Significance
Cultivation-independent sequencing has revealed enormous numbers of bacterial species. Amongst the most enigmatic are members of the Candidate Phyla Radiation, which are estimated to represent over 26% of all bacterial diversity. This group of ultrasmall bacteria are thought to be mostly symbionts; however, understanding these bacteria is hampered by an inability to culture them. Here, we study TM7x, the only strain of the Candidate Phyla Radiation successfully cultured, and demonstrate that it can be virulent and lethal for its bacterial host. The host, however, can rapidly evolve to a state of reduced susceptibility, which results in a long-term parasitic relationship in which both species persist. These findings provide a rare glimpse into the lifestyle and symbiosis of Candidate Phyla Radiation organisms.
Keywords: Saccharibacteria, TM7, oral microbiome, interspecies interaction, candidate phyla radiation
Abstract
Around one-quarter of bacterial diversity comprises a single radiation with reduced genomes, known collectively as the Candidate Phyla Radiation. Recently, we coisolated TM7x, an ultrasmall strain of the Candidate Phyla Radiation phylum Saccharibacteria, with its bacterial host Actinomyces odontolyticus strain XH001 from human oral cavity and stably maintained as a coculture. Our current work demonstrates that within the coculture, TM7x cells establish a long-term parasitic association with host cells by infecting only a subset of the population, which stay viable yet exhibit severely inhibited cell division. In contrast, exposure of a naïve A. odontolyticus isolate, XH001n, to TM7x cells leads to high numbers of TM7x cells binding to each host cell, massive host cell death, and a host population crash. However, further passaging reveals that XH001n becomes less susceptible to TM7x over time and enters a long-term stable relationship similar to that of XH001. We show that this reduced susceptibility is driven by rapid host evolution that, in contrast to many forms of phage resistance, offers only partial protection. The result is a stalemate where infected hosts cannot shed their parasites; nevertheless, parasite load is sufficiently low that the host population persists. Finally, we show that TM7x can infect and form stable long-term relationships with other species in a single clade of Actinomyces, displaying a narrow host range. This system serves as a model to understand how parasitic bacteria with reduced genomes such as those of the Candidate Phyla Radiation have persisted with their hosts and ultimately expanded in their diversity.
The recently discovered Candidate Phyla Radiation comprises more than 26% of bacterial diversity and potentially contains 73 new phyla (1). The members of Candidate Phyla Radiation appear to share unusual characteristics, including small cell size, reduced genomes, and restricted metabolic capacities (2, 3). Strikingly, their genomes reveal a lack of citric acid cycles, respiratory chains, and genes required for amino acid and nucleotide synthesis. Detailed understanding of their biology has been hampered by their recalcitrance to in vitro cultivation.
As the first and only cultivated representative of Candidate Phyla Radiation to date, “Nanosynbacter lyticus” Type-Strain TM7x HMT-952 (4) from the Saccharibacteria (TM7) phylum was coisolated as an obligate epibiotic parasite with its bacterial host Actinomyces odontolyticus, strain XH001, from the human oral cavity (5–7). Specifically, TM7x is found on the outside of host cells and has a confirmed ultrasmall cell size (200–500 nm) along with its very small genome (705 kb and ∼700 genes), which is missing multiple genes encoding essential cellular functions. The observation that TM7x lives on the surface of other bacterial cells, as well as more recent evidence that members of the OD1 phylum can live intracellularly (8), has led to the hypothesis that most of the Candidate Phyla Radiation organisms have evolved a symbiotic lifestyle with other organisms. The coculture of TM7x and XH001 therefore represents a powerful model system to better understand the Candidate Phyla Radiation.
Bacteria belonging to Saccharibacteria are commonly found in many environments (9, 10), including the human microbiome (11–14), as revealed by 16S rRNA gene sequencing and metagenomics. Indeed, there is increasing evidence that Saccharibacteria are associated with multiple mucosal diseases such as inflammatory bowel disease and periodontitis (15–18). For example, the relative abundance of several Saccharibacteria species (spp.) are increased in patients with periodontitis while Actinomyces spp., a highly prevalent member of supragingival and subgingival plaque, is decreased (17). The observed tight physical association between cells of TM7x and XH001 bacteria could potentially impact both organisms’ physiology, their ecological function in oral microbiome, as well as their role in oral health and diseases (19).
Previous observations suggested that strain TM7x is parasitic in nature and that there is a general stress response to TM7x in the host (5, 6). However, the detailed parasitic mechanism on its host and the dynamics of the relationship remain poorly understood. The most comparable ecology to TM7x among well-studied bacteria is the predatory lifestyle of Micavibrio and Vampirovibrio, which sit on the outside of their bacterial hosts, leach cytoplasmic content, and rapidly kill them (20–23). In our experiment, we studied N. lyticus strain TM7x with its host A. odontolyticus strain XH001 (which we will subsequently refer to as TM7x/XH001) in coculture to ask whether N. lyticus has a comparably negative effect on its host. Applying various assays tailored toward Saccharibacteria, we carefully monitored the growth dynamics and host physiology changes within the TM7x/XH001 coculture.
Results
The Coculture of TM7x and XH001 Maintains a Reservoir of Uninfected XH001 Cells.
The coculture of TM7x and XH001 (TM7x/XH001) provides an opportunity to study a live member of the Saccharibacteria (TM7). The original cultivation of these bacteria from the human oral cavity (5) required a number of passages of both species in coculture, which raises the possibility that de novo evolution in the laboratory affects the relationship between the two species. We explore and discuss this possibility further below, but the ability to perform a detailed study of the effects of a live Saccharibacteria strain on a host is unprecedented. We therefore performed a detailed microscopic study of the coculture dynamics at the population level.
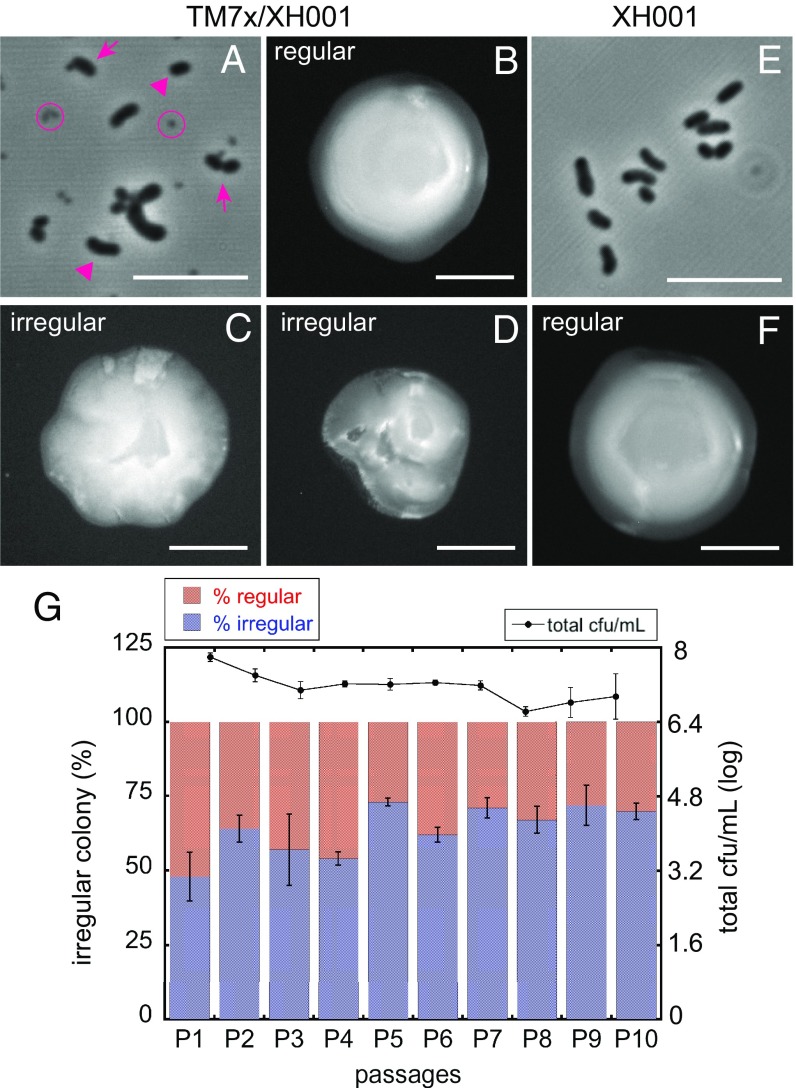
Imaging TM7x/XH001 coculture revealed the existence of three subpopulations of cells: TM7x-infected XH001, TM7x-uninfected XH001, and free-floating independent TM7x (Fig. 1A and SI Appendix, Fig. S1A). Plating the TM7x/XH001 coculture on blood agar generated two colony types: regular and irregular (Fig. 1 B–D). Regular colonies are characterized by even circles with a rounded convex (umbonate) surface, while irregular colonies have rough edges and/or a nonconvex surface. Since TM7x is an obligate epibiont, free-floating TM7x cells cannot grow on blood agar. In contrast, imaging and plating the XH001 monoculture showed only uninfected XH001 cells and resulted in 100% regular colonies (Fig. 1 E and F). These results suggest that infected and uninfected hosts give rise to irregular and regular colonies, respectively, which is further supported by our PCR and colony-culturing analysis showing that a majority of irregular colonies were TM7x positive while regular colonies were PCR negative (SI Appendix, Fig. S1 B and C). Using the irregular and regular colonies as a proxy for infection readout, we followed the population dynamics of the coculture over several passages (Fig. 1G). Interestingly, the coculture maintained relatively stable subpopulation of TM7x-infected and TM7x-uninfected host cells as shown by the fluctuating but overall stable ratio of regular to irregular colonies over 10 passages (48–72% irregular colonies), supporting that the coculture always maintained a reservoir of uninfected XH001 cells.
Fig. 1.
TM7x/XH001 coculture contains heterogeneous populations. Phase-contrast image (A) and colony morphology (B–D) of TM7x/XH001 coculture reflect three subpopulations of cells: TM7x-infected XH001 (arrows and irregular colonies), TM7x-uninfected XH001 (arrowheads and regular colonies), and free-floating TM7x (circles). In contrast, XH001 monoculture displayed only TM7x-uninfected XH001 cells (E) and regular colonies (F). (Scale bars: phase contrast, 5 μm; colonies, 1 mm.) (G) Three independent cocultures were passaged for 10 d, and the irregular colony percentage was determined daily. The bar graph is the percent regular (red) or irregular (blue) colonies, whereas the line graph is the total cfu/mL (log scale). The error bars indicate the SD of the triplicate cultures.
TM7x Infection Reduces Host Growth and Inhibits Cell Division.
The unexpected existence of a stable proportion of uninfected hosts gives rise to two key hypotheses to explain the population dynamics of the two species: One hypothesis is that infected host cells divide, generating both TM7x-infected and TM7x-uninfected daughter cells, thus contributing to the pool of uninfected host cells. Alternatively, it is possible that the cell division of TM7x-infected hosts is severely inhibited, and only uninfected host cells divide to provide hosts for future TM7x infection as well as to replenish the uninfected host reservoir. To distinguish between these possibilities, we imaged the monoculture and coculture cells in real time at the single-cell level and monitored cell growth and cell division.
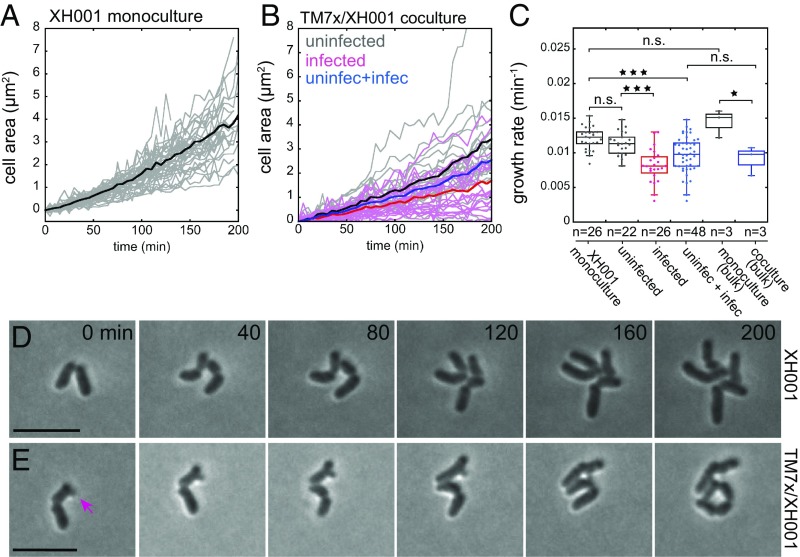
We tracked the growth of individual cells in the images by measuring the cell area and determined the growth rate by fitting the single-cell tracking with an exponential growth equation, to give an estimated growth rate of 0.012 ± 0.002 min−1 for monoculture cells (Fig. 2 A and C and SI Appendix, Fig. S2A). Surprisingly, the uninfected hosts in the coculture grew similar to XH001 monoculture cells with no significant difference in growth rate, 0.011 ± 0.002 min−1 (Fig. 2 B and C). In contrast, the infected hosts (0.008 ± 0.002 min−1) had significantly lower growth rate compared with uninfected hosts from the monocultures or cocultures (Fig. 2 B and C and SI Appendix, Fig. S2B). Furthermore, when we compared combined average growth rate of both infected and uninfected host cells from the same coculture (0.009 ± 0.003 min−1) to that of a XH001 monoculture, we observed smaller but still significantly different growth rate (Fig. 2C, blue plot). These computed growth rates at the single-cell level agreed well with the bulk assay measurements of the entire cultures: monoculture (0.014 ± 0.002 min−1) and coculture (0.009 ± 0.002 min−1) (Fig. 2C and SI Appendix, Fig. S2C). It should be noted that because our coculture consists of both infected and uninfected hosts, and the bulk measurement cannot distinguish between these two cell types within the coculture, we compared the bulk measurement of coculture to that of combined infected and uninfected cells from the single-cell imaging (Fig. 2C, blue plots). These findings argue that physical interaction between the two bacteria is key in TM7x’s ability to inhibit its host’s growth.
Fig. 2.
Single-cell live imaging reveals XH001 growth and cell division. The cell area of individual XH001 cells in monocultures (A) or cocultures (B) were tracked during the live imaging. For the monoculture (A), 26 individual XH001 cells (gray traces) were tracked from six independent experiments, and the average is shown in dark black trace. Each trace was fitted with exponential growth equation to determine growth rate and summarized in C (SI Appendix, Fig. S2A). For coculture (B), TM7x-uninfected (gray lines, n = 22) and TM7x-infected (pink lines, n = 26) hosts were tracked from seven independent experiments. The average growth of uninfected (dark black), infected (dark red), or both (dark blue) are shown. Growth rates were determined and summarized in C (SI Appendix, Fig. S2B). (C) Growth rate summary of single-cell or bulk cell growth measurements (SI Appendix). Each dot reflects growth rate from each trace in A and B or SI Appendix, Fig. S2C. Time-lapse images of monoculture (D) and coculture (E) cells are shown with 40-min interval. (E) TM7x-infected (arrow) and uninfected daughter cells divide differently. (Scale bars: 5 μm.) *P < 0.05, ***P < 0.0001; n.s., not significant.
While monitoring the cell growth, we also observed that TM7x inhibited cell division of XH001. The majority of the monoculture XH001 cells replicated by binary fission (Fig. 2D), with cell division occurring very quickly, comparable to the milliseconds “popping” mechanism of Staphylococcus aureus (24). A small portion of the XH001 monoculture cells also replicated by a previously hypothesized Actinomyces cell division mechanism, where the cells initially grew to a long filament and then fragmented into multiple smaller cells (SI Appendix, Fig. S2D) (25). Within the coculture, however, while uninfected host cells divided the same as monoculture cells (SI Appendix, Fig. S2E), TM7x-infected hosts showed minimal cell division. The cell division defects of the infected cells were most obvious when two newly divided daughter cells, one infected and the other not, were monitored (Fig. 2E). While the uninfected daughter cell underwent two cell divisions, the infected daughter did not divide over the same time period. Our available imaging resolution cannot accurately capture TM7x cell division, thus we do not discuss it in this paper. Combined effects of slow growth and severely inhibited cell division led to longer (SI Appendix, Fig. S2F) and branched host cells (SI Appendix, Fig. S2G). This is consistent with our previously published measurements, where we reported predominantly longer, branched cells in coculture (6).
Confirming the cell division inhibition, 87.8% of the infected hosts that we tracked did not divide during our observation (200 min), while 95.1% and 97.4% of the uninfected hosts from coculture and monoculture, respectively, divided at least once (SI Appendix, Fig. S3A). In addition, membrane staining showed a significantly increased number of cells with a visible septum in coculture compared with monoculture XH001 cells (SI Appendix, Fig. S3 B and C). Some cells in coculture had more than one septum, which we did not observe in monoculture cells. Supporting the cell division arrest and abnormal cell septum formation, our recent transcriptomic study revealed that many of the XH001 cell division genes were significantly down-regulated in coculture compared with monoculture (SI Appendix, Fig. S3D) (5). These experiments suggest that the infected cells have a relatively minor contribution to the production of new hosts in coculture, such that host population dynamics are dominated by the replication of uninfected cells. All these features are similar to parasitic castration where the parasite, TM7x in this case, severely blocks replication but partially reduces growth of its host and, therefore, benefits from the host’s longevity (26, 27). This leads to a long-term stable coculture that can be passaged indefinitely.
Naïve Host Cells Develop Reduced Susceptibility to Virulent TM7x Infection.
Our detailed study of the TM7x/XH001 coculture suggests a lifestyle in which these ultrasmall bacteria chronically infect a host cell in a castration-like manner that severely inhibits replication but allows growth to continue at a reduced rate. As discussed above, however, by necessity the coculture of TM7x/XH001 must be passaged several times to establish a viable and stable experimental system where host and TM7 are in balance. This raises the question of whether the behavior of TM7x in this coculture is similar to that in a natural setting. Fluorescence in situ hybridization (FISH) imaging of Saccharibacteria in human saliva shows very similar infection pattern to that of coculture where, critically, only one or a few Saccharibacteria cells are on the outside of Actinomyces host cells (SI Appendix, Fig. S3E). While not definitive, these data are consistent with the inference that what we are observing in coculture is recapitulating what occurs in the human oral cavity.
To further investigate the initiation of attachment and growth as well as the development of the coculture stability observed, we exposed a naïve population of the host cells to TM7x. Specifically, by naïve host we mean a second A. odontolyticus isolate, XH001n, which was independently isolated from the same human subject and cultured without TM7x in the laboratory. XH001n was sequenced and confirmed to be genetically identical with XH001 with a small number of differences at the nucleotide level, which will be discussed later. This allowed us to separate any effects of host evolution in continuous coculture from the behavior of TM7x and XH001 in initial primary coculture. However, since naïve XH001n was isolated from the same subject where TM7x-infected XH001 was isolated, we cannot exclude the possibility that the lineage may have been previously infected in the oral cavity. We also cannot accurately determine if TM7x in the laboratory coculture had varied genetically from what was likely present in the oral cavity. Nevertheless, as described below, the cocultivation of TM7x with naïve host cells rapidly evolved to a system of long-term parasitism seen both in oral microbial communities and the original coculture.
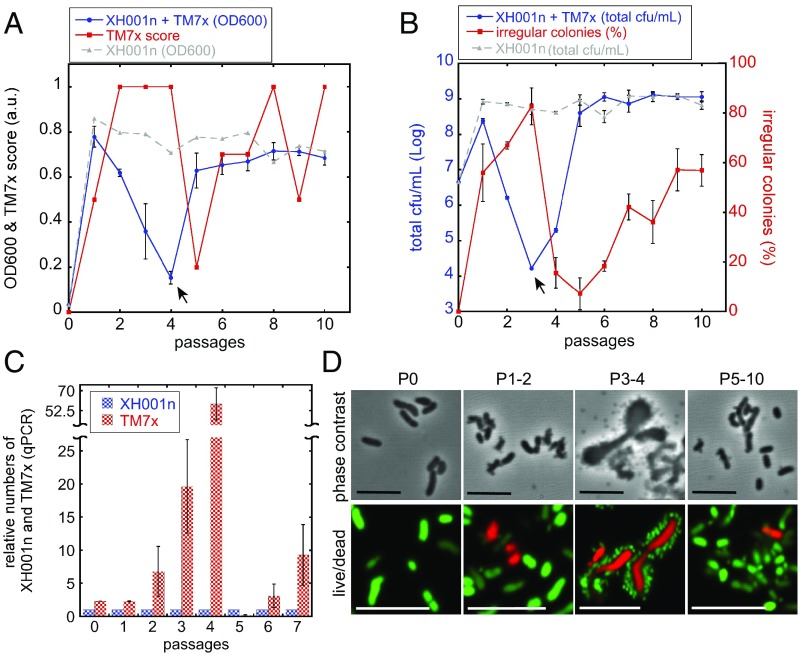
To investigate the initial infection, we first devised a method to isolate free-floating TM7x from the coculture (SI Appendix, Fig. S4 A and B). We then enumerated the isolated TM7x via a previously developed virus counting method with slight modifications (SI Appendix, Fig. S4 C–E) (28). To test if free-floating TM7x can reinfect naïve hosts, we carried out a reinfection assay where freshly isolated TM7x was mixed with naïve XH001n cells in a 3:1 (TM7x:XH001n) ratio (Fig. 3 A and B). We passaged the mixed culture every 24 h and at each passage monitored the growth and the dynamic behavior between TM7x and XH001n using: (i) optical (cell) density at 600 nm (OD600) and TM7x score by phase-contrast microscopy (Fig. 3A and SI Appendix, Fig. S5A); (ii) total colony forming units per milliliter of media (total cfu/mL) and the percentage of irregular colonies from the total cfu/mL (Fig. 3B); and (iii) the ratio of TM7x per XH001n cells revealed by qPCR (Fig. 3C). Both OD600 and the total cfu/mL reflect the growth of XH001n cells since the smaller TM7x cells did not significantly contribute to the OD600 reading (SI Appendix) and the free-floating TM7x cells did not form colonies. TM7x score, irregular colony percentage, and TM7x/XH001n ratio (qPCR) indicate the overall TM7x abundance, infected population, and number of TM7x per XH001n, respectively.
Fig. 3.
Naïve hosts develop reduced susceptibility to TM7x. Isolated free-floating TM7x (SI Appendix, Fig. S4) was mixed with naïve XH001n strain at 3:1 ratio (TM7x:XH001n). The growth of XH001n was monitored by optical density at 600 nm (A) and the total cfu/mL (blue solid line, circles) (B). The growth of TM7x was monitored by TM7x score (A), irregular colony percentage (red solid line, squire) (B), and qPCR (ratio between TM7x to XH001n) (C). The control experiments with no TM7x is shown in dashed gray lines (triangle, A and B). After mixing the cells, they were passaged every 24 h (x axis). (B) Total cfu/mL and irregular colony percentage are on the right and left y axis, respectively. The XH001n “growth crash points” are indicated by the black arrows (A and B). Error bars indicate the SD of the triplicate measurements. (D) Phase-contrast (Top row) and live/dead (Bottom row) imaging of reinfection experiment at indicated passages. (Scale bars: 5 μm.)
As revealed in Fig. 3, during the early passages (1–4) of the coculture of TM7x and naïve XH001n, TM7x was able to infect and proliferate by all measurements. At passage 4, both TM7x score and cell ratio by qPCR showed higher TM7x, but irregular colony percentage was low, suggesting that the majority of the TM7x cells were free floating. By contrast, over the same period, XH001n growth first increased, then as the TM7x population reached its maximum, XH001n population growth crashed, reaching its lowest point at passage 4 by OD600 and passage 3 by total cfu/mL (Fig. 3 A and B). Consistent with the population crash, live/dead staining showed that nearly all hosts were dead while TM7x cells were live at passages 3 and 4 (Fig. 3D). In addition, imaging revealed that many TM7x cells were infecting single hosts at this passage (Fig. 3D), with qPCR estimating >50 TM7x per host in the population (Fig. 3C). These severely infected host cells showed no growth under the single-cell live imaging, confirming the live/dead stain data where these cells were dead (SI Appendix, Fig. S5B). However, the host cells also did not appear to lyse while imaged, which contrasts with predatory bacteria and lysogenic phages that lyse their host to propagate (SI Appendix, Fig. S5C). Irregular colony percentage also reached >90% during the growth crash (Fig. 3B), much higher than our previously established coculture (Fig. 1G). In summary, our data suggest that the virulent infection of TM7x drives the killing of naïve XH001n cells and the collapse of the host population.
After the growth crash point, however, the host population started to recover and, by passage 5, cell numbers reached those of a control culture without TM7x (Fig. 3 A and B). As the XH001n started to recover, the TM7x score, irregular colony percentage, and the cell ratio by qPCR dropped and then increased while XH001n maintained its growth. By passage 10, we observed a coculture with characteristics similar to our initial established coculture involving XH001, with three subpopulations without severe infection by TM7x. We believe at these later passages that the XH001n develops a reduced susceptibility to TM7x overgrowth and killing and forms a stable long-term parasitic lifestyle with TM7x that is essentially the same as the coculture we described in Figs. 1 and 2.
Host-Reduced Susceptibility to TM7x Is Driven by Rapid Evolution.
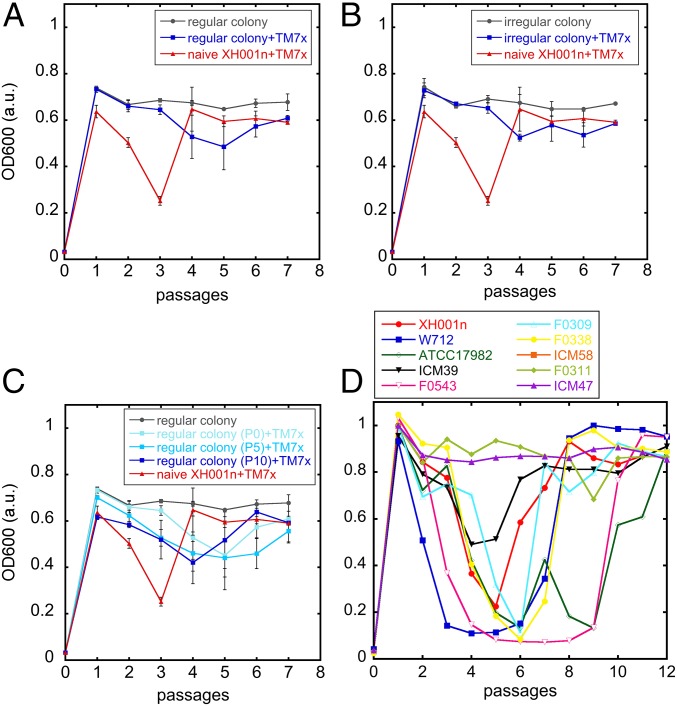
To further investigate how XH001n cells recovered from initial TM7x-induced growth crash and prevented virulent infection, we harvested and grew independent regular and irregular colonies, three of each, from TM7x/XH001n coculture (passage 6) that recovered from a reinfection assay that had growth crash at passage 3. By PCR, the cultures derived from regular colonies showed no TM7x, while those from irregular colonies had TM7x (SI Appendix, Fig. S6A). The cultures derived from regular and irregular colonies also displayed normal growth that resembled the growth of monocultures and cocultures, respectively (SI Appendix, Fig. S6D). When free-floating isolated TM7x cells were added to the cultures derived from these regular and irregular colonies, no obvious growth crash nor severe infection was observed, although we still saw regular and irregular colonies become infected by TM7x at the reduced level similar to the established coculture from Fig. 1 (Fig. 4 A and B and SI Appendix, Fig. S6 B and C). We did see a slight decrease in the cell growth at the later passages, but this was likely due to the fact that a portion of the cells became infected by TM7x and switched to the slow-growing elongated cells.
Fig. 4.
Host-reduced susceptibility to TM7x evolves rapidly. The reinfection assay was carried out with regular (blue squares, A and C) or irregular colony (blue squares, B) cells from the recovered reinfection assay. (C) Regular colonies were passaged 5 (∼50 generations) or 10 (∼100 generations) times before they were added to reinfection assay. Gray (circles) or red (triangles) lines are control experiments of colony alone or naïve host cells with TM7x, respectively (A–C). (D) A growth crash is seen in many, but not all, susceptible host Actinomyces species in clade 2 (SI Appendix, green in SI Appendix, Fig. S7A).
Since regular colonies from the TM7x/XH001n coculture are not infected with TM7x, we passaged these cells every day for a total of 5 (∼50 generations) and 10 (∼100 generations) days before reinfecting them with TM7x. Interestingly, these passaged cells still maintained their reduced susceptibility to severe infection by TM7x and displayed no growth crash change between passage 0 and 10 (Fig. 4C), suggesting the reduced susceptibility is likely due to a stable genetic change. To confirm this, we sequenced the genome of three independent naïve XH001n monocultures and three independent TM7x/XH001n cocultures that were recovered (passage 6) from the reinfection assay and performed a detailed genomic comparison (SI Appendix, Fig. S6E). Compared with naïve XH001n, nine XH001n genes with single-nucleotide changes (resulting in amino acid change) and one gene with single-nucleotide deletion (resulting in frame shift) were unique to all replicates of the TM7x-exposed XH001n (SI Appendix, Table S1). These mutations can be consistently detected within all three postcrash cocultures with frequencies between 11.2% and 54.8% (SI Appendix, Table S1). The data are consistent with parallel host evolution driven by the presence of TM7x. Five of these genes were membrane transporter genes or their regulator (PadR is a transcriptional regulator for multidrug resistance pumps) (29). Two genes were involved in DNA replication during cell division, and two more genes were involved in tRNA function or synthesis. The last gene encoded a putative secreted protein. Seven of the nine mutations fell within the putative functional domain of the gene, suggesting that these could be loss of function mutations (SI Appendix, Table S1). Although the detailed functions and their involvement in TM7x/XH001n interaction require further investigation, the large number of mutations associated with transporters raises the intriguing possibility that the host lineages may have evolved to limit the resources provided to TM7x or that transporters may be attachment sites for TM7x.
TM7x Rapidly Forms Stable Associations with a Number of Actinomyces Species.
Finally, we tested the ability of TM7x to form associations with a range of potential hosts. Isolated TM7x cells were added to strains from multiple Actinomyces species and common oral bacterial species from other genera (strains from The Forsyth Institute strain collection). We found that TM7x was able to establish a stable association with several Actinomyces species that are closely related to A. odontolyticus in clade 2 (SI Appendix, Fig. S7A). In some cases, it appeared that a stable association formed immediately, but in most, TM7x again drove a population crash followed by rapid recovery as observed in XH001n (Fig. 4D). This suggests that TM7x has a relatively narrow host range, but it nevertheless has the potential to rapidly establish stable relationships with strains of a number of species. TM7x was not able to form a stable association with Actinomyces species from clade 1 or with species outside the genus Actinomyces (SI Appendix, Fig. S7A). We note that host range determined in this study is slightly different and more thorough than our previously published findings (5), since we have found that multiple passaging is required to establish this stable relationship in coculture.
It is intriguing to speculate that the Actinomyces species in clade 1, which do not support TM7x growth, may not be susceptible to TM7x infection due to variation or mutations in the genes from SI Appendix, Table S1. Comparative pangenomic analysis of the two clades, however, revealed that: (i) Six of the 10 genes were universally conserved in clade 2 (growth supporting) while none of these were completely conserved in clade 1 (nongrowth supporting); (ii) 4 of the 10 genes were only found in clade 2; and (iii) 2 genes were universally conserved in clade 2 with no close homologs in clade 1 (SI Appendix, Fig. S7B). The simplest inference from the above gene distribution is that genes missing from clade 1 and only found in clade 2 may be involved in stable growth support or binding of TM7x.
Discussion
To date, N. lyticus strain TM7x is the only cultivated Candidate Phyla Radiation organism, which provides a unique window into the lifestyle of this enigmatic group. By following population and single-cell dynamics, we found that N. lyticus is capable of a stable long-term relationship with its hosts. This extended parasitic association is also consistent with direct FISH imaging of the human saliva. In addition, the stable association of Saccharibacteria (TM7) species with other endogenous human oral bacterial species appears to have a long history, as Saccharibacteria have been detected in the microbiome of hunter-gatherers and Neanderthals (30, 31).
We were surprised then to discover that N. lyticus strain TM7x drove a population crash in naïve host cells that had not previously experienced TM7x in the laboratory setting. Moreover, this population crash was associated with a virulent phenomenon whereby many TM7x cells infected each host cell and rapidly killed them. However, we also found that the host cell population rapidly recovered over successive passaging via reduced susceptibility. This likely occurs through a genetic change in the host as XH001n cells keep their reduced susceptibility for over 100 generations without TM7x present (Fig. 4C), and we observed multiple mutations in the evolved host cells (SI Appendix, Table S1). However, without appropriate targeted genetic mutations in these genes, we cannot rule out the possibility that the phase variation and epigenetics may also contribute. What drives the host reduced susceptibility to TM7x? Several mechanisms might help keep TM7x parasitism under control. Decreased host and parasite binding is a strong candidate since, in the stable coculture, we do not see severe infection wherein high numbers of TM7x cover hosts. Accordingly, the transporters in SI Appendix, Table S1 could represent TM7x docking sites, and the mutations in these genes may lead to a decrease in numbers or change in the properties of exposed regions to limit binding. Mutated transport and regulatory genes may also limit TM7x growth after binding by changing the availability, production, or transport of nutrients from the host. Whatever the case, the observed reduced susceptibility was not sufficient to prevent TM7x infection, but it did prevent severe infection and force TM7x to establish a stable relationship observed in the original coculture.
Most predatory bacteria and phage have evolved to kill their hosts quickly and efficiently (32, 33). In contrast, our study suggests that even when TM7x cells exert virulent killing of naïve hosts, the process occurs relatively more slowly than predatory bacteria or phage killing. It does not rapidly lyse the entire culture, suggesting a major difference between the lifestyle behavior of N. lyticus and predatory bacteria or lytic phages. In addition, predatory bacteria typically have large genomes, whereas phage are more similar to Candidate Phyla Radiation organisms in that they are reliant upon prokaryotic hosts due to their reduced biosynthetic capacity. Our data also illustrate the bacterial host range by a member of the Saccharibacteria. Although not exhaustively tested on all oral genera, the range of bacteria that could act as hosts for N. lyticus strain TM7x appears narrower than is typical of predatory bacteria, which can kill diverse species. The fact that TM7x stably infects only a specific clade of Actinomyces is intriguing and may indicate that Saccharibacteria and possibly other Candidate Phyla Radiation species are relatively specialist parasites.
In summary, our work suggests that Saccharibactria species have evolved a unique parasitic lifestyle characterized by the long-term stable infection of bacterial hosts. We also find that they are capable of more virulent infections against naïve hosts, but the host can develop reduced susceptibility to control the virulent infection. This evolutionary process may have influenced the macroevolution of the Candidate Phyla Radiation (34). Parasites that virulently infect a new host risk reducing host number so far as to prevent their own continued transmission (35). Despite an initial crash in the host population, N. lyticus strain TM7x escapes this outcome in our experiments as its host evolved reduced susceptibility to TM7x. Importantly, this is partial, rather than complete, protection thereby allowing the parasite to persist in a community. Thus, as the diversity of the non-Candidate Phyla Radiation Bacteria expanded, a parasite could have potentially tagged along in many lineages. Whether these parasites would adapt to the new association via additional genetic mutations, potentially leading to the observed diversity of the Candidate Phyla Radiation, remains an intriguing question that warrants further investigation.
Materials and Methods
Detailed experimental procedures are provided in SI Appendix. To interrogate such a unique coculture system containing an ultrasmall obligate epibiotic bacteria that cannot be grown as a pure monoculture, we needed to develop multiple assays. We used differential colony morphology of XH001 on blood agar plate to quantify infected vs. uninfected hosts. This adaptation of the colony morphology results correlated well with other assays such as TM7x scoring and qPCR results. We also developed a method to isolate pure TM7x to target only free-floating TM7x within the coculture. The isolated TM7x were added back to the naïve XH001n and passaged every 24 h (reinfection assay) to mimic continuous culture without depletion of nutrient. Genome sequencing of the cultures were carried out using paired-end 300-bp sequencing on an Illumina Miseq and assembly with SPAdes 3.9.
Supplementary Material
Acknowledgments
Research reported in this publication was supported by The National Institute of Dental and Craniofacial Research of the National Institutes of Health (NIH) under Awards 1R01DE023810, 1R01DE020102, 1R01DE026186 (to X.H., J.S.M., and W.S.); R37DE016937 and DE024468 (to F.E.D.); and F32DE025548-01, 5T90DE022734-04, and 1K99DE027719-01 (to B.B.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Footnotes
Conflict of interest statement: W.S. is a part-time chief science officer of C3 Jian, Inc., which has licensed technologies from the University of California Regents that could be indirectly related to this research project.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1810625115/-/DCSupplemental.
References
- 1.Castelle CJ, Banfield JF. Major new microbial groups expand diversity and alter our understanding of the tree of life. Cell. 2018;172:1181–1197. doi: 10.1016/j.cell.2018.02.016. [DOI] [PubMed] [Google Scholar]
- 2.Luef B, et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat Commun. 2015;6:6372. doi: 10.1038/ncomms7372. [DOI] [PubMed] [Google Scholar]
- 3.Brown CT, et al. Unusual biology across a group comprising more than 15% of domain bacteria. Nature. 2015;523:208–211. doi: 10.1038/nature14486. [DOI] [PubMed] [Google Scholar]
- 4.Mclean J, et al. 2018. Evidence of independent acquisition and adaption of ultra-small bacteria to human hosts across the highly diverse yet reduced genomes of the phylum Saccharibacteria. bioRxiv:10.1101/258137.
- 5.He X, et al. Cultivation of a human-associated TM7 phylotype reveals a reduced genome and epibiotic parasitic lifestyle. Proc Natl Acad Sci USA. 2015;112:244–249. doi: 10.1073/pnas.1419038112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bor B, et al. Phenotypic and physiological characterization of the epibiotic interaction between TM7x and its basibiont actinomyces. Microb Ecol. 2016;71:243–255. doi: 10.1007/s00248-015-0711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McLean JS, et al. Draft genome sequence of Actinomyces odontolyticus subsp. actinosynbacter strain XH001, the basibiont of an oral TM7 epibiont. Genome Announc. 2016;4:e01685-15. doi: 10.1128/genomeA.01685-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong J, Qing Y, Guo X, Warren A. “Candidatus Sonnebornia yantaiensis”, a member of candidate division OD1, as intracellular bacteria of the ciliated protist Paramecium bursaria (Ciliophora, Oligohymenophorea) Syst Appl Microbiol. 2014;37:35–41. doi: 10.1016/j.syapm.2013.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Dinis JM, et al. In search of an uncultured human-associated TM7 bacterium in the environment. PLoS One. 2011;6:e21280. doi: 10.1371/journal.pone.0021280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rheims H, Spröer C, Rainey FA, Stackebrandt E. Molecular biological evidence for the occurrence of uncultured members of the actinomycete line of descent in different environments and geographical locations. Microbiology. 1996;142:2863–2870. doi: 10.1099/13500872-142-10-2863. [DOI] [PubMed] [Google Scholar]
- 11.Gao Z, Tseng CH, Pei Z, Blaser MJ. Molecular analysis of human forearm superficial skin bacterial biota. Proc Natl Acad Sci USA. 2007;104:2927–2932. doi: 10.1073/pnas.0607077104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marcy Y, et al. Dissecting biological “dark matter” with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eckburg PB, et al. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bik EM, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci USA. 2006;103:732–737. doi: 10.1073/pnas.0506655103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duran-Pinedo AE, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 2014;8:1659–1672. doi: 10.1038/ismej.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffen AL, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abusleme L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuehbacher T, et al. Intestinal TM7 bacterial phylogenies in active inflammatory bowel disease. J Med Microbiol. 2008;57:1569–1576. doi: 10.1099/jmm.0.47719-0. [DOI] [PubMed] [Google Scholar]
- 19.Baker JL, Bor B, Agnello M, Shi W, He X. Ecology of the oral microbiome: Beyond bacteria. Trends Microbiol. 2017;25:362–374. doi: 10.1016/j.tim.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dashiff A, Junka RA, Libera M, Kadouri DE. Predation of human pathogens by the predatory bacteria Micavibrio aeruginosavorus and Bdellovibrio bacteriovorus. J Appl Microbiol. 2011;110:431–444. doi: 10.1111/j.1365-2672.2010.04900.x. [DOI] [PubMed] [Google Scholar]
- 21.Davidov Y, Huchon D, Koval SF, Jurkevitch E. A new alpha-proteobacterial clade of Bdellovibrio-like predators: Implications for the mitochondrial endosymbiotic theory. Environ Microbiol. 2006;8:2179–2188. doi: 10.1111/j.1462-2920.2006.01101.x. [DOI] [PubMed] [Google Scholar]
- 22.Lambina VA, Afinogenova AV, Romaĭ Penabad S, Konovalova SM, Pushkareva AP. Micavibrio admirandus gen. et sp. nov. Mikrobiologiia. 1982;51:114–117. [PubMed] [Google Scholar]
- 23.Soo RM, Woodcroft BJ, Parks DH, Tyson GW, Hugenholtz P. Back from the dead; the curious tale of the predatory cyanobacterium Vampirovibrio chlorellavorus. PeerJ. 2015;3:e968. doi: 10.7717/peerj.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou X, et al. Bacterial division. Mechanical crack propagation drives millisecond daughter cell separation in Staphylococcus aureus. Science. 2015;348:574–578. doi: 10.1126/science.aaa1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaal KP, Yassin AA. Genus I. Actinomyces. Bergey’s Manual of Systematic Bacteriology. Springer; Heidelberg: 2012. pp. 42–109. [Google Scholar]
- 26.Lafferty KD, Kuris AM. Trophic strategies, animal diversity and body size. Trends Ecol Evol. 2002;17:507–513. [Google Scholar]
- 27.Lafferty KD, Kuris AM. Parasitic castration: The evolution and ecology of body snatchers. Trends Parasitol. 2009;25:564–572. doi: 10.1016/j.pt.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Diemer GS, Kyle JE, Stedman KM. 2012 Counting viruses using polycarbonate Track Etch membrane filters as an alternative to Anodisc membrane filters. Available at web.pdx.edu/∼kstedman/. Accessed May 7, 2016.
- 29.Huillet E, Velge P, Vallaeys T, Pardon P. LadR, a new PadR-related transcriptional regulator from Listeria monocytogenes, negatively regulates the expression of the multidrug efflux pump MdrL. FEMS Microbiol Lett. 2006;254:87–94. doi: 10.1111/j.1574-6968.2005.00014.x. [DOI] [PubMed] [Google Scholar]
- 30.Weyrich LS, et al. Neanderthal behaviour, diet, and disease inferred from ancient DNA in dental calculus. Nature. 2017;544:357–361. doi: 10.1038/nature21674. [DOI] [PubMed] [Google Scholar]
- 31.Adler CJ, et al. Sequencing ancient calcified dental plaque shows changes in oral microbiota with dietary shifts of the Neolithic and Industrial revolutions. Nat Genet. 2013;45:450–455e1. doi: 10.1038/ng.2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sockett RE. Predatory lifestyle of Bdellovibrio bacteriovorus. Annu Rev Microbiol. 2009;63:523–539. doi: 10.1146/annurev.micro.091208.073346. [DOI] [PubMed] [Google Scholar]
- 33.Ofir G, Sorek R. Contemporary phage biology: From classic models to new insights. Cell. 2018;172:1260–1270. doi: 10.1016/j.cell.2017.10.045. [DOI] [PubMed] [Google Scholar]
- 34.Stanley SM. A theory of evolution above the species level. Proc Natl Acad Sci USA. 1975;72:646–650. doi: 10.1073/pnas.72.2.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson RM, May RM. The population dynamics of microparasites and their invertebrate hosts. Philos Trans R Soc B. 1981;291:451–524. doi: 10.1098/rstb.2014.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.