Significance
Fragile X syndrome (FXS) is the most common form of inherited intellectual disability and autism. FXS results from the loss of functional fragile X mental retardation protein (FMRP), an RNA binding protein involved in translational regulation. However, the impact of FMRP on gene expression has not been evaluated comprehensively. Here, we present simultaneous high-resolution ribosome profiling and RNA-sequencing data from the same wild-type and FMRP-deficient adult neural stem cells. We find remarkable and heretofore unknown forms of regulation by FMRP critical for neural differentiation. Importantly, our data also show that FMRP controls RNA expression in six distinct ways. Thus, we have uncovered a molecular foundation for pathophysiologies associated with FXS.
Keywords: fragile X syndrome, neural stem cells, translation, ribosome profiling, neural differentiation
Abstract
Fragile X syndrome (FXS) is caused by the loss of fragile X mental retardation protein (FMRP), an RNA binding protein whose deficiency impacts many brain functions, including differentiation of adult neural stem cells (aNSCs). However, the mechanism by which FMRP influences these processes remains unclear. Here, we performed ribosome profiling and transcriptomic analysis of aNSCs in parallel from wild-type and Fmr1 knockout mice. Our data revealed diverse gene expression changes at both mRNA and translation levels. Many mitosis and neurogenesis genes were dysregulated primarily at the mRNA level, while numerous synaptic genes were mostly dysregulated at the translation level. Translational “buffering”, whereby changes in ribosome association with mRNA are compensated by alterations in RNA abundance, was also evident. Knockdown of NECDIN, an FMRP-repressed transcriptional factor, rescued neuronal differentiation. In addition, we discovered that FMRP regulates mitochondrial mRNA expression and energy homeostasis. Thus, FMRP controls diverse transcriptional and posttranscriptional gene expression programs critical for neural differentiation.
Fragile X syndrome (FXS) is the most common form of inherited intellectual disability, which occurs in about 1 in 4,000 males and 1 in 6,000 females. FXS patients display a wide spectrum of autistic features, including cognitive, behavioral, and social deficits (1). Nearly all FXS cases are caused by a CGG trinucleotide repeat expansion in the 5′ UTR of the fragile X mental retardation 1 (FMR1) gene, which leads to hypermethylation of the promoter region (2) and consequent transcriptional silencing and loss of the encoded protein, fragile X mental retardation protein (FMRP). FMRP is an RNA binding protein that is highly expressed in the brain and largely functions as a translational repressor (3). In addition to FMR1, other mutations underlying autism spectrum disorders occur in genes that signal to the translational apparatus, such as TSC complex subunit 2 (TSC2) and phosphatase and tensin homolog (PTEN) (4, 5). It is therefore apparent that maintenance of translational homeostasis is essential for proper neural functions such as synaptic efficacy (6). Indeed, a number of phenotypes of the Fmr1 knockout (KO) mouse are rescued by depleting a variety of translational activators, likely through rebalancing the translational homeostasis (7, 8).
Neurogenesis persists throughout life in two germinal zones: the subgranular zone in the dentate gyrus (DG) of the hippocampus and the subventricular zone of the lateral ventricles (9). The newborn neurons in the DG integrate into the existing circuitry of the hippocampus, which is crucial for cognitive function and implicated in both neurodevelopmental and neuropsychiatric disorders (9). We have shown that selective deletion of FMRP from adult neural stem cells (aNSCs) leads to impaired performance on hippocampus-dependent learning tasks; conversely, restoration of FMRP specifically in aNSCs rescues these learning deficits in FMRP-deficient mice (10). FMRP deficiency in mouse aNSCs leads not only to aberrant activation and increased proliferation of aNSCs, but also to reduced neural differentiation (11, 12). Although several FMRP-regulated genes and pathways have been identified in aNSCs, genome-wide gene expression changes resulting from FMRP deficiency remain unknown. Moreover, how gene expression programs are compromised in FMRP-deficient aNSCs remains elusive.
Several studies using high-throughput technologies, such as cross-linking immunoprecipitation (CLIP) coupled with sequencing (13–15), have identified partially overlapping lists of FMRP target mRNAs. However, those approaches suffer from the heterogeneity of brain tissue, the lack of sufficient sensitivity, and the inability of capturing simultaneous changes in mRNA and translation.
To obtain an in-depth understanding of gene regulation by FMRP, we performed simultaneous ribosome profiling and RNA sequencing (RNA-seq) on Fmr1 KO aNSCs. Our data revealed six distinct forms of FMRP regulation at mRNA and translation levels that are involved in mitosis, neurogenesis, synaptic, and mitochondrial function. Interestingly, many mitosis and neurogenesis genes were dysregulated primarily at the mRNA level, whereas numerous synaptic genes were mostly dysregulated at the translation level. We discovered that the expression changes of many FMRP target mRNAs were buffered at the translational level, whereby ribosome association with mRNA is offset by altered transcript levels. We also found a previously unknown role of FMRP in stem cell maintenance and metabolic regulation. Thus, we have uncovered gene groups with distinct modes of dysregulation that might be responsible for various aspects of FXS pathophysiology.
Results
Ribosome Profiling Reveals Diverse Changes of Gene Expression in Fmr1 KO aNSCs.
To identify the FMRP-controlled gene expression program in mouse aNSC differentiation, we isolated aNSCs from the DG of Fmr1 KO mice and their wild-type (WT) littermates and cultured them as relatively homogenous neurospheres (16). Four biological replicates of these cultures were analyzed by ribosome profiling, a genome-wide method that monitors mRNA translation by sequencing ribosome-protected fragments (RPFs) (Fig. 1A) (17). Translational efficiency (TE) was calculated by normalizing RPFs with mRNA as measured by RNA-seq performed in parallel. As expected, most RPFs were mapped to the same frame as the annotated coding sequences (CDSs), indicating that they faithfully reflect the distribution of ribosomes during translation (SI Appendix, Fig. S1 A–E and Table S1).
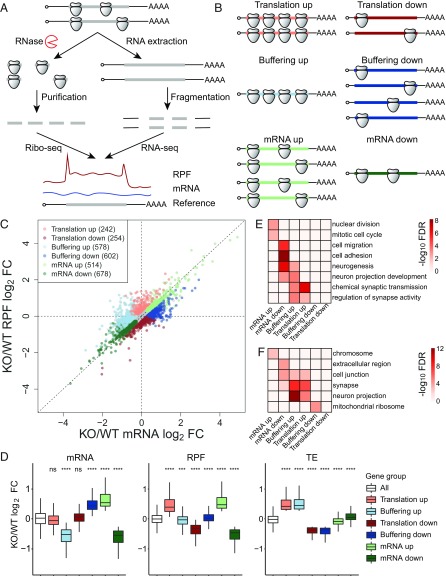
Fig. 1.
Ribosome profiling reveals diverse changes of gene expression in Fmr1 KO aNSCs. (A) Schematic diagram of the experimental procedures for ribosome profiling. (B) Schematic diagram of six regulatory groups captured by ribosome profiling. (C) Scatter plot of expression changes of mRNA levels and RPFs. Dysregulated mRNAs in the absence of FMRP are classified into six regulatory groups as shown in B; 12,502 genes past filtering are used for the scatter plot. Absolute fold change (FC) > 1.2, nominal P < 0.05, and false discovery rate (FDR) = 0.042 by permutation test. (D) Box plots of expression changes at different levels across six regulatory groups that visualize the medians. The lower and upper hinges correspond to the first and third quartiles. The whiskers extend from the hinges to the largest and smallest values no further than 1.5-fold of interquartile range. Outliers are not shown. Gene expression changes of each regulatory group were compared with those of all genes used for differentially expressed gene analysis (ns, not significant; ***P < 0.001; ****P < 0.0001; Wilcoxon rank sum test after multiple test correction with the Bonferroni method). (E) Top GO terms of biological process enriched in each regulatory group. The enrichment significance (−log10 FDR) is color coded. (F) Top GO terms of cellular component enriched in each regulatory group. See also SI Appendix, Figs. S1 and S2 and Datasets S1–S3.
Several relationships between mRNA and RPF changes can be computed with ribosome profiling data (Fig. 1B) (18). When RPFs are up or down but with little change in RNA levels, TEs change accordingly. TE can also vary when mRNA levels are up or down but are accompanied by no significant change in RPFs. Translational “buffering” also occurs when significant mRNA changes are accompanied by offsetting alterations in RPFs. In these cases, buffering describes steady-state gene expression without implying a cause-and-effect relationship between ribosome loading and RNA levels (19). Lastly, mRNA levels and RPFs may change concordantly without statistically distinguishable differences in TE.
We utilized anota2seq software to separate the buffering groups from the conventional translation group. In contrast to conventional analysis methods based on the log ratios between RPF and mRNA, anota2seq uses analysis of partial variance, which estimates translational activity independent of mRNA levels (20). Among 12,502 detectable genes, we identified 242 in the “translation up” group, 254 in the “translation down” group, 578 in the “buffering up” group, 602 in the “buffering down” group, 514 in the “mRNA up” group, and 678 in the “mRNA down” group (absolute fold change > 1.2, nominal P < 0.05, and false discovery rate = 0.042 by permutation test) (Fig. 1C, SI Appendix, Fig. S1F, and Dataset S1). The expression changes of each group match the above definitions, confirming that the classification algorithms are able to separate the buffering groups from the translation groups (Fig. 1D). We performed gene ontology (GO) analysis with terms of biological process (Fig. 1E and Dataset S2) and cellular component (Fig. 1F and Dataset S3) to determine whether specific biological functions were enriched in each regulatory gene group. Genes related to nuclear division and mitotic cell cycle were enriched in the mRNA up group, which is consistent with the excessive proliferation of Fmr1 KO aNSCs (10–12). In contrast, genes related to cell adhesion and neurogenesis were enriched in the mRNA down group, underlying the defective neural differentiation of Fmr1 KO aNSCs (10–12). Surprisingly, synaptic genes were enriched in both translation up and buffering up groups, with increased TEs regardless of mRNA changes. In WT undifferentiated aNSCs, synaptic gene expression was low (SI Appendix, Fig. S1G), which might represent a transcriptionally primed status as shown for radial glial precursors in embryonic murine cortex (21).
We explored several mRNA features that could potentially confer distinct expression changes to the different regulatory groups, including lengths, GC content, and secondary structures of 5′ UTRs, CDSs, and 3′ UTRs (SI Appendix, Fig. S2). The buffering up and mRNA down groups had significantly longer 5′ and 3′ UTRs, which correlated with decreased mRNA abundance in Fmr1 KO. Conversely, the buffering down group had significantly shorter 3′ UTRs, which correlated with increased mRNAs. Translation down and buffering down groups had significantly shorter CDSs, which correlated with decreased TEs (SI Appendix, Fig. S2A).
Mitosis and Neurogenesis Genes Are Dysregulated at the mRNA Level in Fmr1 KO aNSCs.
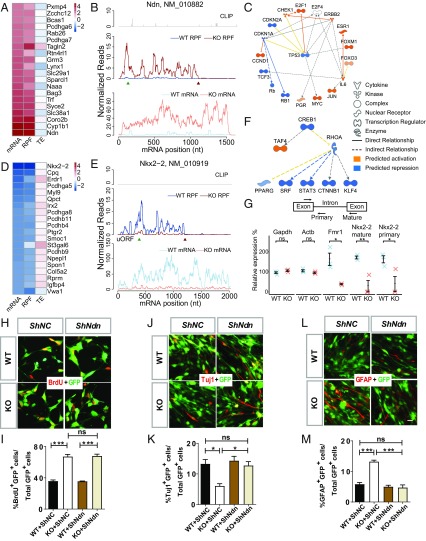
In Fmr1 KO aNSCs, many mitosis-related genes exhibited increased mRNA and RPFs with little change of TE (Fig. 2 A and B). We sought to find upstream regulators responsible for the proliferation defects in the mRNA up group by using Ingenuity Pathway Analysis (IPA) software, which uncovered a network of several transcription factors in mitosis (Fig. 2C). Here, P53 serves as the hub, and its activity is predicted to be inhibited in Fmr1 KO aNSCs, consistent with our previous result (11). In addition, several neurogenesis-related genes in the mRNA down group showed both reduced mRNAs and RPFs; their TEs were slightly increased without passing the significance cutoff (Fig. 2 D and E). Based on the mRNA down genes, we also identified an upstream regulatory network consisting of neurogenesis transcriptional factors predicted to be repressed (Fig. 2F), which could be involved in neural differentiation defects.
Fig. 2.
Mitosis and neurogenesis genes are dysregulated at the mRNA level in Fmr1 KO aNSCs. (A) Heatmap of expression changes (log2 fold change KO/WT) for the top 20 genes in the mRNA up group. (B) Read distributions on the Ndn gene of mRNA up group. FMRP CLIP tags (Top) (13) and normalized RPF reads (Middle) and mRNA reads (Bottom) averaged across all replicates are plotted along the mRNA nucleotide positions, with green and red triangles for annotated start and stop codons, respectively. (C) Transformation-related protein 53 (TP53) upstream network of increased mRNAs predicted by IPA. (D) Heatmap of expression changes (log2 fold change KO/WT) for the top 20 genes in the mRNA down group. (E) Read distributions on the Nkx2-2 gene of mRNA down group. (F) cAMP responsive element binding protein 1 (CREB1) upstream network of decreased mRNAs predicted by IPA. (G, Top) Schematic diagram of the primer design to measure the abundance of primary and mature transcripts. (G, Bottom) qPCR validation of mRNA changes for selected genes. Data are presented as mean ± SEM (n = 3; ns, not significant; *P < 0.05; **P < 0.01; two-tailed Student’s t test). (H) Acute knockdown of Ndn by shNdn did not affect the proliferation rate of Fmr1 KO aNSCs as assessed by BrdU incorporation under proliferating conditions followed by quantitative analysis (I). (J) Acute knockdown of Ndn by shNdn rescued neuronal differentiation phenotypes of Fmr1 KO aNSCs as assessed by β-tubulin III (Tuj1; a neuronal marker) followed by quantitative analysis (K). (L) Acute knockdown of Ndn using shNdn rescued astroglial differentiation phenotypes of Fmr1 KO aNSCs as assessed by glial fibrillary acidic protein (GFAP; an astroglial marker) followed by quantitative analysis (M). (H, J, and L) Red for BrdU, Tuj1, or GFAP; green for shNdn or shNC viral-infected cells. (Scale bar, 20 µm.) (I, K, and M) n = 3; ns, not significant; *P < 0.05; ***P < 0.001; two-way ANOVA. Data are presented as mean ± SEM. See also SI Appendix, Fig. S3.
The changes of steady-state mRNA levels can be achieved by transcriptional or posttranscriptional mechanisms; to distinguish between them, we used qRT-PCR with exon–exon primer pairs to detect mature transcripts and with exon–intron primers for primary transcripts. Using NK2 homeobox 2 (Nkx2-2) as an example of mRNA down group, similar reductions were observed for both primary and mature transcripts, suggesting a transcriptional repression (Fig. 2G). Although NECDIN (Ndn) (an example of mRNA up group) is an intronless gene and, thus, not suitable for this method, we confirmed increased Ndn mRNA by qRT-PCR (SI Appendix, Fig. S3A). Because we did not detect differences in Ndn mRNA stability with an actinomycin D chase assay (SI Appendix, Fig. S3B), we infer that this mRNA is transcriptionally up-regulated in Fmr1 KO aNSCs.
To assess the biological implications of genes dysregulated at the mRNA level, we assessed the function of Ndn, which is critical for progenitor cell proliferation and differentiation during brain development (22). We investigated the impact of Ndn knockdown on aNSC proliferation as assessed by 5-bromodeoxyuridine (BrdU) pulse-labeling and differentiation as assessed by cell lineage-specific antibodies: β-tubulin III for neurons; glial fibrillary acidic protein for astrocytes. Acute knockdown of Ndn (by shNdn) did not affect cell proliferation in either WT or Fmr1 KO aNSCs (Fig. 2 H and I and SI Appendix, Fig. S3C) but rescued neuronal differentiation (Fig. 2 J and K) and astroglial differentiation (Fig. 2 L and M) in Fmr1 KO aNSCs. The shNdn treatment restored the differentiation of Fmr1 KO aNSCs to that of WT (lenti-shNC–treated) aNSCs (Fig. 2 K and M). Thus, reducing Ndn rescued differentiation deficits of FMRP-deficient aNSCs.
FMRP CLIP Targets Have Increased TEs in Fmr1 KO aNSCs.
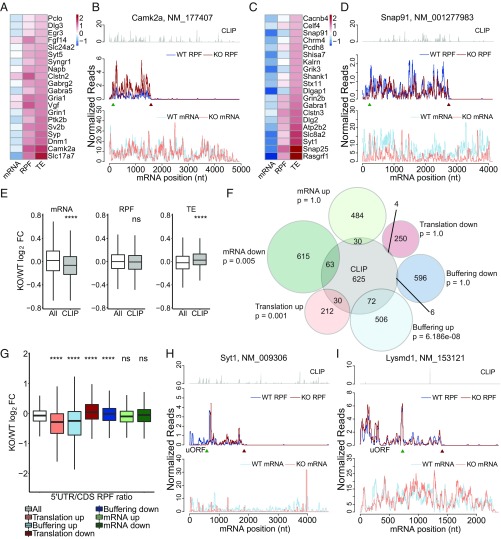
Current models suggest that FMRP represses the translation elongation of specific mRNAs essential for synaptic plasticity and other neural functions (3, 7, 8, 13). Thus, we focused on the translation up and buffering up groups with increased TEs. Although mRNAs encoding synaptic proteins in the translation up group changed only marginally, their RPFs increased, leading to significantly elevated TEs (Fig. 3 A and B). Unlike the translation up group, synaptic genes in the buffering up group had significantly decreased mRNAs, with negligible change of RPFs (Fig. 3 C and D).
Fig. 3.
FMRP CLIP targets have increased TEs in Fmr1 KO aNSCs. (A) Heatmap of expression changes [log2 fold change (FC) KO/WT] for the top 20 synaptic genes in the translation up group. (B) Read distributions on the calcium/calmodulin-dependent protein kinase II alpha (Camk2a) gene of translation up group. (C) Heatmap of expression changes (log2FC KO/WT) for the top 20 synaptic genes in the buffering up group. (D) Read distributions on the synaptosomal-associated protein 91 (Snap91) gene of buffering up group. (E) Box plots of expression changes at different levels for FMRP CLIP targets compared with those of all genes used for differentially expressed gene (DEG) analysis (ns, not significant; ****P < 0.0001; Wilcoxon rank sum test after multiple test correction with the Bonferroni method). CLIP genes are top FMRP targets identified in ref. 13. (F) Overlap between FMRP CLIP genes and genes in each regulatory group. Statistical significance is calculated with a hypergeometric test with the Bonferroni correction. See also SI Appendix, Table S2. (G) Box plots of changes of 5′ UTR/CDS read ratios in different regulatory groups compared with those of all genes used for DEG analysis (ns, not significant; ***P < 0.001; ****P < 0.0001; Wilcoxon rank sum test after multiple test correction with the Bonferroni method). (H) Read distributions on the synaptotagmin I (Syt1) gene with decreased 5′ UTR/CDS ratio. (I) Read distributions on the LysM and putative peptidoglycan-binding domain-containing 1 (Lysmd1) gene with increased 5′ UTR/CDS ratio. See also SI Appendix, Fig. S4.
To evaluate the expression changes of direct targets of FMRP in aNSCs, we compared our data to those of the FMRP CLIP dataset from WT mouse cortex (13), reasoning that mRNA expressed in both models would be conserved substrates of FMRP activity that are shared across different developmental stages and neural lineages. As a group, the top FMRP CLIP targets had significantly lower mRNA levels but the same RPF levels, resembling the buffering up group (Fig. 3E). We found significant overlaps between FMRP CLIP genes and genes in the translation up or the buffering up group, indicating that direct targets of FMRP had increased TEs in Fmr1 KO aNSCs (Fig. 3F and SI Appendix, Table S2).
Because CLIP experiments failed to identify consensus FMRP binding sites in whole mouse cortex (13), we first stratified FMRP-regulated mRNAs into the six regulatory groups outlined in Fig. 1. For the mRNAs with FMRP CLIP tags, we identified previously reported TAC and TGGA motifs (14, 23). In addition, we identified several motifs that we surmise may be responsible for the diverse gene expression changes (SI Appendix, Fig. S4A).
Several studies have shown that upstream ORFs (uORFs), which mediate downstream ORF translation (24), are prevalent in the mammalian genome (25) and are important for synaptic plasticity (26). Because ribosome profiling does not capture scanning 43S preinitiation complexes and because uORFs are usually inhibitory for main CDS translation, we used the ratios between the reads on the 5′ UTR and CDS as a measurement of uORF translation. Interestingly, we found decreased 5′ UTR/CDS ratios for the translation up and buffering up groups in Fmr1 KO aNSCs, which is consistent with their increased translational activities (Fig. 3 G and H). In contrast, the translation down and buffering down groups had significantly increased 5′ UTR/CDS ratios in Fmr1 KO aNSCs, consistent with the inhibitory function of uORFs (Fig. 3 G and I). Because the internal read ratio between 5′ UTR and CDS of the same mRNA is independent of the TE calculation, the results confirm that those identified genes indeed underwent dysregulated translation in the absence of FMRP. In addition, we found no statistically significant correlation between KO/WT changes of 5′ UTR/CDS RPF ratio and the length or complexity of the 5′ UTR, suggesting that the altered uORF translation in Fmr1 KO aNSCs cannot be simply explained by these features of 5′ UTR sequences (SI Appendix, Fig. S4 B and C).
Compromised Expression of Nuclear-Encoded Mitochondrial Protein mRNAs Is Correlated with Disturbed Energy Homeostasis in Fmr1 KO aNSCs.
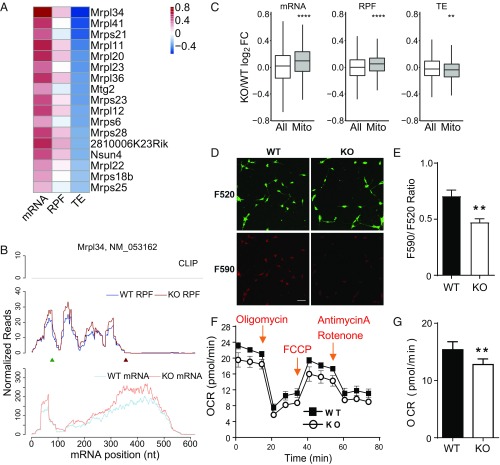
We found a substantial number of genes with decreased TEs in Fmr1 KO aNSCs (Fig. 1C). For the buffering down group, we noticed a statistically significant enrichment of mitochondrial ribosomal protein genes (Fig. 1F). The increase of RPFs was smaller than the increase of mRNA, resulting in decreased TE (Fig. 4 A and B). To evaluate whether this is a general phenomenon, we compared the expression of genes encoding proteins with mitochondrial function to that of all the genes (27). The buffering down pattern was maintained for nearly all annotated nuclear-encoded mitochondrial genes, raising the possibility of defective energy metabolism in Fmr1 KO aNSCs (Fig. 4C). FMRP has been implied to play a role in energy metabolism and mitochondrial function in a Drosophila model (28) but it has not been studied in mouse aNSCs. Thus, we first examined the mitochondrial membrane potential (MMP) in aNSCs with a JC-10 cationic lipophilic fluorescent assay (Fig. 4D). The ratio of fluorescence emission at 590 nm to fluorescence emission at 520 nm (F590/F520) directly correlates with the MMP. Our analysis found a significantly decreased F590/F520 ratio in the Fmr1 KO aNSCs compared with WT aNSCs, indicating a less polarized MMP (a sign of poorly functioning mitochondria) in Fmr1 KO aNSCs (Fig. 4E). Next, we performed a bioenergetic profile of aNSCs using the Agilent Seahorse XF-24 Analyzer (Fig. 4F). Compared with WT, Fmr1 KO aNSCs displayed a significantly lower aerobic respiration as measured by decreased O2 consumption rate (OCR), indicating mitochondrial respiration dysfunction (Fig. 4G). Thus, FMRP deficiency in aNSCs compromises expression of nuclear-encoded mitochondrial protein mRNAs, as well as energy homeostasis.
Fig. 4.
Compromised expression of nuclear-encoded mitochondrial protein mRNAs is correlated with disturbed energy homeostasis in Fmr1 KO aNSCs. (A) Heatmap of expression changes [log2 fold change (FC) KO/WT] for mitochondrial ribosomal protein mRNAs in the buffering down group. (B) Read distributions on the mitochondrial ribosomal protein L34 (Mrpl34) gene of buffering down group. (C) Box plots of expression changes at different levels for nuclear-encoded mitochondrial protein mRNAs compared with those of all genes used for differentially expressed gene analysis (**P < 0.01; ****P < 0.0001; Wilcoxon rank sum test after multiple test correction with the Bonferroni method). “Mito” genes are selected based on the MitoCarta2.0 database (27). (D and E) JC-10 assay and quantification of mitochondrial membrane potential in WT and Fmr1 KO aNSCs. (D) aNSCs were stained with JC-10 solution for 30 min at 37 °C. (Scale bar, 20 µm.) (E) The mitochondrial membrane potential was assessed by quantifying the ratio between intensity of red fluorescence (590 nm) and green fluorescence (520 nm). n = 3; **P < 0.01; Student’s t test was used for data analyses. Data are presented as mean ± SEM. (F and G) Bioenergetic profile of aNSCs by using an Agilent Seahorse XF-24 Analyzer. The OCR is measured before the addition of drugs (basal OCR) and then after the addition of the indicated drugs: Oligomycin is an ATP synthase inhibitor, and the reduction in OCR after oligomycin indicates the amount of O2 consumed for mitochondrial ATP generation. Carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) uncouples ATP synthesis from the electron transport chain (ETC). After FCCP, the maximum capacity of the mitochondria to use oxidative phosphorylation (maximal OCR) is revealed. Rotenone (complex I inhibitor) and antimycin A (complex III inhibitor) together render a complete shutdown of the ETC. Spare respiratory capacity is the difference between maximal OCR and basal OCR and, as such, is an indicator of how close to its bioenergetic limit the cell is functioning (47). n = 3; **P < 0.01; Student’s t test was used for data analyses. Data are presented as mean ± SEM.
Discussion
aNSCs represent an exceptional model for studying molecular functions of FMRP because their high homogeneity eliminates the variances of gene expression common among mixed cell types. Fmr1 KO aNSCs also exhibit defined and consistent cellular phenotypes that serve as functional readouts. Using translating ribosome affinity purification (TRAP), a recent study found altered translation of 120 mRNAs and down-regulated expression of FMRP target mRNAs as a group in Fmr1 KO CA1 pyramidal neurons (29, 30). The authors were not able to distinguish the contributions made by possible altered mRNA levels due to a lack of RNA-seq data versus ribosome packing density; thus, they could not compute TE. Moreover, TRAP-seq is generally unable to delineate the number of ribosomes bound to an mRNA, further complicating determinations of TE. In contrast, through our ribosome profiling data, we could faithfully calculate TE and found that FMRP-regulated mRNAs fall into six distinct classes based on their differential expression: translation up, translation down, buffering up, buffering down, mRNA up, and mRNA down.
In the adult hippocampus, quiescent NSCs dynamically integrate both extrinsic and intrinsic signals to either maintain their dormant state or become activated and give rise to intermediate progenitor cells (IPCs), which subsequently differentiate into postmitotic neurons or astrocytes (31, 32). The regulation of NSC activation plays a key role in neural regeneration, functions, plasticity, and disease (33). We previously found that a loss of FMRP led to greater activation and proliferation of aNSCs in vivo that can be recapitulated by neuroprogenitors/neurospheres isolated from WT and Fmr1 KO mice in vitro (10–12, 34). We also published that small molecules, including one that targets the MDM2-P53 pathway, can correct proliferation and differentiation of cultured Fmr1 KO neurospheres as well as in vivo NSC proliferation and cognitive impairment of Fmr1 KO mice (10, 11, 35). Therefore, our previous studies have demonstrated that, at least in the case of FXS, neurospheres derived from WT and KO adult DG are good models for in vivo NSC phenotypes. In the present study, Fmr1 KO aNSCs exhibited increased proliferation and astroglial production but reduced neuronal differentiation (Fig. 2 H–M). More importantly, P53 was predicted to be repressed in Fmr1 KO aNSCs using IPA (Fig. 2C), which was consistent with our previous study. Thus, our data not only confirmed a previous finding of a pathway regulating aNSC proliferation and differentiation (P53) observed in the nascent NSCs in the adult brain, but also uncovered factors that control differentiation as well as energy balance in aNSCs.
In contrast to the relatively large changes of translational activities in other models such as mammalian target of rapamycin signaling in cancer cells (36), the changes in TEs in Fmr1 KO aNSCs were more modest. This was not unexpected, given that general protein synthesis in Fmr1-deficient hippocampus is only ∼15% greater than in WT (7). One possible explanation for the relatively small change in gene expression in the Fmr1 KO cells is that the aNSCs adapt to the chronic loss of FMRP by compensatory mechanisms at the transcription, RNA stability, or translational level. Perhaps more importantly, it is increasingly appreciated that brain and neurons are particularly vulnerable to disruption of RNA-binding protein dosage and dynamics (37, 38). Thus, subtle changes of translational activities for hundreds of mRNAs might collectively contribute to the dysregulated neural differentiation and function in Fmr1 KO aNSCs.
The transcription factor NECDIN plays an essential role in neuron/glia lineage determination of aNSCs. Because Ndn mRNA is not identified by FMRP CLIP, it may not be a direct target of this protein. It is possible that excess translation of epigenetic and/or transcriptional regulators indirectly induces the transcription of Ndn, similar to what has been shown for bromodomain containing 4 (Brd4), a chromatin reader targeted by FMRP (39). In this case, Brd4 protein levels were significantly higher in Fmr1 KO mouse brain and neurons, which resulted in widespread changes in chromatin regulation and aberrant gene expression. More importantly, inhibition of Brd4 function alleviated many phenotypes associated with FXS.
Many synaptic genes were expressed at low levels but with increased TEs in Fmr1 KO aNSCs. One intriguing explanation is that WT aNSCs are transcriptionally primed for neuronal differentiation but are kept in check translationally by FMRP to maintain “stemness”, a critical feature of stem cells. In Fmr1 KO aNSCs, this delicate transcription–translation coupling is disrupted, resulting in uncontrolled proliferation and aberrant differentiation. Similar aberrant up-regulation of neuronal genes has been found in methyl-CpG binding domain protein 1–deficient aNSCs displaying impaired neuronal differentiation and neurogenesis (40).
Our data unveil a translational buffering mechanism by FMRP. Many synaptic genes in the buffering up group had increased TEs but decreased mRNAs. How FMRP regulates mRNA levels in aNSCs is unclear. Mechanistically, FMRP could control mRNA stability independent of its translational activity through interactions with N6-methyladenosine (m6A) modifications (41) as well as microRNAs (42). Alternatively, increased translation might result in a secondary destabilization of mRNA, as the two processes are tightly coupled (43). Another finding is that there are opposite changes of RPFs on uORFs and main CDSs for all of the gene groups with differences in TEs. Based on this observation, we hypothesize that uORF-mediated regulation of main CDS initiation could influence FMRP-regulated translation as much as modulation of polypeptide elongation (7, 13). Mechanistically, FMRP could regulate uORF translation either indirectly through eukaryotic translation initiation factor alpha phosphorylation (24) or directly through binding to m6A modifications (44). The detailed mechanisms remain to be explored.
Lastly, the mitochondrial defects in Fmr1 KO aNSCs revealed by both the gene expression analysis and bioenergetic profiling resulted in lower oxidative phosphorylation activity. Although our data do not prove a direct causal relationship between the compromised expression of nuclear-encoded mitochondrial protein mRNAs and impaired mitochondrial function, it is possible that the defective synthesis of mitochondrial proteins results in reduced MMP and aerobic respiration, which in turn leads to increased mitochondrial mRNA levels through a feedback mechanism. Although each mitochondrial mRNA has a small change in TE, when all of the affected mitochondrial mRNAs are considered, the aggregated changes are likely to have a profound effect on mitochondrial function. Recent results have suggested crucial roles of mitochondria in the maintenance of pluripotency and differentiation (45). Stem cells rely more on glycolytic metabolism, while cell differentiation is often associated with increased mitochondrial oxidative phosphorylation. Thus, the repressed oxidative phosphorylation in Fmr1 KO aNSCs might contribute to the overproliferation defects.
In summary, we present a comprehensive analysis of gene expression changes at both the mRNA and translation levels in FMRP-deficient aNSCs. Our in-depth analysis has parsed high-confidence consensus motifs for FMRP activity in distinct groups of mRNA that have diverse expression changes. In addition to uncovering a key transcription factor that governs aNSC differentiation, our investigations show that far from being a relatively straightforward translation factor, FMRP regulates gene expression at multiple levels.
Materials and Methods
Ribosome Profiling.
Adult-derived NSCs were isolated from the DG of 8- to 10-wk-old male Fmr1 KO mice and WT littermate controls based on our published method (16). Cycloheximide (CHX) was added into cultured aNSCs to a final concentration of 100 μg/mL, and cells were incubated at 37 °C for 10 min to allow the penetration of neurosphere clusters and the stabilization of ribosomes. Next, cells were washed twice with ice-cold PBS containing 100 μg/mL CHX, pelleted, and immediately stored at −80 °C. Cells were lysed in polysome lysis buffer by trituration through a 25-gauge needle. For the RNA-seq, 500 ng of purified cytoplasmic RNA was used for the library preparation with the Ovation RNA-Seq System V2. For the ribosome profiling, cleared lysates were digested with RNases A and T1 at 25 °C for 30 min and separated by sedimentation through sucrose gradients. Monosome fractions were identified, pooled, and extracted with TRIzol LS. Ribosome profiling libraries were prepared following the published protocols (46) and sequenced with Illumina NextSeq.
Read Mapping.
For the ribosome profiling data, individual samples were separated based on the barcode sequences. Adaptor sequences were removed with cutadapt. Trimmed reads were quality filtered and mapped to the mouse rRNA and tRNA references with Bowtie2. Unmapped reads were next mapped to the mm10 mouse genome with Tophat2. PCR duplicates were marked based on the Unique Molecular Identifier sequences, and only uniquely mapped reads without duplicates were retained with samtools for the downstream analysis. For the RNA-seq data, reads unmapped to rRNA and tRNA were mapped to mm10 genome with Tophat2. RPF length distribution, P-site offsets, and frame preference were calculated with plastid. Counts at each nucleotide position were extracted using P sites of RPFs and the 5′ end of mRNA reads with +11 offset. For all of the transcript-level analysis, the most abundant transcript isoform was selected for each gene.
Differential Translation Analysis.
Uniquely mapped reads were mapped to RefSeq (v69) mouse coding sequences with RSEM to quantify the gene expression. Genes were filtered with a minimum of 10 average reads across all replicates, and then the read counts were batch corrected with sva. Batch-corrected counts were normalized with the trimmed mean of M values method and used to identify differentially expressed genes with anota2seq. GO analysis was performed with DAVID 6.8. Upstream regulators were inferred by IPA.
Proliferation and Differentiation Assays.
Proliferation and differentiation of aNSCs were analyzed as described (11). To study cell proliferation, we dissociated NSCs with trypsin and plated them on 24-well plates with poly-l-ornithine and laminin-coated coverslips at a density of 1 × 105 cells per well in proliferation medium. At 18 h postplating, BrdU was added to the culture medium for 6 h. aNSCs were then washed with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. Fixed cells were pretreated with 1 M HCl for 30 min at 37 °C and washed with borate buffer (pH 8.5) for 30 min, followed by the standard immunocytochemical protocol (12). For the differentiation assay, at 24 h postplating, cells were changed into differentiation medium for 4 d. Upon fixation with 4% paraformaldehyde, the coverslips were subjected to our standard immunohistochemistry protocol (12).
JC-10 Assay.
The mitochondrial membrane potential in WT and Fmr1 KO aNSCs was determined by JC-10 Mitochondrial Membrane Potential Assay Kit (Abcam) following the manufacturer’s protocol. Briefly, aNSCs were stained with JC-10 solution for 30 min at 37 °C. After adding buffer B, aNSCs were imaged using an A1RSi confocal microscope system (Nikon) with a 20× objective. At least 50 cells were randomly selected from each cell line, and the intensities of F520 and F590 were measured after subtracting background pixel intensity in the same image using ImageJ software (NIH). The average intensity from each cell line (at least 50 cells) was counted as n = 1 for statistical analysis. The mitochondrial membrane potential was assessed by quantifying the ratio between red fluorescence (590 nm) and green fluorescence (520 nm) intensities.
OCR Analysis.
OCR analysis was performed using the Seahorse XF Cell Mito Stress Test Kit and an Agilent Seahorse XF-24 Analyzer based on the protocol provided by the vendor. Briefly, we plated 1 × 104 aNSCs per well in a 96-well plate. At 24 h postplating, we changed the medium to the assay medium provided in the kit and preincubated cells for 1 h before the assay. The next steps were performed following the protocol provided in the kit without alteration. The data were generated and analyzed by the XF-24 Analyzer.
Extended experimental details are described in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Elisa Donnard and Manuel Garber (University of Massachusetts Medical School) and Zhisong He (Max Planck Institute for Evolutionary Anthropology) for assisting in the computational analysis. This work was supported by NIH Grants U54HD082013 and R01NS079415 (to J.D.R.); R01MH080434, R01MH078972, and R21NS095632 (to X.Z.); and U54HD090256 (to the Waisman Center, University of Wisconsin–Madison); the University of Wisconsin Vilas Trust (Kellett Mid-Career Award), the University of Wisconsin–Madison, and the Wisconsin Alumni Research Foundation (X.Z.); the Jenni and Kyle Professorship (X.Z.); the Charles H. Hood Foundation (J.D.R.); and Matching Funds of Tianjin University of Traditional Chinese Medicine Grant 60106/YL18020259 (to Y.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE112502).
See Commentary on page 12086.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1809588115/-/DCSupplemental.
References
- 1.Santoro MR, Bray SM, Warren ST. Molecular mechanisms of fragile X syndrome: A twenty-year perspective. Annu Rev Pathol. 2012;7:219–245. doi: 10.1146/annurev-pathol-011811-132457. [DOI] [PubMed] [Google Scholar]
- 2.Nelson DL, Orr HT, Warren ST. The unstable repeats–Three evolving faces of neurological disease. Neuron. 2013;77:825–843. doi: 10.1016/j.neuron.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frazier TW, et al. Molecular and phenotypic abnormalities in individuals with germline heterozygous PTEN mutations and autism. Mol Psychiatry. 2015;20:1132–1138. doi: 10.1038/mp.2014.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Auerbach BD, Osterweil EK, Bear MF. Mutations causing syndromic autism define an axis of synaptic pathophysiology. Nature. 2011;480:63–68. doi: 10.1038/nature10658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelleher RJ, 3rd, Bear MF. The autistic neuron: Troubled translation? Cell. 2008;135:401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 7.Udagawa T, et al. Genetic and acute CPEB1 depletion ameliorate fragile X pathophysiology. Nat Med. 2013;19:1473–1477. doi: 10.1038/nm.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Richter JD, Bassell GJ, Klann E. Dysregulation and restoration of translational homeostasis in fragile X syndrome. Nat Rev Neurosci. 2015;16:595–605. doi: 10.1038/nrn4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jessberger S, Gage FH. Adult neurogenesis: Bridging the gap between mice and humans. Trends Cell Biol. 2014;24:558–563. doi: 10.1016/j.tcb.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Guo W, et al. Ablation of Fmrp in adult neural stem cells disrupts hippocampus-dependent learning. Nat Med. 2011;17:559–565. doi: 10.1038/nm.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y, et al. MDM2 inhibition rescues neurogenic and cognitive deficits in a mouse model of fragile X syndrome. Sci Transl Med. 2016;8:336ra61. doi: 10.1126/scitranslmed.aad9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo Y, et al. Fragile x mental retardation protein regulates proliferation and differentiation of adult neural stem/progenitor cells. PLoS Genet. 2010;6:e1000898. doi: 10.1371/journal.pgen.1000898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Darnell JC, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ascano M, Jr, et al. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maurin T, et al. HITS-CLIP in various brain areas reveals new targets and new modalities of RNA binding by fragile X mental retardation protein. Nucleic Acids Res. 2018;46:6344–6355. doi: 10.1093/nar/gky267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo W, Patzlaff NE, Jobe EM, Zhao X. Isolation of multipotent neural stem or progenitor cells from both the dentate gyrus and subventricular zone of a single adult mouse. Nat Protoc. 2012;7:2005–2012. doi: 10.1038/nprot.2012.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016;165:22–33. doi: 10.1016/j.cell.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McManus CJ, May GE, Spealman P, Shteyman A. Ribosome profiling reveals post-transcriptional buffering of divergent gene expression in yeast. Genome Res. 2014;24:422–430. doi: 10.1101/gr.164996.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Larsson O, Sonenberg N, Nadon R. Identification of differential translation in genome wide studies. Proc Natl Acad Sci USA. 2010;107:21487–21492. doi: 10.1073/pnas.1006821107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zahr SK, et al. A translational repression complex in developing mammalian neural stem cells that regulates neuronal specification. Neuron. 2018;97:520–537.e6. doi: 10.1016/j.neuron.2017.12.045. [DOI] [PubMed] [Google Scholar]
- 22.Huang Z, Fujiwara K, Minamide R, Hasegawa K, Yoshikawa K. Necdin controls proliferation and apoptosis of embryonic neural stem cells in an oxygen tension-dependent manner. J Neurosci. 2013;33:10362–10373. doi: 10.1523/JNEUROSCI.5682-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson BR, Chopra P, Suhl JA, Warren ST, Bassell GJ. Identification of consensus binding sites clarifies FMRP binding determinants. Nucleic Acids Res. 2016;44:6649–6659. doi: 10.1093/nar/gkw593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S, et al. Global mapping of translation initiation sites in mammalian cells at single-nucleotide resolution. Proc Natl Acad Sci USA. 2012;109:E2424–E2432. doi: 10.1073/pnas.1207846109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Di Prisco GV, et al. Translational control of mGluR-dependent long-term depression and object-place learning by eIF2α. Nat Neurosci. 2014;17:1073–1082. doi: 10.1038/nn.3754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calvo SE, Clauser KR, Mootha VK. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weisz ED, et al. Loss of Drosophila FMRP leads to alterations in energy metabolism and mitochondrial function. Hum Mol Genet. 2018;27:95–106. doi: 10.1093/hmg/ddx387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomson SR, et al. Cell-type-specific translation profiling reveals a novel strategy for treating fragile X syndrome. Neuron. 2017;95:550–563.e5. doi: 10.1016/j.neuron.2017.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heiman M, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Christian KM, Song H, Ming GL. Functions and dysfunctions of adult hippocampal neurogenesis. Annu Rev Neurosci. 2014;37:243–262. doi: 10.1146/annurev-neuro-071013-014134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kempermann G, Song H, Gage FH. Neurogenesis in the adult hippocampus. Cold Spring Harb Perspect Biol. 2015;7:a018812. doi: 10.1101/cshperspect.a018812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–545. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo W, et al. Inhibition of GSK3β improves hippocampus-dependent learning and rescues neurogenesis in a mouse model of fragile X syndrome. Hum Mol Genet. 2012;21:681–691. doi: 10.1093/hmg/ddr501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, et al. Reducing histone acetylation rescues cognitive deficits in a mouse model of fragile X syndrome. Nat Commun. 2018;9:2494. doi: 10.1038/s41467-018-04869-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsieh AC, et al. The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conlon EG, Manley JL. RNA-binding proteins in neurodegeneration: Mechanisms in aggregate. Genes Dev. 2017;31:1509–1528. doi: 10.1101/gad.304055.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gennarino VA, et al. A mild PUM1 mutation is associated with adult-onset ataxia, whereas haploinsufficiency causes developmental delay and seizures. Cell. 2018;172:924–936.e11. doi: 10.1016/j.cell.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Korb E, et al. Excess translation of epigenetic regulators contributes to fragile X syndrome and is alleviated by Brd4 inhibition. Cell. 2017;170:1209–1223.e20. doi: 10.1016/j.cell.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jobe EM, et al. Methyl-CpG-binding protein MBD1 regulates neuronal lineage commitment through maintaining adult neural stem cell identity. J Neurosci. 2017;37:523–536. doi: 10.1523/JNEUROSCI.1075-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edupuganti RR, et al. N6-methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat Struct Mol Biol. 2017;24:870–878. doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edbauer D, et al. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron. 2010;65:373–384. doi: 10.1016/j.neuron.2010.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richter JD, Coller J. Pausing on polyribosomes: Make way for elongation in translational control. Cell. 2015;163:292–300. doi: 10.1016/j.cell.2015.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou J, et al. N6-methyladenosine guides mRNA alternative translation during integrated stress response. Mol Cell. 2018;69:636–647.e7. doi: 10.1016/j.molcel.2018.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu X, et al. Mitochondrial regulation in pluripotent stem cells. Cell Metab. 2013;18:325–332. doi: 10.1016/j.cmet.2013.06.005. [DOI] [PubMed] [Google Scholar]
- 46.Heyer EE, Ozadam H, Ricci EP, Cenik C, Moore MJ. An optimized kit-free method for making strand-specific deep sequencing libraries from RNA fragments. Nucleic Acids Res. 2015;43:e2. doi: 10.1093/nar/gku1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nicholls DG, et al. Bioenergetic profile experiment using C2C12 myoblast cells. J Vis Exp, 2010:2511. doi: 10.3791/2511. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.