Significance
The airborne microbiome is a topic of high interest in ecology, biogeochemistry, environmental and human health, among other fields. Large amounts of microorganisms are continuously exchanged among ecosystems and continents through the atmosphere, with unpredictable consequences for ecosystem services and biogeochemical cycling. Using wet bioaerosols collected fortnightly over a 7-y period at a mountaintop Long-Term Ecological Research Network site, we generated the most comprehensive long-term monitoring of the airborne microbiome reported to date. We found nonrandom recurrent interannual dynamics coupled to air circulation regimes, with a complex composition of original tracked sources. We unveil the factors influencing seasonal distributions with consistent evidence regarding the recurrence of microbes with a potential forensic signature. We highlight the need for a connected global sampling network for the airborne microbiome.
Keywords: LTER, aeroplankton, bioaerosols, intercontinental dispersal, microbial forensics
Abstract
Airborne microbes (bacteria, archaea, protists, and fungi) were surveyed over a 7-y period via high-throughput massive sequencing of 16S and 18S rRNA genes in rain and snow samples collected fortnightly at a high-elevation mountain Long-Term Ecological Research (LTER) Network site (LTER-Aigüestortes, Central Pyrenees, Spain). This survey constitutes the most comprehensive mountain-top aerobiology study reported to date. The air mass origins were tracked through modeled back-trajectories and analysis of rain water chemical composition. Consistent microbial seasonal patterns were observed with highly divergent summer and winter communities recurrent in time. Indicative microbial taxa were unveiled as a forensic signature, and ubiquitous taxa were observed as common atmosphere inhabitants, highlighting aerosols as a potentially successful mechanism for global microbial dispersal. Source-tracking analyses identified freshwater, cropland, and urban biomes as the most important sources for airborne bacteria in summer, while marine and forest biomes prevailed in winter, in agreement with air mass retrotrajectories and the prevailing general and regional atmospheric circulation.
Microorganisms from different sources (e.g., deserts, oceans, forests, agricultural and urban areas) are continuously injected into high atmospheric altitudes as aerosol particles, remaining suspended for days and being transported by wind over long distances before being washed out by precipitation (rain and snow) or dry deposition (1). The airborne microbes that successfully travel among continents are more diverse than previously suspected (2–5), including pathogens (6, 7). Hence, aerosols may represent a potentially widespread mechanism for global microbial dispersal. Since high amounts of microbes are injected into the atmosphere annually, and aerosol loads are potentially increasing due in part to the deforestation and transformation of Earth’s surface into deserts, the need to establish a global sampling network to monitor airborne microbes has been recently encouraged (8). The increase in global microbial mass movements may exacerbate changes in ecosystem services and biogeochemical cycling in several unpredictable ways (9).
The airborne microbiome is a very dynamic assemblage, highly influenced by the local environment. Aeroplankton assemblages collected at low-elevation sites are within the atmospheric boundary layer (turbulent air mixing) and show temporal changes at scales ranging from months (10–12) to a few hours (13), with the local influence modulated by sampling location and land use around the sampling site (14, 15). Spatial surveys of airborne microbe distribution shows geographic patterns in both terrestrial and marine environments (16, 17). However, substantial differences in the airborne biological composition are observed when the aeroplankton is collected directly from the troposphere (e.g., by aircraft) (18–20), although homogeneous distribution has recently been noted in the atmosphere up to 12 km (21). Interestingly, tropospheric microbial communities are much closer to those recovered from high-elevation sites, such as on-ground passive collectors located in high mountain areas, than to lowland sites. The matching is even greater for wet aerosols than for dry deposition, probably due to the cleansing of the atmosphere by rain and snow. Thus, although sampling the free troposphere may provide a more accurate view of the long-range transport of microbes while avoiding ground surface contamination (8, 19), on-ground sampling of washed-out aerosols in high-elevation mountains would be a cost-effective method of monitoring the long-term intercontinental exchange of the airborne microbiome.
In the present investigation, we evaluated the most comprehensive temporal monitoring of atmospheric microbes (bacteria, archaea, protists, and fungi) carried out to date, on rain and snow samples collected fortnightly in a high-elevation Long-Term Ecological Research (LTER) Network site in the Central Pyrenees (northeastern Spain) over a 7-y period. We applied high-throughput sequencing techniques on 16S and 18S rRNA gene amplicons to characterize the airborne microbiome present in wet depositions. Air mass origins were tracked after modeled back-trajectories and analysis of water chemical composition. The dataset is unique and improves our current understanding of aerial microbiome dynamics and the factors influencing the distribution of atmospheric microbes. Our study unveils consistent evidence regarding the recurrence of microbes with a forensic signature, and on the ubiquity of microbes and their ability to remain in the atmosphere.
Results
Air Mass Pathways and Chemical Characterization.
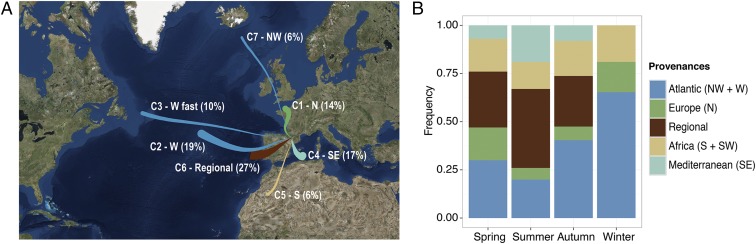
The general airflow pathways for all precipitation events, collected at the LTER-Aigüestortes (LTER-AT) sampling station, clustered into seven main air trajectories according to wind speed and direction (Fig. 1A). Predominant airflows regimes were Northern (cluster 1), Western (cluster 2 and cluster 3 related to fast-moving Western trajectories), Southeastern (cluster 4), South (cluster 5), Regional recirculation (cluster 6), and Northwestern (cluster 7). The Atlantic-influenced airflows (i.e., all Western transports), accounted for 35% of the tracked back-trajectories. Transports from the South contributed up to 23% and included air flows from both North Africa (Sahel) and Mediterranean regions. Air flows from continental Europe with Northern origin were the least frequent (14%).
Fig. 1.
(A) Cluster centroids and relative frequencies of back-trajectories associated with each cluster during the 2007–2013 survey period. (B) Frequency of clusters associated to each provenance by season. See also SI Appendix, Fig. S1.
The air retrotrajectories were further grouped into five main geographic provenances—Atlantic, Mediterranean, Africa, Europe, and Regional—that showed consistent seasonal patterns (Fig. 1B and SI Appendix, Fig. S1). According to tracking frequencies, autumn and winter precipitation were coming mostly from the Atlantic (40–64%), whereas summer rain was mostly related to Regional recirculation (41%), and spring showed both provenances equally. South precipitation (North Africa and Mediterranean) was more frequent in summer (33%).
After factor analysis, the chemical component present in the rain/snow samples dataset was combined into three main factors that explained 29.3%, 27.5%, and 18.5% of the total variance, respectively (SI Appendix, Table S1). Factor one was positively correlated with concentrations of NO3−, SO42−, NH4+, DOC, and Mg2+ (in order of relevance). We interpreted this factor as indicative of atmospheric pollution. Factor two showed very high values for pH, ANC, DIC, Ca2+, and Mg2+, thus representing an alkalinity-acidity axis indicating the influence in rainwater of solutes from carbonate-rich lithological areas. The high Cl− and Na+ loads of the third factor indicated marine influence. Interestingly, seasonal analysis showed that factor one predominated in spring and summer depositions, while factor two had the lowest values in winter (P < 0.001, ANOVA and Tukey’s HSD test; SI Appendix, Fig. S2). The third factor did not show any seasonal trend, as marine aerosols from different origins were common at all times of the year.
Interannual Structure and Composition of Airborne Microbes.
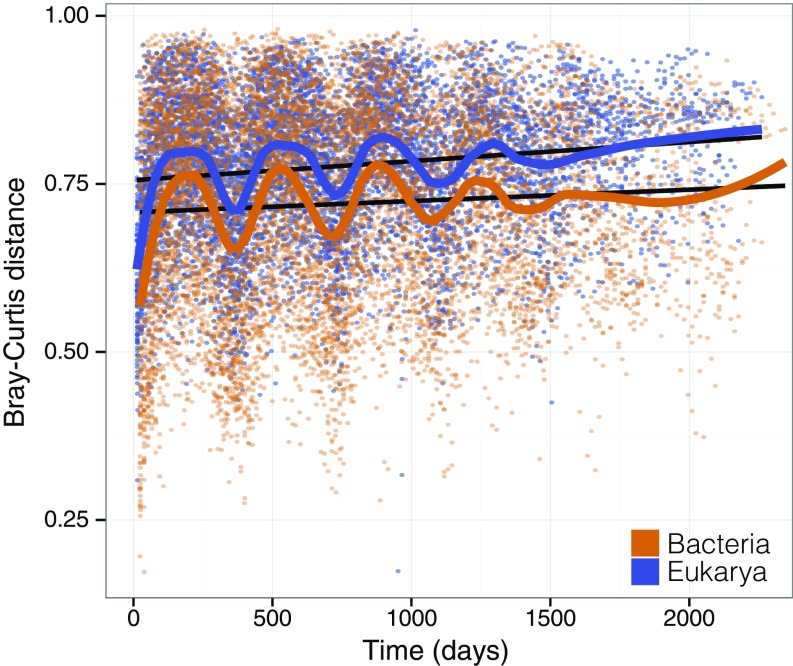
For the complete dataset, we observed a significant temporal decay pattern for eukaryotes but not for bacteria (Mantel test, rM = 0.14; P < 0.01; linear trend in black in Fig. 2). Overall, this temporal tendency indicated greater airborne assemblage dissimilarity for eukaryotes (mostly fungi) than for bacteria. In addition, similarities in the airborne microbiome of both bacteria and eukaryotes were strongly related to season (local polynomial trends in Fig. 2) irrespective of the year [R = 0.24 and 0.29 for bacteria and eukaryotes, respectively; P < 0.001, analysis of similarities (ANOSIM)]. Interestingly, more similar bacterial communities were observed during the summer compared with winter, and the opposite was found for eukaryotes (P < 0.001, multiple-comparison test after Kruskal–Wallis ANOVA; SI Appendix, Fig. S3).
Fig. 2.
Dissimilarity distances against time for the airborne microbiome components. Interannual trends (fitted linear regression, black lines) and seasonal trends (local polynomial regression, loess; orange, bacteria; blue, eukaryotes) are shown. A significant interannual trend (Mantel test, rM = 0.14; P < 0.01) was found for eukaryotes (mostly fungi), but not for bacteria.
Alpha diversity indices [Shannon, phylogenetic diversity (PD), and richness] of bacterial versus eukaryal communities were positively associated, with the highest correlation values found in winter (Pearson’s ρ = 0.71 and 0.75 for richness and PD, respectively; P < 0.001; SI Appendix, Fig. S4C). Airborne bacterial communities showed a consistent seasonal trend, with the highest diversity indices found in winter (P < 0.001, Kruskal–Wallis test; SI Appendix, Fig. S4A). Eukaryotes also showed greater Shannon diversity in autumn and winter than in summer (P < 0.001, Kruskal–Wallis test), although richness did not change seasonally, and phylogenetic distances among airborne protists and fungi were slightly lower in winter (P < 0.05, Kruskal–Wallis test; SI Appendix, Fig. S4B).
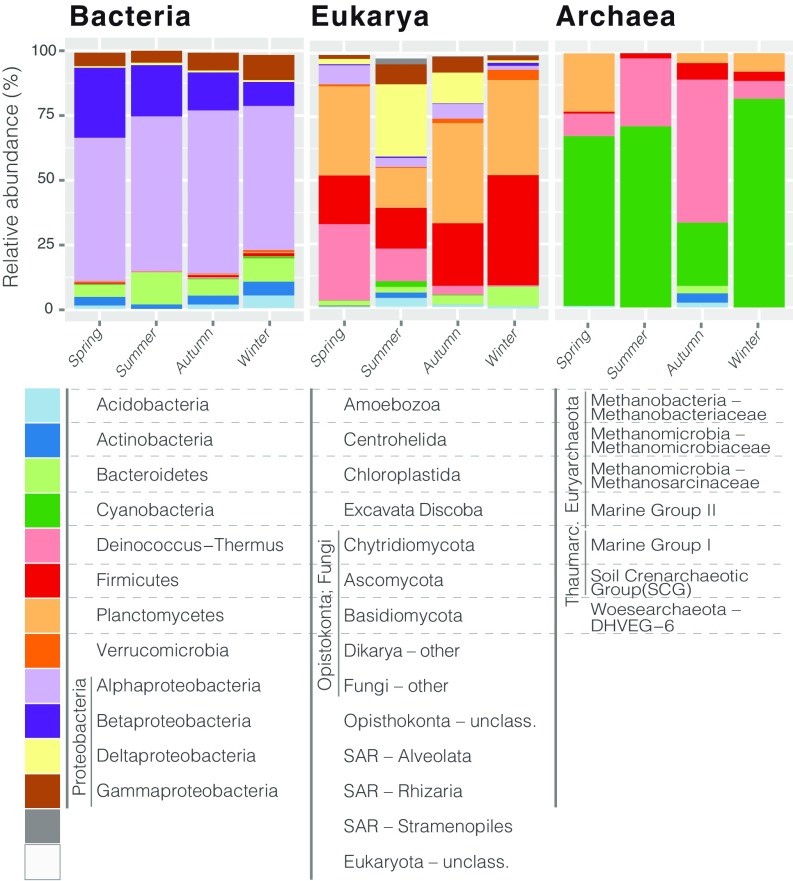
The taxonomic composition analysis showed Alphaproteobacteria in >58% of the total bacterial sequence reads, followed by Betaproteobacteria (18%, ranging from 27% in spring to 9% in winter), Bacteroidetes (8%), and Gammaproteobacteria (6%) (Fig. 3). Overall, we identified approximately 1,200 airborne bacterial genera, although only 12 of them were widespread in almost all samples (>90%). Some of these ubiquitous genera were found to be rather abundant and were putatively identified as the core airborne bacteria in the dataset; these included Sphingomonas, Methylobacterium, Massilia, Pseudomonas, Polaromonas, Acidiphilium, Ramlibacter, Mucilaginibacer, Hymenobacter, and Noviherbaspirillum, among others (SI Appendix, Fig. S5).
Fig. 3.
Relative seasonal abundance of the most abundant (>1% relative abundance) microbiome components.
Fungi averaged >75% of the eukaryote reads (Fig. 3). Basidiomycota and Ascomycota were the most frequently detected (average values of 32% and 26%, respectively), whereas airborne Chytridiomycota reached 30% of the sequence pool in spring. Within protists, Alveolata and Rhizaria were found in higher proportions in summer (28.4% and 7.8%, respectively). Overall, most of the microbial eukaryotes were found in only ∼10% of samples, and ∼7% were ubiquitous microbes (detected in >90% of samples), mostly Fungi but also Colpodea (Ciliophora, Alveolata) and Cercozoa (Rhizaria) (SI Appendix, Fig. S5).
Archaea was detected in approximately 45% of samples, predominantly in autumn, winter, and spring. The average relative abundance per sample reached only 0.3% among prokaryotes, with a maximum of 4% in winter samples. Marine Group II (Euryarchaeota) was the most frequently detected in winter, spring, and summer (average value of 75%), whereas Marine Group I (AOA Thaumarchaeota) reached 55% of the archaeal sequence pool in autumn (Fig. 3).
Seasonal Aeroplankton Variability and Indicator Taxa.
Unexpectedly, consistent seasonal microbial signals were observed in the interannual aeroplankton dynamics. Burkholderiales and Sphingomonadales reached higher relative abundances in spring, whereas Rhodospirillales and Acidobacteriales, among others repeatedly peaked in winter, Cytophagales and Bdellovibrionales repeatedly peaked in summer, and Rhizobiales repeatedly peaked in autumn (SI Appendix, Fig. S6A). The same signals accounted for airborne microbial eukaryotes. Chytridiomycota consistently peaked in spring, and Ascomycota and Basidiomycota consistently peaked in fall or winter, with Tremellales also peaking in spring (SI Appendix, Fig. S6B). Trebouxiophyceae (Chlorophyta) were also observed at greater abundance in winter. Colpodea (Ciliophora) and Gracilipodida (Amoebozoa) were significantly more abundant in summer.
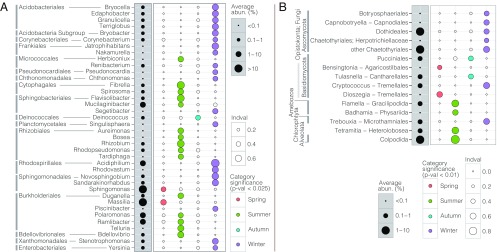
Indicator value (IndVal) analyses unveiled consistent seasonal microbial genera indicators with abundances and recurrences unevenly distributed over time. Thus, Sphingomonas (Sphingomonadales) and Massilia (Burkholderiales) consistently showed higher signals for spring, Polaromonas and Ramlibacter (Burkholderiales) and Mucilaginibacter (Sphingobacteriales) showed higher signals for summer, and Acidiphilium (Rhodospirillales) showed higher signals for winter (Fig. 4A). Interestingly, all the Ascomycota showed higher signals for winter, and different Basidiomycota showed higher signals for spring, autumn, or winter (Fig. 4B). Conversely, several protist groups with potential for cyst formation (e.g., Gracilipodida, Physariida, Heterolobosea, Colpodida), showed higher IndVal indices in summer, and Microthamniales (Trebouxia) had higher signals in winter.
Fig. 4.
Bacterial (A) and eukaryal (B) indicator taxa (mostly at the genus level) from bioaerosols among seasons. See also SI Appendix, Fig. S6.
Aeroplankton Novelty.
Most of the airborne operational taxonomic units (OTUs) showed a high percentage of identity to already-described microbial sequences. Up to 78% of the bacterial OTUs were 97% identical to previous sequences, and 49% had >99% identity, suggesting high cosmopolitanism. For eukaryotes, 52% were 97% identical, and 11% had >99% identity. No difference in bacterial novelty among seasons was detected (P > 0.05, Kruskal–Wallis test). Conversely, microbial eukaryotes showed significantly lower mean identities in summer (P < 0.001, ANOVA with Tukey’s post hoc test). Overall, we observed the lowest novelty to be positively correlated with occurrence, that is, the higher the frequency, the lower the novelty (P < 0.001, Spearman correlation test; SI Appendix, Fig. S7).
Environmental Predicted Source for Airborne Microbes.
The most frequent sources of the Environmental Ontology (ENVO) terms matched soil, sediment, lake, sea, biofilm, rhizosphere, organ, watercourse, and forest. Some of these sources showed varying predominance according to season (P < 0.01, multiple-comparison test after Kruskal-Wallis ANOVA; SI Appendix, Fig. S8A). In summer, airborne bacteria were more closely related to aquatic, cropland, and urban biomes. Conversely, in winter the predominant bacteria were more closely related to terrestrial, forest, and marine biomes. Bacteria from the Mediterranean Sea were significantly more abundant in summer than in spring, while desert and mangrove biomes showed the lowest prevalence in spring. Significant seasonal differences were found for microbial eukaryotes from freshwater, which were more abundant in summer, while forest-related biomes predominated in fall (SI Appendix, Fig. S8B). Marine sources did not show seasonal preferences.
Discussion
Air Mass Origin Drives the Seasonality of Aeroplankton.
Our long-term investigation consistently showed evidence of recurrent interannual dynamics of the airborne microbiome coupled to general and regional atmospheric circulation. Aeroplankton communities tended to more closely resemble one another in the same seasons annually. Aeroplankton communities in summer and winter were the most strongly differentiated. This interannual variability was also found in the chemical composition of wet depositions (rain and snow) and provenance of the air masses. The frequency and provenance of the air masses reported here were in agreement with the general precipitation regimes in the Mediterranean area (22). The signal of terrestrial aerosols in summer was also supported by the Saharan dust intrusions to the Pyrenees region, which had the greatest frequency during late spring and summer (May–September) for the period 2007–2013 (SI Appendix, Fig. S9). This highly segregated temporal pattern shows that the long-range transport of aeroplankton could influence ground surface environments in a highly predictable manner. A comprehensive understanding of the processes that control the variability of aeroplankton communities is certainly needed to foresee microbial colonization and their associated potential risks for health, ecosystem services, and biogeochemical cycling (9).
Predicted ENVO sources showed major differences between summer and winter, in agreement with the general provenance of air masses. Bacteria from freshwater biomes, as well as cropland and urban biomes, dominated the airborne communities in summer in which aerosols of regional origin also prevailed. The marine source dominated in winter, in agreement with the Atlantic origin of most aerosols. Bacteria from the forest biome, being more abundant in winter, would be indicative of the presence of bacteria commonly found in the leaf surface environment. However, the abundance of phyllosphere bacteria would be expected during spring to summer months, when leaf biomass is higher (23). This unexpected result may suggest a long-distance intercontinental phyllosphere bacteria transport, probably from the North American Taiga. Curiously, seasonal differences in the predicted environmental sources of eukaryotes were much less marked than those for bacteria. Only eukaryotes from freshwater biomes dominated in summer, in agreement with bacterial counterparts. Overall, fewer biomes were associated with eukaryotes compared with bacteria. However, this result was likely biased by underrepresentation of eukaryotes in public databases. Despite procedural limitations, we could associate the main biomes of the ENVO to airborne bacteria, revealing the atmosphere as a heterogeneous environment that combines microbes from widely distributed sources. Consequently, air could constitute a potential habitat for generalists capable of dealing with a wide range of environmental conditions (24).
The chemistry of precipitation and the origin of aerosols, together with the potential sources of microbes, explain most of the temporal variability observed among aeroplankton communities. All support the origin of air particles as the main driver of change for the long-range transported airborne microbiome. Therefore, changes in air mass circulation regimes will directly affect the expected dispersal of microorganisms. In fact, general transport regimes compared for two 10-y consecutive periods within the years 1984–2009 in the study area reported decreases in Atlantic advections and increases in African and European air flow (22). This observation suggests changes in the dispersal potential and fate of aeroplankton that can occur in a relatively short period (10 y).
Role of Environmental Factors in Filtering Airborne Microbial Communities.
The sources of aerosols are thought to drive the composition of airborne microbes, but environmental factors and stress adaptation can also shape the communities (12). Diversity measures derived from our dataset may support this idea. Airborne bacterial communities showed lower ecological diversity and were generally more similar to one another in summer than in the other seasons. This is likely due to the higher radiation and dryness levels, which could act as environmental filters by eroding nonresistant microbes during atmospheric long-range transport (4). We could not determine whether such selective pressures shaped the lower diversity values for the eukaryal communities in summer. However, these communities would largely correspond to the dispersion of spores and cysts, and thus rather high tolerance to stress could be argued, although the viability of encysting structures has been a matter of debate (25). In addition, we observed that the proportions of dominant fungal airborne communities decreased considerably in summer. The seasonal variability of fungal dispersal has been associated with relative humidity and rain frequencies (11). In fact, some groups of Ascomycota and Basidiomycota (including some Agaricomycetes) have been observed to actively discharge their spores under humid conditions (26). Therefore, the greater dryness that characterizes the Mediterranean summers could lead to a decline in fungal dispersal. In contrast, the proportions of some varieties of high-ranking eukaryal groups increased in summer.
The Atmospheric Environment as a Global Habitat and Dispersal Medium for Microbes.
Abundant taxa were recurrently detected in the long-term monitoring of aeroplankton (i.e., potential core species). They were considered consistently ubiquitous taxa that thus would commonly inhabit the atmosphere. The temporal variability observed in the origin of aerosols supports the idea that some aerial microbes may remain metabolically active (27). Some of these taxa had already been isolated in the air (28, 29). Methylobacterium and Sphingomonas include well-known and metabolically versatile microbes able to use C1–C4 carbon compounds usually present in the atmosphere. Species affiliated to Hymenobacter and Ramlibacter are adapted to desiccation conditions and high levels of radiation. Such traits could help these recurrent airborne bacteria survive and outcompete other bacteria (18). Conversely, the picture may be substantially different for recurrent eukaryotes (mostly fungi), which are airborne only in a dispersal phase (i.e., spores, cysts), making their recurrence closely dependent on the frequency of dispersion rather than on their ability to develop in the atmosphere. The enrichment in wet deposition could be related to the fact that fungi and fungal spores may act as cloud condensation nuclei and/or are actively discharged during precipitation (30).
Interestingly, ubiquitous airborne Methylobacteriaceae and Oxalobacteraceae found in the Mediterranean area also have been identified as core families of the upper troposphere in the United States (18). They also are commonly found in ground surface and indoor environments on different continents (e.g., refs. 16 and 31). These observations support the recurrent long-range dispersal of microbes, as well as their capacity to globally colonize and influence distant environments. The low novelty of airborne microbes reported in our study also supports a cosmopolitan distribution. Most ubiquitous microbes were highly related to known species, in agreement with the dominance of typical culturable airborne bacteria (24). Even through that the atmosphere may be one of the microbiological frontiers yet to be explored (8), we believe that it is more a global-scale interconnector of environments than a redoubt of highly novel microbes.
Seasonal Predictable Taxa and Forensic Signatures.
Indicator taxa were observed mostly in summer and winter, as would be expected for the two most extreme seasons, a result that was highly consistent over the 7-y study. Thus, these taxa are valuable as microbial forensic signals, an area recently highlighted as an international research priority (32). Members affiliated with known Acidobacteriales and Acidiphilum, with affinity for acidic conditions, were taxa indicators in winter according to the acidity of depositions for this period. Interestingly, Ramlibacter was one of the most abundant and recurrent taxa in summer, specifically related to a single OTU with 99.6% identity to the type strain Ramlibacter tataouinensis TTB310. This betaproteobacterium was first isolated from meteorite fragments buried in the sands of a desert near Tataouine, Tunisia, showing a unique cell cycle specifically adapted for living in hot, dry desert environments (33). Given that summer is the season with both more precipitation of southern origin and greater frequency of Saharan dust intrusions, we suggest the use of this taxon as a signature of African dust incoming. For the eukaryal counterparts, one of the most abundant and recurrent taxa in summer was related to Colpodida, an Alveolata with encysting capacity that dominates in arid and semiarid soils (34). Conversely, saprotrophic fungi and biotrophic plant pathogens were mostly enriched in wet depositions from autumn to spring. However, the relative abundance of Dothideomycetes (Botryosphaeriales, Capnodiales, and Dothideales) should be interpreted with caution because of the multicellularity of their spores (30).
Interestingly, some indicator taxa—mostly within the low mean abundance range—were closely related to animal pathogens. This represents an opportunity to predict the temporal dynamics of airborne pathogens and merits more in-depth study. This is the case for Corynebacterium, Stenotrophomonas, and Yersinia (potential obligate human pathogens); Renibacterium (a potential fish pathogen) preferentially found in winter; and the fungi Cryptococcus, a known opportunistic pathogen (35) that is also recurrent in winter. Interestingly, the highest signals of potential pathogenic microbes were recurrently found in winter.
Concluding Remarks.
Overall, our present study shows that the composition of the long-range airborne microbiome is closely related to the origin of air masses and thus to the source of aerosols. This was driven by the general and regional air mass regimes in the Mediterranean area, although a similar pattern could be acting globally in accordance with the global recurrent movement of air masses. The air mass regimes are predictable within a defined climatologic framework, and thus in a global climate change scenario, changes in air mass circulation will directly affect the expected dispersal of the airborne microbiome, with unforeseeable consequences. Because air mass regimes can experience substantial changes in 10 y (22), the monitoring of airborne microorganisms is warranted (as with chemical air quality standards), given the potential biosphere risks. We highlight the need for a high-frequency and connected global sampling network strategy for the airborne microbiome.
Methods
Bioaerosol Collection.
The sampling site was located at ∼1,800 MASL on a rocky landscape within the protected area of the Aigüestortes i Estany de Sant Maurici National Park (42°33′N 0°53′E; Pyrenees, northeastern Spain) and next to an automatic weather station of the LTER-AT node site (parcsnaturals.gencat.cat/en/aiguestortes/coneixeu-nos/ambits_o_linies_de_treball/node_lter/). The ground surrounding the station is snow-covered for several months of the year. Composite samples of atmospheric precipitation (both rain and snow) were collected twice monthly over 7 y, from May 15, 2007, to October 15, 2013. Approximately 150 samples were collected, as detailed in SI Appendix. DNA was extracted using the Mobio PowerSoil DNA Isolation Kit (Mobio Laboratories). PCR and high-speed multiplexed SSU rRNA gene Illumina MiSeq sequencing were carried out for 16S and 18S rRNA genes (regions and primers listed in SI Appendix) following the methods of the Research Technology Support Facility at Michigan State University (https://rtsf.natsci.msu.edu/). The chemical composition of water samples was determined as detailed in SI Appendix.
Back-Trajectory Analysis.
The air mass origins were tracked using the National Oceanic and Atmospheric Administration HYSPLIT trajectory model and Global Data Assimilation System meteorological data (36). For each precipitation event, the 72-h backward trajectory at 3,000 MASL was computed. Each backward trajectory consisted of 12 endpoints corresponding to the air mass location at 6-h intervals and characterized by geographic coordinates (longitude, x; latitude, y; and altitude, z) (37). All trajectories were grouped per season using the clustering module implemented in HYSPLIT 4.0.
Sequence Processing.
Raw rRNA gene sequences were processed using the UPARSE pipeline (SI Appendix). OTUs (97% identity) were taxonomically assigned with SILVA_119. A total of 3,630 prokaryote and 5,814 eukaryote OTUs were compiled. Genetic novelty and ENVO terms (SI Appendix) were determined after BLASTn closest-match analyses against the GenBank database (search conducted in November 2015). The entire genetic dataset is available in the National Center for Biotechnology Information Sequence Read Archive under accession no. PRJEB14358.
Statistical Analyses.
Factor analysis using Varimax rotation (SI Appendix) was carried out using the S-Plus software package (Insightful) on chemical data. Community ecology parameters were analyzed with the R vegan and ggplot2 (38) packages (www.r-project.org). Distance-based community analyses were calculated with Bray–Curtis dissimilarities after Hellinger standardization (39). Correlations between community similarity patterns were calculated by Mantel tests with the Spearman method. ANOSIM was based on 1,000 permutations to test for significant differences (40). The multivariate homogeneity of group dispersions (variances) was previously checked to rule out heteroscedasticity among groups (permutest.betadisper function) (41).
The Quantitative Insights Into Microbial Ecology (QIIME) toolkit (42) was used for rarefied alpha-diversity metrics, including OTU richness and Shannon and Faith PD indices. The IndVal index (R labdsv package) was used to identify microbial taxa as “indicator species.” The index combines relative abundance (tied to the concept of specificity) and relative frequency of occurrence (i.e., fidelity) (43). Hypothesis contrast tests were carried out with the stats package.
Supplementary Material
Acknowledgments
We thank the authorities of the Aigüestortes and Estany de Sant Maurici National Park for sampling facilities in protected areas and support, members of the High Mountain Research Center, University of Barcelona for sampling and access to laboratory and logistical facilities, and X. Torras for help with Fig. 1. This research is included within the global surveillance activities of the Long-Term Ecological Research node Aigüestortes (LTER-AT) and was supported by Grant ECOSENSOR-BIOCON 04/009 from BBVA Foundation; Grants AERBAC 079/2007, AERBAC-2 178/2010, and DISPERSAL 829/2013 from the Spanish Office for the Environment’s National Parks Research Network (OAPN; to E.O.C.); and the Catalan Government’s Support Program for Consolidated Research Groups Grant GECA 2017 SGR 910.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. PRJEB14358).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812826115/-/DCSupplemental.
References
- 1.Burrows SM, Elbert W, Lawrence MG, Pöschl U. Bacteria in the global atmosphere, part 1: Review and synthesis of literature data for different ecosystems. Atmos Chem Phys. 2009;9:9263–9280. [Google Scholar]
- 2.Griffin DW. Atmospheric movement of microorganisms in clouds of desert dust and implications for human health. Clin Microbiol Rev. 2007;20:459–477. doi: 10.1128/CMR.00039-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hervàs A, Camarero L, Reche I, Casamayor EO. Viability and potential for immigration of airborne bacteria from Africa that reach high mountain lakes in Europe. Environ Microbiol. 2009;11:1612–1623. doi: 10.1111/j.1462-2920.2009.01926.x. [DOI] [PubMed] [Google Scholar]
- 4.Smith DJ, Griffin DW, McPeters RD, Ward PD, Schuerger AC. Microbial survival in the stratosphere and implications for global dispersal. Aerobiologia. 2011;27:319–332. [Google Scholar]
- 5.Smith DJ, et al. Intercontinental dispersal of bacteria and archaea by transpacific winds. Appl Environ Microbiol. 2013;79:1134–1139. doi: 10.1128/AEM.03029-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown JKM, Hovmøller MS. Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science. 2002;297:537–541. doi: 10.1126/science.1072678. [DOI] [PubMed] [Google Scholar]
- 7.Polymenakou PN. Atmosphere: A source of pathogenic or beneficial microbes? Atmosphere. 2012;3:87–102. [Google Scholar]
- 8.Smith DJ. Aeroplankton and the need for a global monitoring network. Bioscience. 2013;63:515–516. [Google Scholar]
- 9.Zhu Y-G, et al. Microbial mass movements. Science. 2017;357:1099–1100. doi: 10.1126/science.aao3007. [DOI] [PubMed] [Google Scholar]
- 10.Bertolini V, et al. Temporal variability and effect of environmental variables on airborne bacterial communities in an urban area of Northern Italy. Appl Microbiol Biotechnol. 2013;97:6561–6570. doi: 10.1007/s00253-012-4450-0. [DOI] [PubMed] [Google Scholar]
- 11.Bowers RM, et al. Seasonal variability in bacterial and fungal diversity of the near-surface atmosphere. Environ Sci Technol. 2013;47:12097–12106. doi: 10.1021/es402970s. [DOI] [PubMed] [Google Scholar]
- 12.Franzetti A, Gandolfi I, Gaspari E, Ambrosini R, Bestetti G. Seasonal variability of bacteria in fine and coarse urban air particulate matter. Appl Microbiol Biotechnol. 2011;90:745–753. doi: 10.1007/s00253-010-3048-7. [DOI] [PubMed] [Google Scholar]
- 13.Fierer N, et al. Short-term temporal variability in airborne bacterial and fungal populations. Appl Environ Microbiol. 2008;74:200–207. doi: 10.1128/AEM.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bowers RM, McLetchie S, Knight R, Fierer N. Spatial variability in airborne bacterial communities across land-use types and their relationship to the bacterial communities of potential source environments. ISME J. 2011;5:601–612. doi: 10.1038/ismej.2010.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowers RM, et al. Sources of bacteria in outdoor air across cities in the midwestern United States. Appl Environ Microbiol. 2011;77:6350–6356. doi: 10.1128/AEM.05498-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barberán A, et al. Continental-scale distributions of dust-associated bacteria and fungi. Proc Natl Acad Sci USA. 2015;112:5756–5761. doi: 10.1073/pnas.1420815112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayol E, et al. Long-range transport of airborne microbes over the global tropical and subtropical ocean. Nat Commun. 2017;8:201. doi: 10.1038/s41467-017-00110-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DeLeon-Rodriguez N, et al. Microbiome of the upper troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc Natl Acad Sci USA. 2013;110:2575–2580. doi: 10.1073/pnas.1212089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maki T, et al. Variations in airborne bacterial communities at high altitudes over the Noto Peninsula (Japan) in response to Asian dust events. Atmos Chem Phys. 2017;17:11877–11897. [Google Scholar]
- 20.Zweifel UL, et al. High bacterial 16S rRNA gene diversity above the atmospheric boundary layer. Aerobiologia. 2012;28:481–498. [Google Scholar]
- 21.Smith DJ, et al. Airborne bacteria in Earth’s lower stratosphere resemble taxa detected in the troposphere: Results from a new NASA Aircraft Bioaerosol Collector (ABC) Front Microbiol. 2018;9:1752. doi: 10.3389/fmicb.2018.01752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izquierdo R, Avila A, Alarcón M. Trajectory statistical analysis of atmospheric transport patterns and trends in precipitation chemistry of a rural site in NE Spain in 1984–2009. Atmos Environ. 2012;61:400–408. [Google Scholar]
- 23.Bowers RM, McCubbin IB, Hallar AG, Fierer N. Seasonal variability in airborne bacterial communities at a high-elevation site. Atmos Environ. 2012;50:41–49. [Google Scholar]
- 24.Fahlgren C, Hagström A, Nilsson D, Zweifel UL. Annual variations in the diversity, viability, and origin of airborne bacteria. Appl Environ Microbiol. 2010;76:3015–3025. doi: 10.1128/AEM.02092-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Foissner W. Biogeography and dispersal of micro-organisms: A review emphasizing protists. Acta Protozool. 2006;45:111–136. [Google Scholar]
- 26.Elbert W, Taylor PE, Andreae MO, Pöschl U. Contribution of fungi to primary biogenic aerosols in the atmosphere: Wet and dry discharged spores, carbohydrates, and inorganic ions. Atmos Chem Phys. 2007;7:4569–4588. [Google Scholar]
- 27.Womack AM, Bohannan BJM, Green JL. Biodiversity and biogeography of the atmosphere. Philos Trans R Soc Lond B Biol Sci. 2010;365:3645–3653. doi: 10.1098/rstb.2010.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buczolits S, et al. Classification of three airborne bacteria and proposal of Hymenobacter aerophilus sp. nov. Int J Syst Evol Microbiol. 2002;52:445–456. doi: 10.1099/00207713-52-2-445. [DOI] [PubMed] [Google Scholar]
- 29.Weon H-Y, et al. Methylobacterium iners sp. nov. and Methylobacterium aerolatum sp. nov., isolated from air samples in Korea. Int J Syst Evol Microbiol. 2008;58:93–96. doi: 10.1099/ijs.0.65047-0. [DOI] [PubMed] [Google Scholar]
- 30.Woo C, An C, Xu S, Yi S-M, Yamamoto N. Taxonomic diversity of fungi deposited from the atmosphere. ISME J. 2018;12:2051–2060. doi: 10.1038/s41396-018-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Triadó-Margarit X, et al. Bioaerosols in the Barcelona subway system. Indoor Air. 2017;27:564–575. doi: 10.1111/ina.12343. [DOI] [PubMed] [Google Scholar]
- 32.Council NR. Science Needs for Microbial Forensics: Developing Initial International Research Priorities. National Academies Press; Washington, DC: 2014. p. 252. [PubMed] [Google Scholar]
- 33.De Luca G, et al. The cyst-dividing bacterium Ramlibacter tataouinensis TTB310 genome reveals a well-stocked toolbox for adaptation to a desert environment. PLoS One. 2011;6:e23784. doi: 10.1371/journal.pone.0023784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates ST, et al. Global biogeography of highly diverse protistan communities in soil. ISME J. 2013;7:652–659. doi: 10.1038/ismej.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Centers for Disease Control and Prevention 2015 Types of fungal diseases. Available at https://www.cdc.gov/fungal/diseases/index.html. Accessed October 15, 2018.
- 36.Stein AF, et al. NOAA’s HYSPLIT atmospheric transport and dispersion modeling system. Bull Am Meteorol Soc. 2015;96:2059–2077. [Google Scholar]
- 37.Bacardit M, Camarero L. 2009. Fluxes of Al, Fe, Ti, Mn, Pb, Cd, Zn, Ni, Cu, and as in monthly bulk deposition over the Pyrenees (SW Europe): The influence of meteorology on the atmospheric component of trace element cycles and its implications for high mountain lakes. J Geophys Res Biogeosci, doi.org/10.1029/2008JG000732.
- 38.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 39.Legendre P, Gallagher ED. Ecologically meaningful transformations for ordination of species data. Oecologia. 2001;129:271–280. doi: 10.1007/s004420100716. [DOI] [PubMed] [Google Scholar]
- 40.Clarke KR, Warwick RM. Similarity-based testing for community pattern: The two-way layout with no replication. Mar Biol. 1994;118:167–176. [Google Scholar]
- 41.Ramette A. Multivariate analyses in microbial ecology. FEMS Microbiol Ecol. 2007;62:142–160. doi: 10.1111/j.1574-6941.2007.00375.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dufrêne M, Legendre P. Species assemblages and indicator species: The need for a flexible asymmetrical approach. Ecol Monogr. 1997;67:345–366. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.