-seq coverage within the top 5% of mRNAs with increased abundance (beige) and the bottom 5% of mRNAs with decreased abundance (blue). (G and H) The abundance of degradome fragments of the top/bottom 5% most/least DMS reactive transcripts at (G) 42 °C and (H) 22 °C is compared, showing that more reactive transcripts have a higher mean number of degradome fragments.
Next, we investigated possible mechanistic origins of this effect. These results suggested that at least part of the inverse relationship between reactivity and abundance arises from preferential degradation of less structured, highly reactive transcripts. The exosome complex is responsible for one of the major pathways of RNA degradation and is largely conserved throughout eukaryotes. It degrades RNA in a 3′-to-5′ direction (27), and only RNAs with a sufficiently long single-stranded 3′ tail can initiate tunneling through the exosome core (28). Thus, exosome-mediated degradation of unfolded transcripts would be consistent with our observation of heat-induced DMS reactivity increases in 3′UTRs (Fig. 2 E–G). With the notion of 3′ end-initiated degradation of the RNA, we compared the 5% of mRNAs with greatest heat-induced increase or decrease in DMS reactivity and found that the former set of transcripts had significantly greater U content in the final 10 nt of the 3′UTR (SI Appendix, Fig. S5A). This result is consistent with U base-pairing with the adjacent polyA tail that at least partially melts out at 42 °C and facilitates exosome-based degradation.
To test the hypothesis that increased reactivity in the 3′UTR arises from heat-induced unfolding of RNA structure, we chose four 3′UTR sequences and prepared RNAs comprising the last 10 nt of each transcript fused to a 15-nt polyA tail (designated T1–T4; sequences in Materials and Methods). Sequences were chosen from 3′UTR sequences in the top 5% of transcripts with greatest loss in abundance at 42 °C; T1–T4 also had predicted maximal gain in single-strandedness between 22 °C and 42 °C, as derived from free energy estimations at these temperatures, using standard thermodynamic relationships. We assessed the stability of T1–T4 structures by UV-detected thermal denaturation monitored at 260 nm, using in vivo-like monovalent and divalent ion concentrations (Materials and Methods). Plots of fraction folded versus temperature (SI Appendix, Fig. S9) revealed that T2 and T3 (but not T1 and T4) melt with a sigmoidal transition between ∼20 and 40 °C, which are temperatures similar to those used for unstressed and heat-stressed rice, respectively. It is notable that T2 and T3 have the highest U content for the last 10 nt of the transcript, of 6 and 7 Us, respectively, whereas T1 and T4 have lower U content of 2 and 5 Us, respectively. The higher U content in T2 and T3, which comes in two regions of at least two Us each, could drive Watson–Crick base pairing with the polyA tail at 22 °C, which then melts out at 42 °C. These data demonstrate melting of U-rich 3′UTR sequences by 42 °C, which could provide the exosome with access to the 3′ end for degradation.
In addition to degradation from the 3′ end, RNA degradation can occur from the 5′ end, catalyzed in plants by the plant ortholog of XRN1, XRN4, which is a 5′-to-3′ single-stranded exonuclease known to be activated under heat (29). We analyzed the 5′UTRs of rice orthologs of Arabidopsis XRN4-sensitive transcripts (29) and discovered that these transcripts have enriched 5′UTR AU content relative to XRN4-insensitive targets (SI Appendix, Fig. S10). We also observed that the 5% of mRNAs with greatest heat-induced reactivity increase also have enriched AU content at the 5′ end, as well as in the entire 5′UTR (SI Appendix, Fig. S10), which would facilitate enhanced unfolding given the weaker base-pairing of AU versus GC, and thus, degradation at higher temperatures.
To further evaluate our hypothesis of a functional relationship between structure changes in the 5′UTR and transcript abundance, we compared the abundance of degradome fragments of the 5% least- and 5% most-reactive mRNAs, using data from a rice degradome dataset (GSM1040649; Materials and Methods). [By design, degradome libraries are enriched in uncapped mRNAs subject to 5′-to-3′ degradation (30); degradome sequencing thus specifically identifies fragments of degraded mRNA, and so allows an approximate quantification of transcript stability.] At each temperature, we found that the set of transcripts with higher average DMS reactivity had significantly greater abundance of transcript fragments in the degradome (Fig. 4 G and H). This finding suggests that high DMS reactivity transcripts are more susceptible to degradation from 5′ ends. Taken together, our results (Figs. 2 and 4) indicate that melting of both 5′ and 3′ UTRs with heat contributes to mechanisms of selective transcript degradation, and thus transcriptome reprogramming in response to acute heat stress.
Discussion
Recent technical advances have facilitated the field of RNA structural genomics, allowing studies of RNA structure in vivo and genome-wide (31). Although these tools are powerful, there have been very few studies of in vivo structuromes, let alone in response to stress. The Structure-seq methodology (19) allowed us to probe heat-induced structural changes at single-nucleotide resolution in thousands of transcripts simultaneously (Fig. 1), providing a genome-wide perspective on in vivo temperature modulation of RNA structure. Although other mechanisms undoubtedly contribute, our comprehensive structurome, transcriptome, and translatome results are consistent with a major regulatory role in eukaryotes of temperature-modulated mRNA structures that control mRNA abundance, as opposed to control of mRNA translation as in prokaryotes (32).
In prokaryotes, temperature-induced RNA structural changes around the Shine-Dalgarno sequence exert regulatory roles in protein translation (14). In particular, sequences defined as the ROSE element, fourU, and UCCU are prokaryotic 5′UTR RNA thermometers. These motifs sequester the Shine-Dalgarno sequence at low temperatures and melt out at higher temperatures, thus promoting ribosome binding. We found only a few of these sequence candidates in our 5′UTR dataset, and none exhibited unfolding at 42 °C as would be expected for RNA thermometers (SI Appendix, Supporting File). In eukaryotes, the Kozak sequence guides translation initiation. However, only 156 mRNAs containing Kozak sequences were present in both our structurome and Ribo-seq datasets, and these did not exhibit a correlation between DMS reactivity change and heat-induced Ribo-seq signal change in the translatome (SI Appendix, Supporting File). We cannot dismiss the possibility that such correlations might occur for a small set of transcripts under our conditions, or be revealed by a longer duration of heat shock, recognizing that our data and those of others (29) show relatively minor effects of 10 min of heat shock on the translatome. However, our failure to detect prokaryotic-type RNA thermometers suggests that RNA-based temperature-sensing mechanisms of eukaryotes differ markedly from those of prokaryotes. Our experimental and computational conclusions differ from a previous study in which analysis of a single mRNA, Drosophila melanogaster HSP90, suggested that eukaryotes use prokaryotic-type RNA thermometers (33); this comparison illustrates the value of a genome-wide perspective on in vivo RNA structure.
We observe AU richness in both 3′ and 5′ UTRs that exhibit elevated DMS reactivity at 42 °C (SI Appendix, Figs. S5 and S10), consistent with their melting out, and heat-induced DMS reactivity changes show a strong inverse correlation with heat-induced changes in transcript abundance (Fig. 4). These results are suggestive of AU-rich thermometers located in both 5′ and 3′ UTRs, whose melting facilitates RNA degradation; this conclusion is supported by our melts on representative candidates. Consistent with this interpretation, in yeast, mRNAs with a lower in vitro estimated melting temperature declined in abundance under heat shock compared with mRNAs with a higher estimated melting temperature, which was attributed to greater exosome access to unstructured RNA (13).
We also observe evidence for temperature-induced unfolding in 5′UTRs that is associated with mRNA degradation. A previous study on Arabidopsis reported the down-regulation of several thousand mRNAs after heat shock (29). The majority (85%) of the down-regulated transcripts lost down-regulation in an xrn4 mutant (29). Because XRN4 is a single-stranded 5′ to 3′ nuclease, that observation together with our RNA structurome analysis suggest that 5′UTR unfolding facilitates XRN4-mediated degradation, and targeted decay analyses are consistent with this suggestion (SI Appendix, Fig. S10).
Protection from DMS reactivity can be afforded by both base pairing and protein binding; thus, we evaluated the hypothesis that some of the DMS reactivity increases we observed might be a result of heat-induced loss of RNA-binding proteins in UTR regions. Recently, 3′UTR-seq in zebrafish embryos found that AU-rich elements correlated with accelerated degradation after zygotic genome activation (34). In the same study, polyU and UUAG sequences were also associated with delayed degradation of maternal mRNAs early in embryogenesis. In both cases, it was proposed that association with zebrafish mRNA binding proteins, rather than RNA structure, controlled degradation (34). However, a directed analysis of all instances of the UUAG motif in the 3′UTRs of our structurome libraries revealed more instances of no heat-induced change in reactivity (11,861) than either positive (5,157) or negative (3,423) reactivity changes, whereas a change in protein affinity for the binding site should have had a pervasive and uniform signature if protein dissociation was the major causal agent of reactivity changes. We also assessed 3′UTRs in our structurome datasets for the presence of sequences identified as protein-binding mRNA motifs from a PIP-seq analysis in Arabidopsis (35). We found no enrichment of such motifs in regions of the 3′UTR associated with the most increased reactivity on heat exposure, again suggesting that many of the reactivity increases we observe are independent of protein unbinding. Thus, at present, we find no evidence that loss of protein protection has a major contribution to the heat-induced gain in DMS reactivity in rice UTRs, although we acknowledge the current limited nature of data on temperature regulation of the global RNA–protein interactome.
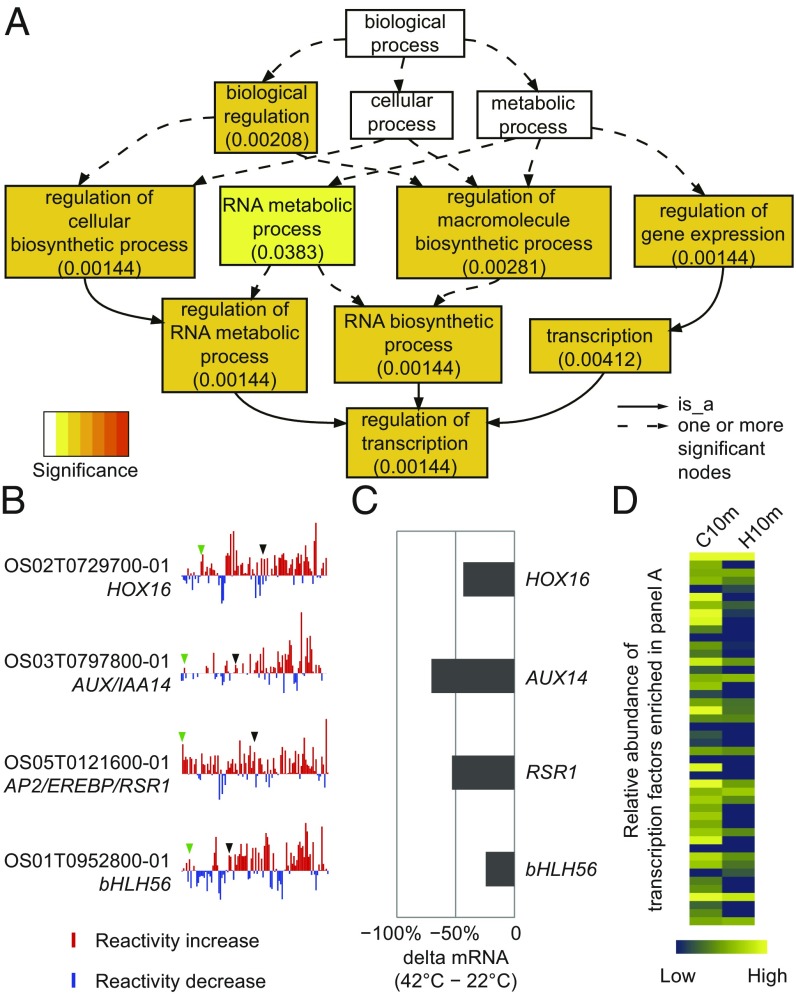
We evaluated the functional roles of mRNAs with elevated DMS reactivity in response to heat shock (Fig. 5). Gene ontology analysis of the 5% of mRNAs with the greatest heat-induced increase in average DMS reactivity showed a significant overrepresentation of genes that function in transcriptional regulation (Fig. 5 A and B). Application of an established assay of mRNA decay (36) confirmed that mRNAs of four transcription factors with dramatic heat-induced DMS reactivity increases exhibited accelerated decay (Fig. 5C), whereas our RNA-seq analyses broadly confirmed that transcription factors in this category rapidly declined in abundance after heat shock (Fig. 5D and SI Appendix, Fig. S11). Our functional (Fig. 5) and biochemical (SI Appendix, Figs. S9 and S11) analyses provide a likely mechanistic underpinning to a previous observation that heat stress reduces transcription factor mRNA abundance in rice floral tissues during anthesis (37), a stage of reproductive development in crops that is particularly sensitive to yield losses after heat stress (1). As transcription factors are master regulators of gene expression, our results may imply a type of widespread hierarchical control of transcriptional regulation mediated by RNA structure change in response to temperature. Interestingly, heat shock transcription factors apparently escape this regulatory mechanism, as those in our dataset show only minor DMS reactivity changes after heat shock (SI Appendix, Table S5).
Fig. 5.
Gene ontology analysis uncovers enrichment of transcription factors in mRNAs with the greatest heat-induced DMS reactivity increases. (A) Enrichment of gene ontology functional categories in the 5% of mRNAs with most elevated DMS reactivity at 42 °C. (B) DMS reactivity profiles for four transcription factors in the “regulation of transcription” category; these show dramatic heat-induced increase in DMS reactivity. For visualization, reactivity differences (42 °C − 22 °C) on all nucleotides in a transcript were placed into 100 bins and averaged within each bin. Green and black arrowheads point to the end of 5′UTR and the start of 3′UTR, respectively. (C) Heat-promoted mRNA decay. Loss in mRNA abundance at 10 min in the presence of cordycepin (42 °C – 22 °C). (D) Transcription factors in the top 5% of transcripts with elevated mRNA DMS reactivity after 10 min of 42 °C heat shock (H10m) show decreased abundance after 10 min heat shock (RNA-seq analysis). Blue and yellow indicate low and high abundance, respectively (SI Appendix, Fig. S11 provides the corresponding RNA-seq heat map at other timepoints).
In summary, given the multifaceted effects of temperature on RNA structure discovered in this in vivo study of RNA structurome modulation by supraoptimal temperatures, we propose that much of the eukaryotic transcriptome functions as an environmental thermosensor. We propose that in eukaryotes, transcripts are dynamically subject to degradation by a molecular mechanism involving heat-induced secondary structure unfolding in AU-rich 5′- and 3′-UTRs. Given that RNA structure can be regulated independent of encoded protein sequence through variation in UTR sequence and synonymous SNPs (38), our observations suggest mechanisms by which rice and other crops could be engineered to better withstand temperature and other stresses.
Materials and Methods
Preparation of Structure-seq and Ribo-seq libraries followed the procedures of Ritchey et al. (19) and Juntawong et al. (39), respectively, with some modifications. RNA-seq library preparation followed the standard Illumina TruSeq RNA Library preparation pipeline. Complete details concerning plant materials, experimental methods, and data analyses are provided in SI Appendix; analyzed library data are provided in Dataset S1.
Supplementary Material
Acknowledgments
We thank Craig Praul (Genomics Core Facility, Penn State University) for high-throughput Illumina sequencing. This work was supported by the National Science Foundation Plant Genome Research Program (NSF-IOS-1339282), and by an Innovation Award from Penn State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo: next-generation sequencing data for the structure-probing libraries (accession no. GSE100714), RNA-seq data (accession no. GSE100713), Ribo-seq data (accession no. GSE102216). Our RNA structure analysis code has been deposited at github (https://github.com/StructureFold/StructureFold and https://github.com/StructureFold2/StructureFold2). Supplementary Dataset S1 contains all the analyzed data (and associated P values) for Structure-seq, Ribo-seq, and RNA-seq datasets.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807988115/-/DCSupplemental.
References
- 1.Bita CE, Gerats T. Plant tolerance to high temperature in a changing environment: Scientific fundamentals and production of heat stress-tolerant crops. Front Plant Sci. 2013;4:273. doi: 10.3389/fpls.2013.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Battisti DS, Naylor RL. Historical warnings of future food insecurity with unprecedented seasonal heat. Science. 2009;323:240–244. doi: 10.1126/science.1164363. [DOI] [PubMed] [Google Scholar]
- 3.Peng S, et al. Rice yields decline with higher night temperature from global warming. Proc Natl Acad Sci USA. 2004;101:9971–9975. doi: 10.1073/pnas.0403720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao C, et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci USA. 2017;114:9326–9331. doi: 10.1073/pnas.1701762114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosová K, Vítámvás P, Prášil IT, Renaut J. Plant proteome changes under abiotic stress–Contribution of proteomics studies to understanding plant stress response. J Proteomics. 2011;74:1301–1322. doi: 10.1016/j.jprot.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Obata T, et al. Metabolite profiles of maize leaves in drought, heat, and combined stress field trials reveal the relationship between metabolism and grain yield. Plant Physiol. 2015;169:2665–2683. doi: 10.1104/pp.15.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kotak S, et al. Complexity of the heat stress response in plants. Curr Opin Plant Biol. 2007;10:310–316. doi: 10.1016/j.pbi.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Bevilacqua PC, Ritchey LE, Su Z, Assmann SM. Genome-wide analysis of RNA secondary structure. Annu Rev Genet. 2016;50:235–266. doi: 10.1146/annurev-genet-120215-035034. [DOI] [PubMed] [Google Scholar]
- 9.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buratti E, Baralle FE. Influence of RNA secondary structure on the pre-mRNA splicing process. Mol Cell Biol. 2004;24:10505–10514. doi: 10.1128/MCB.24.24.10505-10514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kutchko KM, et al. Multiple conformations are a conserved and regulatory feature of the RB1 5′ UTR. RNA. 2015;21:1274–1285. doi: 10.1261/rna.049221.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toscano C, et al. A silent mutation (2939G>A, exon 6; CYP2D6*59) leading to impaired expression and function of CYP2D6. Pharmacogenet Genomics. 2006;16:767–770. doi: 10.1097/01.fpc.0000236331.03681.24. [DOI] [PubMed] [Google Scholar]
- 13.Wan Y, et al. Genome-wide measurement of RNA folding energies. Mol Cell. 2012;48:169–181. doi: 10.1016/j.molcel.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Righetti F, et al. Temperature-responsive in vitro RNA structurome of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 2016;113:7237–7242. doi: 10.1073/pnas.1523004113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kortmann J, Narberhaus F. Bacterial RNA thermometers: Molecular zippers and switches. Nat Rev Microbiol. 2012;10:255–265. doi: 10.1038/nrmicro2730. [DOI] [PubMed] [Google Scholar]
- 16.Ding Y, et al. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature. 2014;505:696–700. doi: 10.1038/nature12756. [DOI] [PubMed] [Google Scholar]
- 17.Wan Y, et al. Landscape and variation of RNA secondary structure across the human transcriptome. Nature. 2014;505:706–709. doi: 10.1038/nature12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spitale RC, et al. Structural imprints in vivo decode RNA regulatory mechanisms. Nature. 2015;519:486–490. doi: 10.1038/nature14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchey LE, et al. Structure-seq2: Sensitive and accurate genome-wide profiling of RNA structure in vivo. Nucleic Acids Res. 2017;45:e135. doi: 10.1093/nar/gkx533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deng H, et al. Rice in vivo RNA structurome reveals RNA secondary structure conservation and divergence in plants. Mol Plant. 2018;11:607–622. doi: 10.1016/j.molp.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leamy KA, Assmann SM, Mathews DH, Bevilacqua PC. Bridging the gap between in vitro and in vivo RNA folding. Q Rev Biophys. 2016;49:e10. doi: 10.1017/S003358351600007X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schymanski SJ, Or D, Zwieniecki M. Stomatal control and leaf thermal and hydraulic capacitances under rapid environmental fluctuations. PLoS One. 2013;8:e54231. doi: 10.1371/journal.pone.0054231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tinoco I, Jr, Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 24.Wu X, Bartel DP. Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell. 2017;169:905–917.e11. doi: 10.1016/j.cell.2017.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krajewski SS, Narberhaus F. Temperature-driven differential gene expression by RNA thermosensors. Biochim Biophys Acta. 2014;1839:978–988. doi: 10.1016/j.bbagrm.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 26.McClure BA, Guilfoyle T. Characterization of a class of small auxin-inducible soybean polyadenylated RNAs. Plant Mol Biol. 1987;9:611–623. doi: 10.1007/BF00020537. [DOI] [PubMed] [Google Scholar]
- 27.Lykke-Andersen S, Tomecki R, Jensen TH, Dziembowski A. The eukaryotic RNA exosome: Same scaffold but variable catalytic subunits. RNA Biol. 2011;8:61–66. doi: 10.4161/rna.8.1.14237. [DOI] [PubMed] [Google Scholar]
- 28.Bonneau F, Basquin J, Ebert J, Lorentzen E, Conti E. The yeast exosome functions as a macromolecular cage to channel RNA substrates for degradation. Cell. 2009;139:547–559. doi: 10.1016/j.cell.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 29.Merret R, et al. Heat-induced ribosome pausing triggers mRNA co-translational decay in Arabidopsis thaliana. Nucleic Acids Res. 2015;43:4121–4132. doi: 10.1093/nar/gkv234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bevilacqua PC, Assmann SM. Technique development for probing RNA structure in vivo and genome-wide. Cold Spring Harb Perspect Biol. 2018;10:a032250. doi: 10.1101/cshperspect.a032250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mustoe AM, et al. Pervasive regulatory functions of mRNA structure revealed by high-resolution SHAPE probing. Cell. 2018;173:181–195.e18. doi: 10.1016/j.cell.2018.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ahmed R, Duncan RF. Translational regulation of Hsp90 mRNA. AUG-proximal 5′-untranslated region elements essential for preferential heat shock translation. J Biol Chem. 2004;279:49919–49930. doi: 10.1074/jbc.M404681200. [DOI] [PubMed] [Google Scholar]
- 34.Rabani M, Pieper L, Chew GL, Schier AF. A massively parallel reporter assay of 3′ UTR sequences identifies in vivo rules for mRNA degradation. Mol Cell. 2017;68:1083–1094.e5. doi: 10.1016/j.molcel.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gosai SJ, et al. Global analysis of the RNA-protein interaction and RNA secondary structure landscapes of the Arabidopsis nucleus. Mol Cell. 2015;57:376–388. doi: 10.1016/j.molcel.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park SH, et al. Posttranscriptional control of photosynthetic mRNA decay under stress conditions requires 3′ and 5′ untranslated regions and correlates with differential polysome association in rice. Plant Physiol. 2012;159:1111–1124. doi: 10.1104/pp.112.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.González-Schain N, et al. Genome-wide transcriptome analysis during anthesis reveals new insights into the molecular basis of heat stress responses in tolerant and sensitive rice varieties. Plant Cell Physiol. 2016;57:57–68. doi: 10.1093/pcp/pcv174. [DOI] [PubMed] [Google Scholar]
- 38.Solem AC, Halvorsen M, Ramos SB, Laederach A. The potential of the riboSNitch in personalized medicine. Wiley Interdiscip Rev RNA. 2015;6:517–532. doi: 10.1002/wrna.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA. 2014;111:E203–E212. doi: 10.1073/pnas.1317811111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.