Significance
DNA modification gives rise to diverse outcomes, including cancer and cell death. Understanding the chemical and biochemical effects of DNA modification contributes to the fundamental understanding of the etiology and treatment of cancer. 2′-Deoxyguanosine methylation at the N7 position (MdG) is a major mechanism of action of some DNA alkylating agents. We examined MdG in nucleosome core particles (NCPs). Abasic site formation from MdG is suppressed in NCPs. Furthermore, MdG and histone proteins form cross-links [DNA–protein cross-links (DPCs)], a deleterious type of DNA damage. DPCs are also formed in cells treated with monofunctional alkylating agent. DPC formation from MdG is a previously unrecognized process that could have significant effects on cells and may play a role in the cytotoxicity of DNA alkylating agents.
Keywords: DNA damage, DNA alkylation, DNA–protein cross-links, nucleosomes, nucleic acids
Abstract
Monofunctional alkylating agents preferentially react at the N7 position of 2′-deoxyguanosine in duplex DNA. Methylated DNA, such as that produced by methyl methanesulfonate (MMS) and temozolomide, exists for days in organisms. The predominant consequence of N7-methyl-2′-deoxyguanosine (MdG) is widely believed to be abasic site (AP) formation via hydrolysis, a process that is slow in free DNA. Examination of MdG reactivity within nucleosome core particles (NCPs) provided two general observations. MdG depurination rate constants are reduced in NCPs compared with when the identical DNA sequence is free in solution. The magnitude of the decrease correlates with proximity to the positively charged histone tails, and experiments in NCPs containing histone variants reveal that positively charged amino acids are responsible for the decreased rate of abasic site formation from MdG. In addition, the lysine-rich histone tails form DNA–protein cross-links (DPCs) with MdG. Cross-link formation is reversible and is ascribed to nucleophilic attack at the C8 position of MdG. DPC and retarded abasic site formation are observed in NCPs randomly damaged by MMS, indicating that these are general processes. Histone–MdG cross-links were also detected by mass spectrometry in chromatin isolated from V79 Chinese hamster lung cells treated with MMS. The formation of DPCs following damage by a monofunctional alkylating agent has not been reported previously. These observations reveal the possibility that such DPCs may contribute to the cytotoxicity of monofunctional alkylating agents, such as MMS, N-methyl-N-nitrosourea, and temozolomide.
DNA alkylation is a ubiquitous process effected by anticancer agents and mutagens. A variety of exogenous agents that methylate DNA, such as methyl methanesulfonate (MMS) and N-methyl-N-nitrosourea (MNU), are widely used as research tools in cells (1–4). Others, such as temozolomide and dacarbazine, are currently used to treat cancer (5, 6). Although the relative amounts of different types of methylation products from alkylating agents vary, N7-methyl-2′-deoxyguanosine (MdG) is formed in the greatest amounts (60–80%) (7). MdG is also long-lived, with in vivo half-lives on the order of days, providing ample time for it to affect cellular chemistry (8–10). Despite being a major product of methylating agents, MdG itself is often thought of as a nontoxic and nonmutagenic DNA lesion (2, 7). The reactivity of MdG and its biological effects have largely been ascribed to slow hydrolysis (depurination) in duplex DNA resulting in the formation of cytotoxic and mutagenic abasic sites (AP, Fig. 1A). To the best of our knowledge, investigations on MdG reactivity have only been carried out using free DNA (11). We now report that MdG reactivity in nucleosome core particles (NCPs) differs significantly from that in free DNA in two respects. First, hydrolysis is significantly retarded in NCPs. Importantly, we also discovered that MdG directly reacts with histone proteins to form DNA–protein cross-links (DPCs), a deleterious form of DNA damage not previously ascribed to simple monoalkylation adducts. DPC formation between histone proteins and MdG was also observed in MMS-treated V79 Chinese hamster lung cells. Intracellular DPC formation from MdG could have significant biological effects.
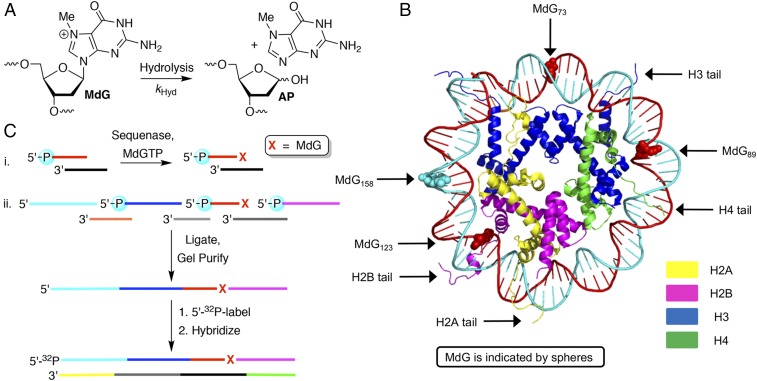
Fig. 1.
N7-Methyl-2′-deoxyguanosine (MdG) hydrolysis and independent incorporation in nucleosome core particles (NCPs). (A) Hydrolysis reaction of MdG. (B) Positions in NCPs at which MdG was site specifically incorporated [Protein Data Bank (PDB) ID code 1kx5]. (C) Methods used for independently generating MdG within 145-bp DNA.
Compared with other DNA lesions, MdG minimally perturbs duplex structure (12). The steric effects of N7-methylation within the major groove are minimal, and the aromatic planar structure is maintained. Although the Watson–Crick hydrogen bonding pattern of dG is retained, recent structural studies raise the possibility that tautomerization encouraged by N7-methylation may increase DNA polymerase promiscuity during replication of templates containing MdG (12). N7-Alkylation encourages secondary reactions, and MdG produces N-methyl formamidopyrimidine (MeFapy•dG) following nucleophilic attack at the C8 position, and abasic sites (AP) via hydrolysis of the glycosidic bond (13). Although MeFapy•dG formation typically occurs at high pH, AP sites are observed in naked DNA under a variety of conditions in multiple sequence contexts. The half-life for depurination ranges from 69 to 192 h under physiologically relevant conditions (pH 7–7.4, 37 °C) (11). Despite the modest rate of MdG hydrolysis, AP formation is believed to be the primary source of cytotoxicity from this lesion (7). However, most studies on MdG have been carried out in free DNA. Human cellular DNA is organized in chromatin and is closely associated with proteins. Histone proteins that make up the octameric core of nucleosomes in chromatin are known to react with other DNA lesions (14–18). For instance, DNA cleavage or excision at AP and oxidized abasic sites is increased as much as 1,500-fold in NCPs compared with free DNA via transient DPC formation (19). Some transient abasic site–histone DPCs ultimately result in histone modifications (15, 19). Other DNA lesions, such as oxidized 8-oxo-7,8-dihydro-2′-deoxyguanosine, or oxanine form stable DPCs (20–22). In the present work, we speculated that the proximity of histone proteins in NCPs may affect MdG reactivity under physiologically relevant conditions, leading to the formation of secondary lesions.
Results
Preparation of NCPs Containing Site-Specific MdG.
NCPs containing MdG at specific sites were designed based on one containing the Widom 601 strong positioning (145 bp) DNA sequence and Xenopus laevis histone proteins, whose X-ray crystal structure has been solved (23, 24). The requisite histone proteins were overexpressed in Escherichia coli and purified as previously described (25). Four positions were selected for MdG incorporation based upon consideration of NCP X-ray crystal structures (Fig. 1B) (24, 26, 27). Positions 89 and 123 are proximal to lysine-rich histone tails that protrude through the octamer in the vicinity of superhelical locations (SHLs) 1.5 and 4.9, respectively. The DNA near SHL 1.5 is notably kinked, and is a hot spot for DNA-damaging molecules (28). DNA near the dyad axis (position 73) is tightly held by the octameric core but is further removed from flexible histone tails. Similarly, position 158 is also distal from any histone tails.
Solid-phase synthesis of oligonucleotides containing MdG is precluded by its proclivity toward depurination and lability to alkaline conditions. Consequently, 145-nt-long oligonucleotides in which MdG was site specifically incorporated were prepared chemoenzymatically and stored at −80 °C (Fig. 1C) (29). The nucleotide triphosphate (MdGTP), prepared from dGTP (30) and purified by anion exchange chromatography, was incorporated at the 3′-terminus of an oligonucleotide hybridized to a complementary template using Sequenase (i, Fig. 1C and SI Appendix, Fig. S1A). The single-nucleotide extended product was purified by denaturing PAGE and subsequently used in a T4 DNA ligase reaction with three or four other oligonucleotides to produce a 145-nt product, which was also gel purified (ii, Fig. 1C and SI Appendix, Fig. S1B). The sequences of these 145-nt products differed from the 601 DNA only at the position of MdG incorporation. Following 5′-32P labeling of the MdG-containing strand, the DNA was hybridized at 25 °C to the complementary 145-mer, which was also prepared via ligation of chemically synthesized oligonucleotides. The NCPs were reconstituted at 4 °C using the salt-dilution method (25). The extent of reconstitution and NCP stability over the course of ensuing reactions, which were as long as 84 h at 37 °C, were established via nucleoprotein gel analysis. Reactions were carried out using NCPs with reconstitution efficiency greater than 95%. The uniformity of the NCPs was established using DNase I footprinting (SI Appendix, Fig. S2).
The integrity of the MdG-containing DNA used to prepare the NCPs was established using a battery of chemical and enzymatic treatments that induce selective effects from MdG and the anticipated products, AP and MeFapy•dG (SI Appendix, Fig. S3). For instance, treatment of the DNA with NaOH cleaves at AP sites but not MdG or MeFapy•dG (31, 32). Similarly, treatment with the base excision repair enzyme, formamidopyrimidine glycosylase (Fpg), cleaves the DNA at MeFapy•dG and AP sites (33). In contrast, sequential treatment of the DNA with human alkyladenine glycosylase (hAAG) and NaOH cleaves the DNA at MdG and AP sites (34). Any dG present due to undetected dGTP impurity in MdGTP or MdG decomposition would be resistant to all of these treatments. We were unable to detect any dG, AP, or MeFapy•dG using these methods and estimate that the nucleosomal DNA is >99% MdG.
MdG Hydrolysis Rate Constants Are Decreased in NCPs.
Depurination of MdG to produce AP sites was measured by taking advantage of the ability to selectively cleave DNA at the latter upon alkaline treatment (0.1 M NaOH, 37 °C, 0.5 h). The rate constant for MdG hydrolysis (kHyd) in free DNA varied almost fourfold depending upon the position (Table 1). However, the corresponding rate constant within the NCP was always lower than in free DNA. The extent to which hydrolysis was retarded within the NCP varied with respect to MdG position but was greater at the positions (89 and 123) where the lesion was most proximal to the highly positively charged histone tails.
Table 1.
MdG hydrolysis in free DNA and NCPs
| kHyd,* ×10−7 s−1 | t1/2,* h | ||||
| MdG position | Free DNA | NCP | Free DNA | NCP | t1/2 (NCP:free DNA) |
| 73 | 17.4 ± 1.1 | 13.9 ± 1.2 | 111 ± 7 | 139 ± 12 | 1.3 |
| 89 | 10.1 ± 0.5 | 3.5 ± 0.2 | 191 ± 8 | 550 ± 27 | 2.9 |
| 123 | 5.7 ± 0.1 | 1.0 ± 0.1 | 338 ± 6 | 1,930 ± 197 | 5.7 |
| 158 | 22.1 ± 2.0 | 9.9 ± 1.0 | 87 ± 8 | 194 ± 20 | 2.2 |
Values are the average ± SD of two experiments, each carried out in triplicate.
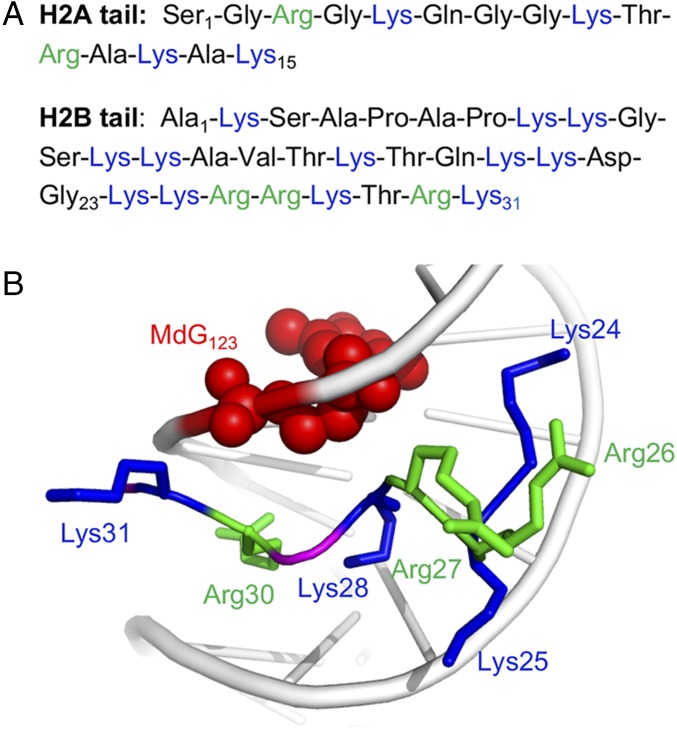
The greatest decrease in hydrolysis rate was detected at position 123 (SHL 4.9), where reaction within the NCP was almost six times slower than in free DNA. An X-ray crystal structure of a NCP containing a different DNA sequence indicates that the H2A and H2B tails are in the vicinity of MdG123 (26). Contributions of these tails to the increased half-life for MdG123 depurination were examined by preparing NCPs containing histone variants (Fig. 2A, Table 2, and SI Appendix, Fig. S4). Removing the entire H2A tail (15 aa) had little if any effect on the half-life for AP formation. The X-ray crystal structure suggests that the longer (31 aa) H2B tail should be more proximal to MdG123. Deletion of the 23 N-terminal amino acids slightly increased the rate constant for hydrolysis. Deleting the remaining 8 aa from the H2B tail, 7 of which are positively charged (Fig. 2A), significantly increased the rate constant describing MdG123 hydrolysis (kHyd, Table 2). The half-life for MdG123 depurination in the NCP lacking the entire H2B tail is less than twofold longer than in free DNA, compared with almost six times longer when the full-length protein is present.
Fig. 2.
Histone variants used to prove MdG123 reactivity in NCPs. (A) N-terminal amino acid sequences of histones H2A and H2B. (B) Proximity of amino acids 24–31 of histone H2B to MdG123 (based on X-ray crystal structure containing native DNA; PDB ID code 1kx5).
Table 2.
Effects of histone variants on MdG123 hydrolysis in NCPs
| Substrate | kHyd,* ×10−7 s−1 | t1/2,* h |
| Free DNA | 5.7 ± 0.1 | 338 ± 6 |
| WT NCP | 1.0 ± 0.1 | 1,930 ± 197 |
| H2A1-15 Del | 1.1 ± 0.1 | 1,750 ± 6 |
| H2B1-23 Del | 1.2 ± 0.1 | 1,600 ± 76 |
| H2B1-31 Del | 3.3 ± 0.1 | 583 ± 10 |
| H2B1-23 Del K24,25,28,31A | 2.7 ± 0.1 | 713 ± 14 |
| H2B1-23 Del R26,27,30A | 1.6 ± 0.1 | 1,200 ± 30 |
Values are the average ± SD of two experiments each carried out in triplicate.
These data suggest that the proximity of positively charged histone tails to MdG123 results in a decrease in hydrolysis rate constant and that the seven positively charged amino acids (four lysines and three arginines) expected to be closest to the alkylated nucleotide are the major contributors (Fig. 2B). To further validate that conclusion, two additional H2B variants were incorporated within NCPs containing MdG123 (Table 2 and SI Appendix, Fig. S4). For ease of preparation, each variant lacked the 23 N-terminal H2B amino acids that have a small effect on hydrolysis. In one variant, the four remaining H2B tail lysines were mutated to alanine (H2B 1–23 Del, K24,25,28,31A), and in the other, alanine was substituted for the three arginines (H2B 1–23 Del, R26,27,30A). The half-life for MdG123 hydrolysis decreased significantly compared with that in the NCP in which the H2B tail (23 N-terminal amino acids) was partially deleted. However, neither NCP yielded a hydrolysis rate constant that was as high as when the entire 31 H2B amino acid tail was removed (H2B 1–31 Del). Overall, site-specific MdG incorporation in NCPs containing wild-type or histone variants reveals that the positively charged environment stabilizes the alkylated nucleotide toward depurination. The magnitude of the effect is proportional to the proximity and magnitude of the positively charged Lys and Arg residues.
MdG Forms Reversible DPCs in NCPs.
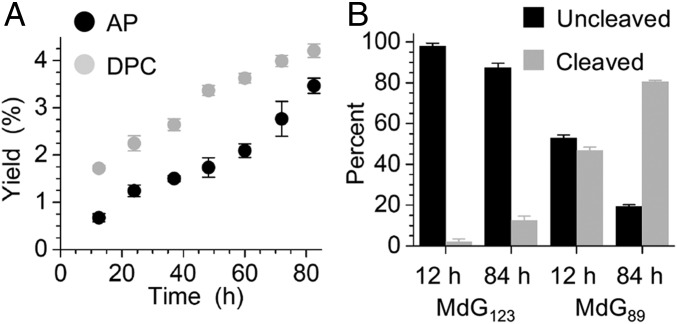
DPCs were readily detected as slower moving products via SDS/PAGE, which were transformed to faster migrating bands following proteinase K treatment. MdG123 is the position where hydrolysis is decreased the most relative to its rate in free DNA. Of the four positions examined, it is also the one in which SDS/PAGE analysis revealed the greatest relative amount of DPCs compared with AP sites. DPCs are formed in amounts slightly higher than AP sites at MdG123 (Fig. 3A). The ratio of DPCs/AP sites produced at MdG89 is slightly less than 1, but the absolute DPC yield is more than twice that at position 123 (SI Appendix, Fig. S5A). In contrast, the NCP containing MdG158, in which the alkylated nucleotide is not close to a flexible histone tail, yields less than 1% DPCs after 84 h (SI Appendix, Fig. S5B). Similarly, although ∼33% of MdG73 is converted to AP in 84 h, the DPC yield at this position is less than 1% (SI Appendix, Fig. S5C). To our knowledge, DPCs have never been reported from MdG or any other alkylated nucleotide produced from a monofunctional alkylating agent. Since MdG is known to undergo nucleophilic attack at the C8 position by hydroxide ion to form MeFapy•dG, we speculated that one or more lysine residues in the proximal H2B tail could react with the alkylated purine at position 123. However, the observed DPCs could also result from AP sites, which are produced upon MdG hydrolysis, and are known to form transient DPCs in NCPs (14).
Fig. 3.
Formation of AP sites and DPCs from MdG in NCPs. (A) Yields of AP and DPCs from MdG123 within NCP as a function of time. (B) Determination of sources of DPCs from NCPs containing MdG89 or MdG123 as a function of time.
The source(s) of the DPCs from MdG89 and MdG123 was determined by taking advantage of the expected differences in structure and reactivity. The DPCs produced from AP sites in NCPs are known to undergo strand scission that is catalyzed by the histone tails, and had previously been examined at position 89 (14, 16). Consequently, the source of DPCs within NCPs containing MdG123 was determined by comparing those obtained from independently generated AP123 with those formed from the alkylated material. Indeed, 100% of the DNA in DPCs isolated from NCPs containing independently generated AP123 was cleaved (SI Appendix, Fig. S6). In contrast, 99% of the DNA in DPCs isolated after incubating NCPs containing MdG123 for 12 h was uncleaved. Even after long reaction times (84 h), when AP from MdG123 hydrolysis will have built up, only 11% of DPCs contained cleaved DNA (Fig. 3B). These data suggest that the DPCs containing uncleaved DNA detected in MdG123-containing NCPs result from the unprecedented reaction between the alkylated nucleotide and histone protein(s), and their yields were determined accordingly (SI Appendix, Fig. S7). A similar analysis of the DPCs produced from MdG89 over the same timescale revealed that a significantly greater fraction of the DPCs were derived from reaction with the abasic site produced upon depurination (Fig. 3B). At early times (12 h), approximately one-half of the DPCs at position 89 were attributed to cross-linking with MdG89. However, at later times as more AP89 builds up, the fraction of DPCs formed from reaction with MdG89 directly decreased to 20%.
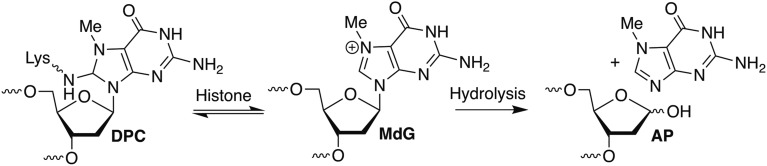
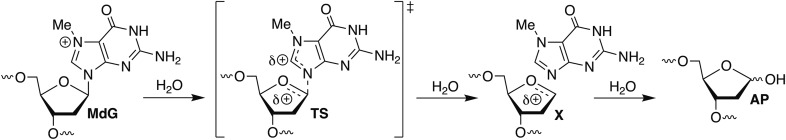
The isolated MdG123 DPC was unstable at 37 °C with a half-life of 10.2 ± 0.3 h (SI Appendix, Fig. S8A). The half-life of the DPC from MdG89 was not determined because of the large amounts of cross-link from AP. The released uncleaved DNA from MdG123 DPCs was stable to NaOH or Fpg, but was cleaved upon sequential treatment with hAAG and NaOH (SI Appendix, Fig. S8B), indicating that the DPC did not decompose via formation of AP or a Fpg substrate, but rather regenerated MdG-containing DNA. The structure of MdG and the reversibility of the process lead us to suggest that the cross-link results from reversible nucleophilic attack at the C8 position and that restoration of aromaticity provides the driving force for release of the otherwise unmodified DNA (Scheme 1).
Scheme 1.
Reactivity of MdG in NCPs.
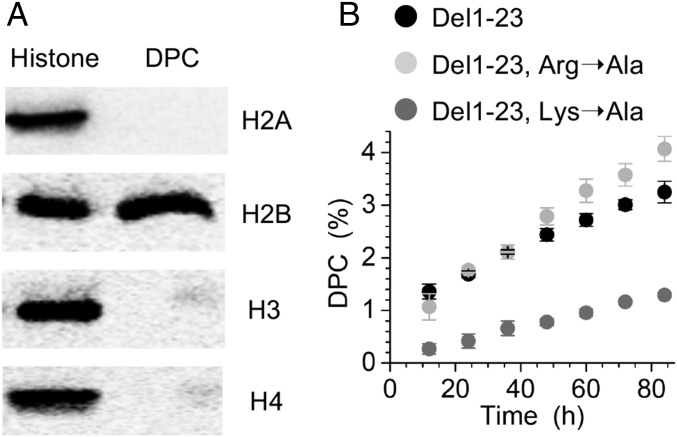
Additional information on the cross-linked proteins was obtained using NCPs containing MdG123 due to their greater homogeneity compared with the products obtained from MdG89. Isolated MdG123 DPCs were thermally decomposed, and the released proteins were detected by Western blotting using histone antibodies, which revealed that H2B was solely responsible for DPC formation with MdG123 (Fig. 4A). This is consistent with the above experiments demonstrating that H2B was responsible for the majority of the decrease in the rate constant for hydrolysis at MdG123 (Table 2). The instability of DPCs prevented us from determining the amino acid residue(s) cross-linked to MdG123 using liquid chromatography (LC)–MS/MS. Instead, the H2B variants lacking the 23 N-terminal amino acids were used for identifying the amino acid(s) responsible for DPC formation (Fig. 4B). Deleting residues 1–23 of H2B slightly decreased the DPC yield compared with wild-type NCP (Figs. 3A and 4B), and substituting alanine for arginine in the remaining region of the H2B tail (H2B 1–23 Del R26,27,30A) had little additional effect on the DPC yield (Fig. 4B). However, the DPC yield decreased by ∼75% when alanines were substituted for lysines (H2B 1–23 Del K24,25,28,31A), indicating that the proximal lysine residues (K24, 25, 28, and 31) accounted for the majority of MdG123 DPCs.
Fig. 4.
Identification of the protein(s) and amino acids responsible for DPC formation with MdG123. (A) Western blot analysis of the proteins released from isolated DPCs. (B) Effects of histone H2B variants on DPC yields as a function of time.
DPC Formation and Decreased Hydrolysis Rates in NCPs Treated with MMS.
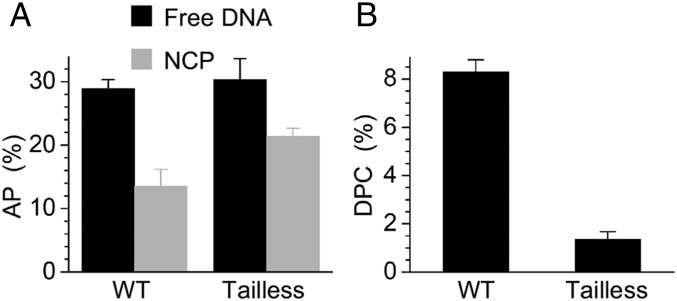
The effects of the NCP environment on MdG reactivity described above were based upon sites purposefully chosen so as to distinguish between alkylated nucleotides whose positions relative to flexible histone tails were understood. Such experiments do not take into account the influence of the core particle structure on the sites of alkylation or the nuances of the NCP structure over the entire 145 bp. The general effects of the NCP environment on MdG reactivity were examined using NCPs composed of the 601 sequence containing only native nucleotides. To compare MdG depurination in free DNA and NCPs, both strands of DNA in the latter were 5′-32P labeled and treated with MMS (10 mM, 1 h, 37 °C). No NCP decomposition was observed after MMS treatment. Following removal of the alkylating agent, a portion of the NCP was extracted with phenol and precipitated to provide free DNA with an identical MdG pattern and levels as in the core particle. Following incubation at 37 °C for 24 h, yields of AP and DPC (per below) in MMS-treated samples were normalized by taking into account the extent of alkylation. The overall AP yield in the NCP was less than one-half that in free DNA (Fig. 5A and SI Appendix, Fig. S9), indicating that the global effect of the NCP environment on MdG hydrolysis is in between the individual sites minimally (MdG73) and maximally (MdG123) affected (Table 1). A comparable experiment using NCPs composed of histone variants in which each protein’s N-terminal tail was deleted supported the important role that these domains play in suppressing MdG hydrolysis (Fig. 5A and SI Appendix, Fig. S9). The yield of AP sites in the tailless NCP increased to >70% that in the corresponding free DNA.
Fig. 5.
Reactivity of MMS-treated free and nucleosomal DNA. (A) Comparison of AP yield after 24 h in free DNA and NCP (with or without N-terminal tails of all four histone proteins). (B) DPC yield in wild-type NCPs and those composed of N-terminal tailless histones. The yields of AP and DPCs were normalized based upon the DNA alkylation level.
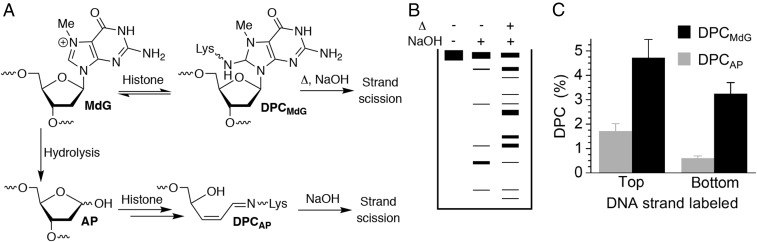
DPC formation was also determined in MMS-treated NCPs following incubation at 37 °C. Although DPCs are the major product from MdG123 (Fig. 3A), AP sites (Fig. 5A) are favored over DPCs (Fig. 5B) in MMS-treated NCPs, and experiments with N-terminal tailless histone proteins corroborate that amino acids in these regions of the proteins are primarily responsible for cross-links. Furthermore, the greater heterogeneity of products in MMS-treated NCPs raises the possibility that the DPCs are formed from AP and MdG. The contributions of AP and MdG to DPCs in MMS-treated NCPs were again determined by taking advantage of their differing chemical properties (Fig. 6). DPCs derived from AP decompose to cleaved DNA upon NaOH (0.1 M, 30 min, 37 °C) treatment, but those derived from MdG require heating (90 °C, 30 min) before NaOH treatment to decompose the cross-link and cleave the DNA. Consequently, aliquots of DPCs isolated by SDS/PAGE from MMS-treated NCPs following 24-h incubation at 37 °C were subjected to these two treatments, and the products were analyzed by denaturing PAGE. The amount of DPCs from AP were quantified following direct NaOH treatment, whereas the amounts of cross-links from MdG were determined by subtracting those attributed to AP from the cleavage observed following sequential heating and alkaline hydrolysis. The uncleaved material in this experiment is ascribed to DPCs involving a nucleotide on the opposite (unlabeled) strand. Hence, the source of DPCs involving each strand was determined separately by 5′-32P labeling the respective 145-mer. These experiments revealed that the top strand (positions 1–145) produced approximately twice the yield of DPCs, but that the bottom strand yielded a greater ratio of cross-links between MdG:AP. Overall, >75% of the DPCs formed in the MMS-treated NCPs were due to direct reaction between a histone protein and MdG (Fig. 6C).
Fig. 6.
Sources of DPCs from MMS-treated NCPs. (A) DPCs involving MdG and AP are distinguishable based upon the different conditions required to cleave their DNA. (B) Denaturing PAGE analysis of isolated DPCs subjected to different treatments reveals cleavage patterns representative of the types of cross-links. (C) DPC yields from MdG and AP in the respective strands of MMS-treated NCPs after 24-h incubation. The DPC yields were normalized based upon the DNA alkylation level in the corresponding strand.
DPC Formation in MMS-Treated Cells.
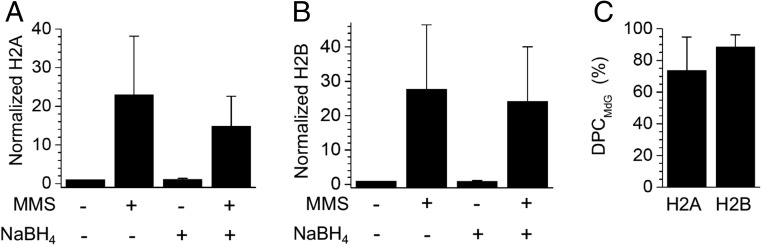
DPC formation was probed in Chinese hamster lung cells (V79) using quantitative proteomics approach incorporating TMT isotope tags (35). V79 cells were used because DPC repair is less efficient in this cell line compared with that in human cells (36). Following MMS treatment (25 mM MMS, 37 °C, 3 h) of V79 cells (in triplicate), genomic DNA containing DPCs was isolated by modified phenol–chloroform methodology (37). Control cells (in triplicate) were treated with buffer only. To identify the proteins within the DPCs, we took advantage of their thermal lability. Equal amounts of DNA containing DPCs from the cells without or with MMS treatment were heated (70 °C, 1 h). Following DNA precipitation with cold ethanol, the released proteins were collected, digested by trypsin, labeled by tandem mass tags (TMTs), and analyzed by nano–HPLC-NSI+-HRMS/MS. We found that both histones H2A and H2B participated in cross-linking to DNA, with DPC levels increasing 23- and 28-fold, respectively, following MMS treatment (Fig. 7 A and B) relative to controls without MMS treatment.
Fig. 7.
DPC formation in MMS-treated V79 cells. The normalized abundance of thermally released H2A (A) and H2B (B) within DPCs isolated from V79 cells. The protein abundance under each condition was normalized to that without MMS and NaBH4 treatment. (C) The percentage of DPCMdG in MMS-treated cells detected as the percentage of the thermally released histone proteins after NaBH4 treatment compared with those released without NaBH4 treatment. The data are the average ± SD of three experiments.
Although these data clearly indicated the formation of DPCs with histone proteins in MMS-treated cells, the products could be attributed to reaction with MdG (DPCMdG) and/or AP sites (DPCAP). DPCMdG and DPCAP were distinguished by utilizing their different thermal stabilities following NaBH4 treatment (SI Appendix, Fig. S10A). NaBH4 reduces the Schiff base within DPCAP, which is then stable upon heating. However, DPCMdG does not react with NaBH4 and the histone proteins are released after heating due to their reversible formation. Suitable conditions were established using isolated DPCs from NCPs containing AP or MdG at position 123 (SI Appendix, Fig. S10B). Following NaBH4 treatment (0.1 M, 4 °C, 1 h) and heating (70 °C, 1 h), no decomposition was observed in DPCAP123 while DPCMdG123 was completely decomposed. A similar strategy was used to determine the amounts of DPCMdG in MMS-treated cells. Genomic DNA isolated from MMS-treated or untreated cells (n = 3) were split into two portions, one of which was treated with NaBH4 to stabilize DPCAP. All four sample groups (control, NaBH4 control, MMS, MMS plus NaBH4) were subjected to quantitative proteomics with TMT tags (Fig. 7C). We found that following NaBH4 treatment, ∼75% of H2A and ∼90% of H2B DPCs were thermally released, indicating that the majority of the DPCs in MMS-treated cells were from MdG, with the remaining conjugates forming at AP sites.
Discussion
DNA alkylation has important biological effects and consequently has been studied for decades. MdG is the major product from reaction of DNA with a variety of methylating agents that act as carcinogens and/or anticancer agents, and persists in cells and the livers of mice and rats. In duplex DNA, MdG tautomers base pair with nucleotides other than dC (12). The stabilized, 2′-fluorinated alkylation product also partially blocks replication by DNA polymerase β (38). However, there is no evidence that tautomerization leads to a decrease in fidelity during bypass by a DNA polymerase. Consequently, its biochemical contribution has been largely attributed to the formation of abasic sites via hydrolysis. The half-life of this process in free DNA varies, but is on the order of days under physiologically relevant conditions, and importantly is on the same timescale as the MdG in vivo lifetime.
The rate constants for hydrolysis (kHyd) in NCPs were compared with those in free DNA. Measuring hydrolysis in MMS-treated NCPs encompasses a heterogeneous collection of alkylated nucleotides with respect to nucleotide identity (dA, dG), position alkylated within a particular nucleotide (e.g., N3, N7), as well as the position within the NCP. We directly compared hydrolysis rates on globally alkylated DNA by carrying out the MMS alkylation on NCPs, followed by liberation of the DNA from an aliquot of the sample. Depurination of the heterogeneously alkylated DNA was approximately one-half as fast within the NCP as in free DNA, suggesting that the positively charged octameric core of histone proteins stabilizes the DNA against hydrolysis. Examining hydrolysis in “synthetic” NCPs containing a single MdG at a specific position indicated that proximity of the positively charged, alkylated nucleotides to the lysine- and arginine-rich, disordered N-terminal histone tails was a major source of the decreased rate constants. MdG positions (e.g., 89, 123) close to where histone tails protrude from the octameric core experienced larger decreases in hydrolysis rate constants than those more distal (e.g., positions 73, 158) when transformed from free DNA to NCPs. Mutagenesis experiments on the histone proteins corroborated the influence of positively charged amino acids proximal to MdG.
The physical basis of the slower MdG depurination is unclear. Hydrolysis of N7-protonated AMP and molecules such as MdG is accepted to occur by a unimolecular mechanism (SN1) in which an oxocarbenium ion (X, Scheme 2) is formed in the rate-determining step (11, 39). Although the overall positive charge is maintained in the rate-determining step, it is dispersed over a greater volume. The rate constants of reactions that undergo charge redistribution in the rate-determining step respond to changes in the polarity of the reaction environment. Increasing the polarity of the reaction environment should stabilize the ground state more than the transition state of the MdG hydrolysis reaction, resulting in a decreased reaction rate constant. The modest increase in the MdG hydrolysis rate constant observed when a single, proximal phosphate was replaced by a neutral methyl phosphonate in free DNA is consistent with this prediction (40). Condensation of DNA into a NCP neutralizes a significant portion of the negative charge in the nucleic acid, creating a less polar environment (41). Hence, we would have expected the rate constant for MdG depurination to be greater in the NCP than in free DNA. Why we see the opposite trend warrants investigation.
Scheme 2.
MdG hydrolysis via a unimolecular (SN1) mechanism.
Given that there is no evidence for MdG being mutagenic, decreased depurination in NCPs (and presumably cells) relative to that in free DNA may reduce the toxicity of the responsible methylation agents by decreasing the yield of cytotoxic and mutagenic AP sites. This point is further accentuated by the observation that the greatest decrease in AP formation rates is observed in the vicinity of the histone tail, because the lysine residues in these same tails are responsible for converting AP sites to single-strand breaks (14, 16).
The long lifetime of MdG provides an opportunity for other chemical pathways to compete with depurination. We discovered that nucleosomal MdG forms reversible DPCs with histones. Isolated MdG-histone DPCs revert to the alkylated DNA over the course of 1–2 d at 37 °C. DPC instability precluded their characterization by standard methods, such as LC-MS/MS analysis of digested material. The proposed structure of the MdG-lysine adduct is based upon DPC reactivity, its reversible formation, and the effects of histone modifications on cross-link yield. DPC formation via nucleophilic attack at the C8 position of MdG is also consistent with the formation of MeFapy•dG at high pH, whose formation is initiated by nucleophilic hydroxide addition to the same position. MS experiments conducted with MMS-treated mammalian cells supported our results, indicating that >75% of DPCs originate from direct attack of the ε-amino group of lysine residues on histones on the C8 of MdG and the remaining DPCs form at abasic sites (Fig. 7).
DPCs are produced by a variety of bifunctional alkylating agents. However, DPCs from DNA alkylated by a monofunctional agent are not well documented. DPCs formed via reaction of DNA with the natural product psorospermin may also arise from the corresponding N7-alkylated 2′-deoxyguanosine (42). Of greater relevance is that, more than 40 years ago, Grunicke et al. (43) separated DNA from proteins via phenol extraction following treatment of Ehrlich ascites tumor cells with bifunctional or monofunctional (MMS) alkylating agents. Both types of alkylating agents gave rise to significant increases in quantities of “DNA resistant to deproteinization” at the aqueous-phenol interface. However, the authors dismissed the possibility that the increased interfacial materials from either family of alkylating agents were due to DPCs because MMS yielded the same result as the bifunctional agent. The results presented herein suggest that Grunicke et al. (43) had indeed observed DPCs from MMS-treated cells.
DPCs are extremely toxic as they can completely block essential DNA transactions, including DNA replication and transcription. DPC formation in cells treated with methylating agents raises the possibility that MdG production is more than a source of abasic sites. If so, one might expect that cells with compromised DPC repair capability would be more susceptible to MMS. DNA-dependent proteases that digest DPCs have been discovered in recent years, but their contribution to repair of MMS-damaged DNA has not been examined (44–48). Although the DPCs in mammalian cells are too large to be repaired by nucleotide excision repair (NER), this pathway contributes to DPC repair in bacteria and possibly following digestion to smaller peptides in other species (49–52). Studies in yeast have shown that deficiencies in NER repair enhance MMS susceptibility (53, 54). The authors provide strong evidence in support of their hypothesis that NER contributes to abasic site repair and resistance to DNA methylation. However, the data cannot exclude contribution of NER to repair of DPCs that are also formed following MMS treatment.
When considered in this context, the observations described above raise the possibility that monofunctional alkylating agents, such as MMS, MNU, and temozolomide may indirectly produce a family of lesions, DPCs, that have not previously been considered to be part of their product profile. DPCs are biologically significant, as they are potent blocks of replication and transcription. DPC formation from MdG and possibly other lesions generated in cells by monofunctional alkylators raises the possibility of alternate sources of the biological effects of such agents. It also raises the question of whether chemists can increase DPC yield in cellular DNA and/or exploit their formation to improve the efficacy of drugs such as temozolomide.
Materials and Methods
Preparation of 145-mer DNA Containing Site-Specific MdG.
Chemically synthesized oligonucleotides (SI Appendix, Fig. S1A) were enzymatically phosphorylated at their 5′-termini in a reaction containing 4 nmol of DNA, 1× PNK buffer, 2 mM ATP, and 50 units of T4 PNK at 37 °C for 4 h. The phosphorylated oligonucleotides were combined with the appropriate scaffolds (6 nmol) (SI Appendix, Fig. S1A) in 1× Sequenase buffer and hybridized by heating at 90 °C for 2 min, followed by slow cooling to room temperature. MdGTP (470 µM) and Sequenase (13 units) were added, and the mixture was incubated at room temperature for 2 h. The reaction mixture was mixed with 95% formamide and purified by a 20% denaturing PAGE gel at 4 °C. The product band was excised from the gel, and the DNA was eluted overnight in 3 mL of elution buffer at 4 °C. The slurry was briefly spun in a centrifuge, and the supernatant was filtered using a Poly-Prep column (Bio-Rad). The DNA was desalted using a C18 Sep-Pak column. The eluted DNA (∼2 nmol) was flash frozen in liquid nitrogen, dried by speed vacuum at low temperature, and resuspended in water. All oligonucleotides (1.5 nmol), except the 5′-terminal fragment and those containing MdG (SI Appendix, Fig. S1B), were separately treated with T4 PNK (50 units) in 1× T4 DNA ligase buffer at 37 °C for 4 h. Every oligonucleotide (1.5 nmol, SI Appendix, Fig. S1B), except that containing MdG and scaffolds (2 nmol, SI Appendix, Fig. S1B) were combined and heated at 95 °C for 2 min, followed by chilling on ice. The 5′-phosphorylated MdG containing oligonucleotide (1.5 nmol) was added, and the mixture was incubated at room temperature for 2 h. T4 DNA ligase (3,600 units) was added, and the mixture was incubated overnight at 16 °C. Following phenol extraction, the supernatant was mixed with 95% formamide and purified by 8% denaturing PAGE gel at 4 °C. The product band was excised from the gel and isolated as described above. The ligated product was concentrated and buffer exchanged extensively with water using a 10K Amicon Ultra membrane at 4 °C. The DNA (∼350 pmol) was stored in H2O at −80 °C.
Determination of AP and DPC Yields from MdG Containing NCPs.
5′-32P–labeled NCPs were incubated at 37 °C. Aliquots were removed at appropriate times, immediately frozen in dry ice, and stored at −80 °C. After the final time point, to determine AP yields, one-half of each aliquot was treated with 100 mM NaOH at 37 °C for 30 min, neutralized with HCl, and treated with proteinase K (8 units) at room temperature for 30 min. The cleaved DNA products were analyzed by 10% denaturing PAGE. To determine the amounts of DPCs, one-half of each aliquot was analyzed by 10% SDS/PAGE at 4 °C.
Distinguishing Between DPCs Formed from AP Sites and MdG by Denaturing PAGE Analysis of DNA.
NCPs containing AP123 were generated (17) and incubated at 37 °C. NCPs containing MdG89 or MdG123 were incubated at 37 °C, and aliquots were removed at the appropriate times. Following analysis by 10% SDS/PAGE at 4 °C, the DPC bands were excised from the gel and eluted at 4 °C for 12 h with 500 µL of buffer containing 10 mM Hepes, pH 7.5, 0.2 M NaCl, and 0.1% SDS. The DPCs were isolated from the gel as described above for DNA concentrated using a 10K Amicon Ultra membrane. Following extensive buffer exchange at 4 °C with 10 mM Hepes buffer (pH 7.5), the DPCs were treated with proteinase K (8 units) at room temperature for 5 min, and the DNA samples were analyzed by 10% denaturing PAGE.
DPC Determination in MMS-Treated Cells.
V79 Chinese hamster lung cells were plated in 15-cm dishes with the density of 3 × 106 and cultured until 90% confluence. The cells were treated with 25 mM MMS for 3 h at 37 °C. Untreated (control) cells were grown side-by-side. Genomic DNA containing DPCs was isolated as described previously (37) and resuspended in Hepes buffer (10 mM, pH 7.5). The DNA concentration was determined by dG analysis by HPLC-UV. DNA (30 µg) was treated with NaBH4 (0 or 0.1 M) at 4 °C for 1 h. The NaBH4-treated samples were quenched with acetic acid. All sample groups (control, NaBH4 control, MMS, MMS plus NaBH4), in triplicate, were subjected to neutral thermal hydrolysis (1 h, 70 °C) followed by ethanol precipitation. The supernatants were collected and dried, and the residues were resuspended in Hepes buffer (100 mM, pH 8.0). The salts were removed by extensive buffer exchange with ammonium bicarbonate buffer (25 mM, pH 8.0) using 3K Amicon filters. The samples were concentrated to a final volume of ∼200 µL. Trypsin (1 µg) was then added directly into the filters, and enzymatic digestion was conducted at 37 °C for overnight. The digested peptides were recovered and concentrated to dryness in glass MS vials. The dried samples were resuspended in Hepes (100 mM, pH 8.0) and labeled by TMT 6-plex reagent (Thermo Scientific). After mixing the labeled peptides, the samples were dried and desalted through a C18 column before nano–LC-MS analysis (Thermo Orbitrap Fusion). The raw data were searched against UniProt Chinese hamster database and the histone protein abundances were quantified with Thermo Scientific Proteome Discoverer 2.1 software.
Supplementary Material
Acknowledgments
We thank Prof. Colin Campbell for V79 cells; Profs. Greg Bowman, Yue Chen, Bertrand Garcia Moreno, and Garegin Papoian for helpful discussions; and Daniel Laverty for critically reading the manuscript. We thank the reviewers for helpful comments and for bringing ref. 42 to our attention. We are grateful for support from the National Institute of General Medical Sciences (Grant GM-063028) and the National Institute of Environmental Health Sciences (Grant ES-023350).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1813338115/-/DCSupplemental.
References
- 1.Lundin C, et al. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci USA. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Groth P, et al. Methylated DNA causes a physical block to replication forks independently of damage signalling, O6-methylguanine or DNA single-strand breaks and results in DNA damage. J Mol Biol. 2010;402:70–82. doi: 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Menke M, Meister A, Schubert I. N-Methyl-N-nitrosourea-induced DNA damage detected by the comet assay in Vicia faba nuclei during all interphase stages is not restricted to chromatid aberration hot spots. Mutagenesis. 2000;15:503–506. doi: 10.1093/mutage/15.6.503. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Stevens MFG, Bradshaw TD. Temozolomide: Mechanisms of action, repair and resistance. Curr Mol Pharmacol. 2012;5:102–114. doi: 10.2174/1874467211205010102. [DOI] [PubMed] [Google Scholar]
- 6.Pretto F, Neri D. Pharmacotherapy of metastatic melanoma: Emerging trends and opportunities for a cure. Pharmacol Ther. 2013;139:405–411. doi: 10.1016/j.pharmthera.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 7.Fu D, Calvo JA, Samson LD. Balancing repair and tolerance of DNA damage caused by alkylating agents. Nat Rev Cancer. 2012;12:104–120. doi: 10.1038/nrc3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Delft JHM, van den Ende AMC, Keizer HJ, Ouwerkerk J, Baan RA. Determination of N7-methylguanine in DNA of white blood cells from cancer patients treated with dacarbazine. Carcinogenesis. 1992;13:1257–1259. doi: 10.1093/carcin/13.7.1257. [DOI] [PubMed] [Google Scholar]
- 9.Frei JV, Swenson DH, Warren W, Lawley PD. Alkylation of deoxyribonucleic acid in vivo in various organs of C57BL mice by the carcinogens N-methyl-N-nitrosourea, N-ethyl-N-nitrosourea and ethyl methanesulphonate in relation to induction of thymic lymphoma. Some applications of high-pressure liquid chromatography. Biochem J. 1978;174:1031–1044. doi: 10.1042/bj1741031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margison GP, Capps MJ, O’Connor PJ, Craig AW. Loss of 7-methylguanine from rat liver DNA after methylation in vivo with methylmethanesulphonate or dimethylnitrosamine. Chem Biol Interact. 1973;6:119–124. doi: 10.1016/0009-2797(73)90078-1. [DOI] [PubMed] [Google Scholar]
- 11.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkylguanine residues in DNA. Chem Res Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 12.Kou Y, Koag MC, Lee S. N7 methylation alters hydrogen-bonding patterns of guanine in duplex DNA. J Am Chem Soc. 2015;137:14067–14070. doi: 10.1021/jacs.5b10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mattes WB, Hartley JA, Kohn KW. Mechanism of DNA strand breakage by piperidine at sites of N7-alkylguanines. Biochim Biophys Acta. 1986;868:71–76. doi: 10.1016/0167-4781(86)90088-6. [DOI] [PubMed] [Google Scholar]
- 14.Sczepanski JT, Wong RS, McKnight JN, Bowman GD, Greenberg MM. Rapid DNA-protein cross-linking and strand scission by an abasic site in a nucleosome core particle. Proc Natl Acad Sci USA. 2010;107:22475–22480. doi: 10.1073/pnas.1012860108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou C, Sczepanski JT, Greenberg MM. Histone modification via rapid cleavage of C4′-oxidized abasic sites in nucleosome core particles. J Am Chem Soc. 2013;135:5274–5277. doi: 10.1021/ja400915w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sczepanski JT, Zhou C, Greenberg MM. Nucleosome core particle-catalyzed strand scission at abasic sites. Biochemistry. 2013;52:2157–2164. doi: 10.1021/bi3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou C, Sczepanski JT, Greenberg MM. Mechanistic studies on histone catalyzed cleavage of apyrimidinic/apurinic sites in nucleosome core particles. J Am Chem Soc. 2012;134:16734–16741. doi: 10.1021/ja306858m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhou C, Greenberg MM. Histone-catalyzed cleavage of nucleosomal DNA containing 2-deoxyribonolactone. J Am Chem Soc. 2012;134:8090–8093. doi: 10.1021/ja302993h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weng L, Greenberg MM. Rapid histone-catalyzed DNA lesion excision and accompanying protein modification in nucleosomes and nucleosome core particles. J Am Chem Soc. 2015;137:11022–11031. doi: 10.1021/jacs.5b05478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X, Muller JG, Ye Y, Burrows CJ. DNA-protein cross-links between guanine and lysine depend on the mechanism of oxidation for formation of C5 vs C8 guanosine adducts. J Am Chem Soc. 2008;130:703–709. doi: 10.1021/ja077102a. [DOI] [PubMed] [Google Scholar]
- 21.Perrier S, et al. Characterization of lysine-guanine cross-links upon one-electron oxidation of a guanine-containing oligonucleotide in the presence of a trilysine peptide. J Am Chem Soc. 2006;128:5703–5710. doi: 10.1021/ja057656i. [DOI] [PubMed] [Google Scholar]
- 22.Nakano T, et al. DNA-protein cross-link formation mediated by oxanine. A novel genotoxic mechanism of nitric oxide-induced DNA damage. J Biol Chem. 2003;278:25264–25272. doi: 10.1074/jbc.M212847200. [DOI] [PubMed] [Google Scholar]
- 23.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 24.Vasudevan D, Chua EYD, Davey CA. Crystal structures of nucleosome core particles containing the “601” strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 25.Dyer PN, et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 2004;375:23–44. doi: 10.1016/s0076-6879(03)75002-2. [DOI] [PubMed] [Google Scholar]
- 26.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 27.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 28.Kuduvalli PN, Townsend CA, Tullius TD. Cleavage by calicheamicin gamma 1I of DNA in a nucleosome formed on the 5S RNA gene of Xenopus borealis. Biochemistry. 1995;34:3899–3906. doi: 10.1021/bi00012a005. [DOI] [PubMed] [Google Scholar]
- 29.Ezaz-Nikpay K, Verdine GL. The effects of N7-methylguanine on duplex DNA structure. Chem Biol. 1994;1:235–240. doi: 10.1016/1074-5521(94)90016-7. [DOI] [PubMed] [Google Scholar]
- 30.Hendler S, Fürer E, Srinivasan PR. Synthesis and chemical properties of monomers and polymers containing 7-methylguanine and an investigation of their substrate or template properties for bacterial deoxyribonucleic acid or ribonucleic acid polymerases. Biochemistry. 1970;9:4141–4153. doi: 10.1021/bi00823a017. [DOI] [PubMed] [Google Scholar]
- 31.Christov PP, et al. Site-specific synthesis and characterization of oligonucleotides containing an N6-(2-deoxy-d-erythro-pentofuranosyl)-2,6-diamino-3,4-dihydro-4-oxo-5-N-methylformamidopyrimidine lesion, the ring-opened product from N7-methylation of deoxyguanosine. Chem Res Toxicol. 2008;21:2324–2333. doi: 10.1021/tx800352a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.San Pedro JMN, Beerman TA, Greenberg MM. DNA damage by C1027 involves hydrogen atom abstraction and addition to nucleobases. Bioorg Med Chem. 2012;20:4744–4750. doi: 10.1016/j.bmc.2012.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tudek B, Van Zeeland AA, Kusmierek JT, Laval J. Activity of Escherichia coli DNA-glycosylases on DNA damaged by methylating and ethylating agents and influence of 3-substituted adenine derivatives. Mutat Res. 1998;407:169–176. doi: 10.1016/s0921-8777(98)00005-6. [DOI] [PubMed] [Google Scholar]
- 34.O’Brien PJ, Ellenberger T. Human alkyladenine DNA glycosylase uses acid-base catalysis for selective excision of damaged purines. Biochemistry. 2003;42:12418–12429. doi: 10.1021/bi035177v. [DOI] [PubMed] [Google Scholar]
- 35.Thompson A, et al. Tandem mass tags: A novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 36.Chesner LN, Campbell C. A quantitative PCR-based assay reveals that nucleotide excision repair plays a predominant role in the removal of DNA-protein crosslinks from plasmids transfected into mammalian cells. DNA Repair (Amst) 2018;62:18–27. doi: 10.1016/j.dnarep.2018.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Groehler AS, IV, Villalta PW, Campbell C, Tretyakova N. Covalent DNA-protein cross-linking by phosphoramide mustard and nornitrogen mustard in human cells. Chem Res Toxicol. 2016;29:190–202. doi: 10.1021/acs.chemrestox.5b00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koag M-C, Kou Y, Ouzon-Shubeita H, Lee S. Transition-state destabilization reveals how human DNA polymerase β proceeds across the chemically unstable lesion N7-methylguanine. Nucleic Acids Res. 2014;42:8755–8766. doi: 10.1093/nar/gku554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parkin DW, Schramm VL. Catalytic and allosteric mechanism of AMP nucleosidase from primary, β-secondary, and multiple heavy atom kinetic isotope effects. Biochemistry. 1987;26:913–920. doi: 10.1021/bi00377a036. [DOI] [PubMed] [Google Scholar]
- 40.Rubinson EH, Christov PP, Eichman BF. Depurination of N7-methylguanine by DNA glycosylase AlkD is dependent on the DNA backbone. Biochemistry. 2013;52:7363–7365. doi: 10.1021/bi401195r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Materese CK, Savelyev A, Papoian GA. Counterion atmosphere and hydration patterns near a nucleosome core particle. J Am Chem Soc. 2009;131:15005–15013. doi: 10.1021/ja905376q. [DOI] [PubMed] [Google Scholar]
- 42.Permana PA, Ho DK, Cassady JM, Snapka RM. Mechanism of action of the antileukemic xanthone psorospermin: DNA strand breaks, abasic sites, and protein-DNA cross-links. Cancer Res. 1994;54:3191–3195. [PubMed] [Google Scholar]
- 43.Grunicke H, et al. Effect of alkylating antitumor agents on the binding of DNA to protein. Cancer Res. 1973;33:1048–1053. [PubMed] [Google Scholar]
- 44.Stingele J, Schwarz MS, Bloemeke N, Wolf PG, Jentsch S. A DNA-dependent protease involved in DNA-protein crosslink repair. Cell. 2014;158:327–338. doi: 10.1016/j.cell.2014.04.053. [DOI] [PubMed] [Google Scholar]
- 45.Stingele J, et al. Mechanism and regulation of DNA-protein crosslink repair by the DNA-dependent metalloprotease sprtn. Mol Cell. 2016;64:688–703. doi: 10.1016/j.molcel.2016.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaz B, et al. Metalloprotease Sprtn/Dvc1 orchestrates replication-coupled DNA-protein crosslink repair. Mol Cell. 2016;64:704–719. doi: 10.1016/j.molcel.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mórocz M, et al. DNA-dependent protease activity of human Spartan facilitates replication of DNA-protein crosslink-containing DNA. Nucleic Acids Res. 2017;45:3172–3188. doi: 10.1093/nar/gkw1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez-Mosqueda J, et al. SPRTN is a mammalian DNA-binding metalloprotease that resolves DNA-protein crosslinks. eLife. 2016;5:e21491. doi: 10.7554/eLife.21491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ide H, Shoulkamy MI, Nakano T, Miyamoto-Matsubara M, Salem AMH. Repair and biochemical effects of DNA-protein crosslinks. Mutat Res. 2011;711:113–122. doi: 10.1016/j.mrfmmm.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 50.Nakano T, et al. Nucleotide excision repair and homologous recombination systems commit differentially to the repair of DNA-protein crosslinks. Mol Cell. 2007;28:147–158. doi: 10.1016/j.molcel.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 51.Baker DJ, et al. Nucleotide excision repair eliminates unique DNA-protein cross-links from mammalian cells. J Biol Chem. 2007;282:22592–22604. doi: 10.1074/jbc.M702856200. [DOI] [PubMed] [Google Scholar]
- 52.Minko IG, et al. Initiation of repair of DNA-polypeptide cross-links by the UvrABC nuclease. Biochemistry. 2005;44:3000–3009. doi: 10.1021/bi0478805. [DOI] [PubMed] [Google Scholar]
- 53.Torres-Ramos CA, Johnson RE, Prakash L, Prakash S. Evidence for the involvement of nucleotide excision repair in the removal of abasic sites in yeast. Mol Cell Biol. 2000;20:3522–3528. doi: 10.1128/mcb.20.10.3522-3528.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Memisoglu A, Samson L. Contribution of base excision repair, nucleotide excision repair, and DNA recombination to alkylation resistance of the fission yeast Schizosaccharomyces pombe. J Bacteriol. 2000;182:2104–2112. doi: 10.1128/jb.182.8.2104-2112.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.