Abstract
Background
Non‐small cell lung cancer (NSCLC) with different EGFR mutation types shows distinct sensitivity to tyrosine kinase inhibitors (TKIs). This study developed a patho‐clinical profile‐based prediction model of TKI‐sensitive EGFR mutations.
Methods
The records of 1121 Chinese patients diagnosed with NSCLC from November 2008 to October 2014 (the development set) were reviewed. Multivariate logistic regression was conducted to identify any association between potential predictors and the classic sensitive EGFR mutations (exon 19 deletion and exon 21 L858R point mutation). A prediction index was created by assigning weighted scores to each factor proportional to a regression coefficient. Validation was made in an independent cohort consisting of 864 patients who were consecutively enrolled between November 2014 and January 2017 (the validation set).
Results
Seven independent predictors were identified: gender (female vs. male), adenocarcinoma (yes vs. no), smoking history (no vs. yes), N stage (N+ vs. N0), M stage (M1 vs. M0), brain metastasis (yes vs. no), and elevated Cyfra 21‐1 (no vs. yes). Each was assigned a number of points. In the validation set, the area under curve of the prediction index appeared as 0.698 (95% confidence interval 0.663–0.733). The sensitivity, specificity, positive and negative predictive values, and concordance were 95.0%, 32.3%, 61.4%, 85.1%, and 65.6%, respectively.
Conclusion
We developed a patho‐clinical profile‐based model for predicting TKI‐sensitive EGFR mutations. Our model may represent a noninvasive, economical choice for clinicians to inform TKI therapy.
Keywords: EGFR, mutation type, non‐small cell lung cancer, prediction model
Introduction
Lung cancer is the most common cancer and the leading cause of cancer death in China.1 Non‐small cell lung cancer (NSCLC) is reported to account for up to 90% of all lung cancer cases.2 Mutation in the EGFR gene is the most common genetic event in NSCLC, especially in adenocarcinoma (AC).3 Thus, EGFR is now one of the main targets of genotype‐directed therapy for NSCLC. Phase III trials have shown that tyrosine kinase inhibitors (TKIs) targeting EGFR, such as gefitinib and erlotinib, could improve the clinical outcomes of patients with advanced EGFR‐mutated NSCLC.4, 5 However, tumors with different EGFR mutation types show distinct sensitivity to TKIs. The exon 19 deletion (del19) and the L858R point mutation of exon 21 are two classic types of TKI‐sensitive mutations. Some mutation types, such as the 20 exon insertion, are resistant to TKIs of all generations.6 Thus, identification of the EGFR mutation type is helpful for accurate selection of TKIs. It is challenging to obtain sufficient tissue for mutation analysis in clinical work. Invasive interventions may be ineffective and unsafe, especially for patients with poor performance status. Some studies have used circulating tumor DNA (ctDNA) in plasma samples as an alternative to test EGFR mutation; however, the results were not always consistent with those from biopsy samples because of the existence of tumor heterogeneity.7, 8, 9 Cost is also an obstacle to generalization of this detection method. A series of epidemiologic studies indicated that the EGFR mutation status of NSCLC patients was related to their patho‐clinical features, including age, gender, smoking history, ethnicity, histological subtype, and stage.10, 11, 12 Girard et al. also built a nomogram to predict the existence of EGFR activating mutations.12 However, there is no patho‐clinical factor‐based model for predicting the mutation types sensitive to TKIs. Therefore, this study aimed to develop such a prediction model of the sensitive del19 and L858R mutations through retrospective analysis of a cohort of NSCLC patients. The model was then validated in an independent cohort.
Methods
Patient selection
The cohort of patients used to develop the model was defined as the development set and included patients who were pathologically diagnosed with NSCLC in the participating centers (the Sun Yat‐sen University Cancer Center and the Guangzhou Institute of Respiratory Health) between 1 November 2008 and 31 October 2014. The exclusion criteria were: (i) no record of EGFR mutation testing, and (ii) prior history of anticancer therapy.
From 1 November 2014 to 31 January 2017, an independent cohort of NSCLC patients was also enrolled from the participating centers and defined as the validation set, which was used to validate the model. The inclusion and exclusion criteria were the same as those of the development set.
The Ethics Committee of the participating centers approved the study. Written informed consent was obtained from all participants included in the study.
Diagnosis and staging
The pathological diagnosis of NSCLC was made via a biopsy of primary lung lesions or metastatic lymph nodes. The pretreatment clinical stage was determined by a computed tomography scan from chest to pelvis, magnetic resonance imaging of the head and neck, and a whole‐body bone scan. Positron emission tomography was performed to confirm suspicious distant metastatic lesions. Serum carcinoembryonic antigen (CEA) and Cyfra 21‐1 levels were routinely tested before treatment.
EGFR mutation analysis
The status and types of EGFR mutation were assessed using the amplification refractory mutation system. The human EGFR gene mutation fluorescence PCR Diagnostic Kit (Amoy Diagnostics, Xiamen, China) was used to identify the 29 most common EGFR mutations from exon 18 to 21, according to the manufacturer's protocol.
Comparability of baseline profiles
Continuous and categorical data was presented as median with range and as number with proportion (%), respectively. Comparison of baseline patho‐clinical profiles between the development and the validation sets was performed using Mann–Whitney U and chi‐square tests for continuous and categorical data, respectively.
Variables and cutoff values
The candidate variables in the model for predicting del19/L858R mutations of the EGFR gene included age, gender (male vs. female), pathology (AC vs. non‐AC), smoking history (yes vs. no), T stage (T1‐2 vs. T3‐4), N stage (N0 vs. N+), M stage (M0 vs. M1), lung metastasis (yes vs. no), brain metastasis (yes vs. no), bone metastasis (yes vs. no), CEA, and Cyfra 21‐1. For ease of use, the continuous variables in the model (age, CEA, and Cyfra 21‐1) were all altered to a binomial form. The cutoff value for age was the median age of the development set, while the cutoff values for CEA and Cyfra 21‐1 were 5.0 and 3.3 ng/mL, respectively, as recommended by the manufacturer of the assay kits (Roche Diagnostics Corp., Tokyo, Japan). The prediction ability of the variables was tested by receiver operating characteristic (ROC) analysis and confirmed by chi‐square test.
Model development
The variables exhibiting statistical significance in ROC analysis and chi‐square test were entered into multivariate logistic regression, which was used by Girard et al. to develop a similar model for predicting EGFR activating mutations of NSCLC.12 We used the backward selection (likelihood ratio) method to select the variables in the prediction model. Odd ratios (ORs) and 95% confidence intervals (CIs) of each variable in the model were calculated. The assignment of points to each variable was made based on a linear transformation of its OR to the corresponding β regression coefficient:
For the users’ convenience, the coefficient of each variable in the model was divided by the lowest β value and rounded to the nearest integer. The prediction index (PI) of a patient was defined as the sum of their points. The best cutoff value of the PI for predicting the del19/L858R mutation was also determined by ROC analysis. Internal validation of this cutoff value was validated using a chi‐square test.
Model validation
The PI for each patient in the validation set was calculated. The prediction efficiency of the model was evaluated using the area under curve (AUC) in ROC analysis. The sensitivity, specificity, positive and negative predictive values, and concordance of the cutoff value established in the development set were also calculated.
All statistical analyses were performed using SPSS version 23.0 (IBM Corp., Armonk, New York, US). A two‐sided P value of < 0.05 was considered to indicate statistical significance. The study design is summarized in Figure 1.
Figure 1.
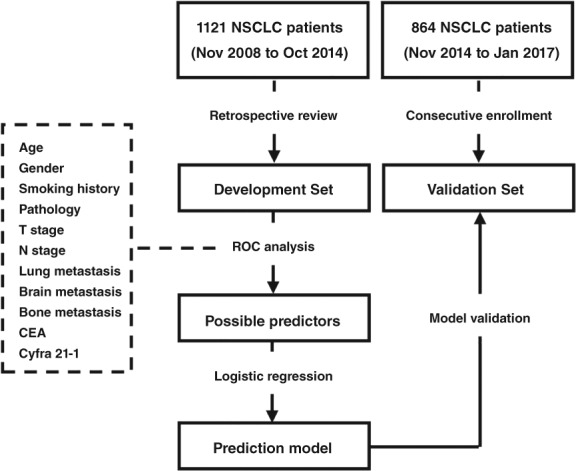
The study process. CEA, carcinoembryonic antigen; NSCLC, non‐small cell lung cancer; ROC, receiver operating characteristic.
Results
Patient enrollment
A total of 1121 patients were enrolled into the development set and 864 consecutive patients into the validation set. There were no differences between the two cohorts in age, gender, pathological classification, smoking history, clinical stage, EGFR mutation types, metastatic sites, or pretreatment serum levels of CEA or Cyfra 21‐1 (Table 1). In other words, the baseline patho‐clinical characteristics were balanced between the two cohorts. The median age of the development set was 61 (range: 22–92) years old, which was defined as the cutoff value of age during the development of our model.
Table 1.
Patho‐clinical profiles of the development and validation sets
| Factors | Development set (n = 1121) | Validation set (n = 864) | P |
|---|---|---|---|
| Age (years) | 61 (22–92) | 61 (19–89) | 0.844 |
| Gender | 0.507 | ||
| Male | 606 (54.1%) | 480 (55.6%) | |
| Female | 515 (45.9%) | 384 (44.4%) | |
| Pathology | 0.397 | ||
| AC | 938 (83.7%) | 735 (85.1%) | |
| Non‐AC | 183 (16.3%) | 129 (14.9%) | |
| Smoking history | 0.445 | ||
| Yes | 270 (24.1%) | 221 (25.6%) | |
| No | 851 (75.9%) | 643 (74.4%) | |
| T stage | 0.753 | ||
| T1–2 | 917 (81.8%) | 702 (81.3%) | |
| T3–4 | 204 (18.2%) | 162 (18.7%) | |
| N stage | 0.517 | ||
| N0 | 596 (53.2%) | 472 (54.6%) | |
| N+ | 525 (46.8%) | 392 (45.4%) | |
| M stage | 0.850 | ||
| M0 | 748 (66.7%) | 580 (67.1%) | |
| M1 | 373 (33.3%) | 284 (32.9%) | |
| Clinical stage | 0.376 | ||
| I | 345 (30.8%) | 282 (32.6%) | |
| II–IV | 776 (69.2%) | 582 (67.4%) | |
| EGFR | 0.699 | ||
| 19del/L858R | 601 (53.6%) | 459 (53.1%) | |
| Other mutation | 28 (2.5%) | 27 (3.1%) | |
| Wild type | 492 (43.9%) | 378 (43.8%) | |
| Lung metastasis | 0.693 | ||
| Yes | 75 (6.7%) | 54 (6.3%) | |
| No | 1046 (93.3%) | 810 (93.8%) | |
| Brain metastasis | 0.215 | ||
| Yes | 134 (12.0%) | 88 (10.2%) | |
| No | 987 (88.0%) | 776 (89.8%) | |
| Bone metastasis | 0.629 | ||
| Yes | 83 (7.4%) | 69 (8.0%) | |
| No | 1038 (92.6%) | 795 (92.0%) | |
| CEA (ng/mL) | 2.45 (0.00–8428.00) | 2.63 (0.00–6188.00) | 0.845 |
| Cyfra 21‐1 (ng/mL) | 0.00 (0.00–335.50) | 0.00 (0.00–317.90) | 0.859 |
Continuous and categorical data are presented as median with range and as number with percentage (%), respectively. AC, adenocarcinoma; CEA, carcinoembryonic antigen.
Possible predictors
The ROC curves of the candidate variables are shown in Figure S1 (see the Supporting information). The variables that exhibited statistical prognostic significance were: age (P = 0.040), gender (P < 0.001), pathology (P < 0.001), smoking history (P < 0.001), N stage (P = 0.011), M stage (P = 0.001), brain metastasis (P = 0.009), and serum Cyfra 21‐1 level (P = 0.003). The ability of these variables to predict the del19/L858R mutation was confirmed by chi‐square test (Table S1).
Model development
Gender (P < 0.001), pathology (P < 0.001), smoking history (P = 0.046), N stage (P = 0.039), M stage (P = 0.017), brain metastasis (P = 0.001), and Cyfra 21‐1 level (P < 0.001) were included as predictors in logistic regression. Age failed to show independent prognostic significance. The assignment of points is shown in Table 2.
Table 2.
Results of the logistic regression
| Variables | β value | OR | 95% CI | P | Points |
|---|---|---|---|---|---|
| Age (years) | |||||
| < 61 vs. ≥ 61 | 0.179 | 1.196 | 0.910–1.572 | 0.199 | NA |
| Gender | |||||
| Female vs. male | 0.866 | 2.376 | 1.751–3.225 | < 0.001 | 3 vs. 0 |
| Pathology | |||||
| AC vs. non‐AC | 2.401 | 10.98 | 6.535–18.52 | < 0.001 | 8 vs. 0 |
| Smoking history | |||||
| No vs. yes | 0.350 | 1.419 | 1.001–2.030 | 0.046 | 1 vs. 0 |
| N stage | |||||
| N+ vs. N0 | 0.290 | 1.337 | 1.018–1.761 | 0.039 | 1 vs. 0 |
| M stage | |||||
| M1 vs. M0 | 0.357 | 1.429 | 1.067–1.916 | 0.017 | 1 vs. 0 |
| Brain metastasis | |||||
| Yes vs. no | 0.781 | 2.183 | 1.361–3.413 | 0.001 | 3 vs. 0 |
| Cyfra 21–1 (ng/mL) | |||||
| <3.3 vs. ≥3.3 | 0.661 | 1.937 | 1.385–2.710 | < 0.001 | 2 vs. 0 |
The assignment of points to the variables was based on a linear transformation to their corresponding β regression coefficients. The coefficient of each variable was divided by 0.290 (the lowest β value in the model) and rounded to the nearest integer. AC, adenocarcinoma; CEA, carcinoembryonic antigen; CI, confidence interval; OR, odds ratio.
The PIs of the patients in the development set were calculated. The median was 12 (range: 0–19) and the cutoff value was 10 (< 10 vs. ≥ 10). The ROC curve of the PI is shown in Figure 2a.
Figure 2.
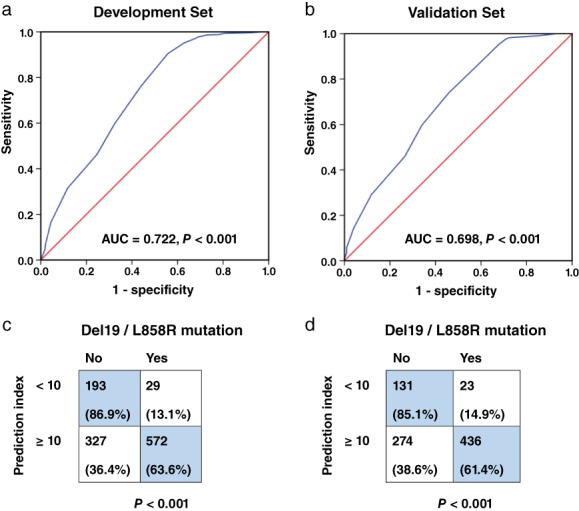
Development and validation of the model. (a,b) Receiver operating characteristic curves of the prediction index (PI) in the development and the validation sets. The area under curves (AUCs) were 0.722 (P < 0.001) and 0.698 (P < 0.001), respectively. (c,d) The PI cutoff value was validated in both sets.
Model validation
The PIs of the patients in the validation set were also calculated. The median in this set was also 12 (range: 0–19). The AUC of the PI was 0.698 (95% CI 0.663–0.733) (Fig 2b). The prediction ability of PI = 10 was confirmed by chi‐square test in both patient sets (Fig 2c,d). The sensitivity, specificity, positive and negative predictive values, and concordance in the validation set were 95.0%, 32.3%, 61.4%, 85.1%, and 65.6%, respectively.
Discussion
EGFR is a member of the receptor tyrosine kinase superfamily. EGFR overexpression is observed in > 50% of NSCLC patients and is thought to cause abnormal cellular proliferation of lung cancer.13 EGFR mutation testing is required before treating NSCLC because of the prominent effects of TKIs, particularly in patients with EGFR mutated tumors. A large‐scale investigation of physicians at general hospitals, chest hospitals, and comprehensive cancer centers located in 12 major cities throughout China found that only 9.6% of patients with advanced NSCLC underwent EGFR mutation testing.14 Tissue accessibility remains the primary barrier. In recent years, ctDNA has emerged as a safe and effective alternative. Nevertheless, high cost and rigorous requirements for lab conditions limit its generalization, especially in developed countries and areas. Hence, prediction based on an easily available patho‐clinical profile is of great clinical value. Many studies have focused on the relationship between patho‐clinical features and EGFR mutation status, but few have combined these features to build a prediction model. Girard et al. integrated age, gender, clinical stage, tobacco consumption, time since quitting smoking, and predominant pathologic subtype to develop the first model to predict EGFR activating mutations in Asian patients with AC.12 Some recent studies also tried to build prediction models of EGFR mutation on the basis of pretreatment radiomic features of NSCLC patients.15, 16, 17, 18
Although the most common mutation types, del19 and L8585R mutations, are sensitive to TKIs, some uncommon types have also been found to be resistant. The exon 20 insertion is the main cause of primary resistance to all frequently used TKIs. The T790M point mutation could confer resistance to TKIs, except osimertinib and avitinib.13, 19 Therefore, it is also important to identify the EGFR mutation type in NSCLC patients. To our knowledge, no model has been created to predict TKI‐sensitive EGFR mutations; therefore, we identified clinical and pathological factors to develop a practical model for predicting the presence of del19 and L8585R mutations in Chinese patients. Our model was based on a relatively large sample size, which is one of the main strengths of our study.
The EGFR mutation rate in our study was 56.2%: 601 (53.6%) and 459 (53.1%) patients exhibited del19 or L8585R mutations in the development and validation sets, respectively. Similar to previous studies, these two types were still the most common EGFR mutations. Through logistic regression, gender (female vs. male, OR 2.376, 95% CI 1.751–3.225), pathology (AC vs. non‐AC, OR 10.98, 95% CI 6.535–18.52), smoking history (no vs. yes, OR 1.419, 95% CI 1.001–2.030), N stage (N+ vs. N0, OR 1.337, 95% CI 1.018–1.761), M stage (M1 vs. M0, OR 1.429, 95% CI 1.067–1.916), brain metastasis (yes vs. no, OR 2.183, 95% CI 1.361–3.413), and serum Cyfra 21‐1 level (< 3.3 vs. ≥ 3.3 ng/mL, OR 1.937, 95% CI 1.385–2.710) were determined as independent predictors of del19 and L8585R mutations.
The classic factors associated with a higher rate of EGFR activating mutations are AC, female gender, and never smoker status. In Chinese patients, the EGFR mutation rate of AC is approximately 52.4%.20 By contrast, EGFR mutations are rarely detected in other pathological types of NSCLC, such as squamous cell carcinoma; only 6.9% of patients with squamous cell carcinoma harbor EGFR mutations.21 Quan et al. enrolled 354 Chinese patients with NSCLC and showed higher EGFR mutation rates in women than in men (60.13% vs. 38.81%; P = 0.029).22 According to data from the CTONG 1506 multicenter survey, tobacco consumption is also negatively associated with EGFR mutations. EGFR mutation rates in never, former, and current smokers are reported as 54.4%, 36.0%, and 30.6% (P < 0.001), respectively.23 Our study further confirmed the association between these factors and the presence of sensitive del19 and L858R mutations. Late N and M stage, and brain metastasis were also independent predictive factors of TKI‐sensitive mutations, consistent with the adverse influence of EGFR on patient prognosis.13 Additionally, Quan et al. and the CTONG 1506 study both reported that age at diagnosis was related to EGFR mutation status. Younger patients (aged < 60–65 years) seemed more likely to have EGFR mutations compared to older patients.22, 23 However, age was not an independent predictor of TKI‐sensitive mutations in our study.
CEA and Cyfra 21‐1 are well‐known serum tumor markers of NSCLC. Several recent studies have assessed their value for predicting EGFR mutation status and the therapeutic effects of TKIs. The serum CEA level in Chinese patients is not only positively associated with EGFR mutation, but also negatively associated with the efficacy of TKI therapy.24, 25 Although data on Chinese patients is lacking, studies performed in other Asian countries have shown that serum Cyfra 21‐1 level was associated with EGFR mutation and tumor response to TKIs.26, 27 Our study indicated that patients with low Cyfra 21‐1 (< 3.3 ng/mL) were more likely to harbor del19 or L858R mutations. However, despite exhibiting predictive value in univariate analyses, serum CEA level did not independently predict del19/L8585R mutation in multivariate logistic regression. These results were inconsistent with those of previous studies. Further research will help to determine the reasons for these differences.
After linear transformation of the ORs, we developed the model in which a PI was used to divide the patients into groups with different probabilities of harboring del19/L858R mutation. The value of the PI for distinguishing these two TKI‐sensitive mutations was validated by a relatively large independent cohort, which is another advantage of our study. The AUC of the PI in ROC analysis was 0.698 (95% CI 0.663–0.733). Its cutoff value from the development set (PI = 10) exhibited excellent sensitivity (95.0%), an ideal negative predictive value (85.1%), an acceptable positive predictive value (61.4%), and concordance of 65.6%.
Indeed, there were still some limitations to our study. First, in NSCLC patients, the proportion of AC is approximately 41.7%.2 The proportion in our study (84.3%) was much higher. This is largely because National Comprehensive Cancer Network clinical practice guidelines recommend routine testing for EGFR mutations in AC.28 Second, some tumor markers, such as CA‐125 and the squamous cell carcinoma antigen, were not included in the model, because of incomplete records of these markers in the development set. Therefore, we propose in generalizing our model before further validation. Additionally, the prediction accuracy of our model was limited by the efficiency of its developing methodology, logistic regression. More sophisticated machine learning techniques, such as random forest and penalized/elastic‐net regression, might help to build models with higher accuracy.
Despite the modest accuracy of the PI, we developed a practical model based on patho‐clinical profiles to predict 19del and L858R mutations in NSCLC patients. Our model contained seven well‐known, easily available predictors and was developed and validated in two independent large cohorts. These findings may provide a noninvasive, economical choice for clinicians to inform TKI therapy, especially when direct analysis of EGFR mutation types is difficult.
Disclosure
No authors report any conflict of interest.
Supporting information
Table S1. Proportion of patients with EGFR del19/L858R mutations in groups divided by different variables.
Figure S1. Receiver operating characteristic curves of the candidate variables. AUC, area under curve; CEA, carcinoembryonic antigen.
Acknowledgment
This study was supported by the Key Science and Technology Project of Guangzhou people's Livelihood (Grant number: 201803010024).
References
- 1. Chen W, Sun K, Zheng R et al Cancer incidence and mortality in China, 2014. Chin J Cancer Res 2018; 30: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Muller DC, Hodge AM, Fanidi A et al No association between circulating concentrations of vitamin D and risk of lung cancer: An analysis in 20 prospective studies in the lung cancer cohort consortium (LC3). Ann Oncol 2018; 29: 1468–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wang S, Wang Z. Meta‐analysis of epidermal growth factor receptor and KRAS gene status between primary and corresponding metastatic tumours of non‐small cell lung cancer. Clin Oncol (R Coll Radiol) 2015; 27: 30–9. [DOI] [PubMed] [Google Scholar]
- 4. Zhong WZ, Wang Q, Mao WM et al Gefitinib versus vinorelbine plus cisplatin as adjuvant treatment for stage II‐IIIA (N1‐N2) EGFR‐mutant NSCLC (ADJUVANT/CTONG1104): A randomised, open‐label, phase 3 study. Lancet Oncol 2018; 19: 139–48. [DOI] [PubMed] [Google Scholar]
- 5. Zhou C, Wu YL, Chen G et al Erlotinib versus chemotherapy as first‐line treatment for patients with advanced EGFR mutation‐positive non‐small‐cell lung cancer (OPTIMAL, CTONG‐0802): A multicentre, open‐label, randomised, phase 3 study. Lancet Oncol 2011; 12: 735–42. [DOI] [PubMed] [Google Scholar]
- 6. Castellanos E, Feld E, Horn L. Driven by mutations: The predictive value of mutation subtype in EGFR‐mutated non‐small cell lung cancer. J Thorac Oncol 2017; 12: 612–23. [DOI] [PubMed] [Google Scholar]
- 7. Weber B, Meldgaard P, Hager H et al Detection of EGFR mutations in plasma and biopsies from non‐small cell lung cancer patients by allele‐specific PCR assays. BMC Cancer 2014; 14: 294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lam DC, Tam TC, Lau KM, Wong WM, Hui CK et al Plasma EGFR mutation detection associated with survival outcomes in advanced‐stage lung cancer. Clin Lung Cancer 2015; 16: 507–13. [DOI] [PubMed] [Google Scholar]
- 9. Keppens C, Palma JF, Das PM et al Detection of EGFR variants in plasma: A multilaboratory comparison of a real‐time PCR EGFR mutation test in Europe. J Mol Diagn 2018; 20: 483–94. [DOI] [PubMed] [Google Scholar]
- 10. Shi Y, Au JS, Thongprasert S et al A prospective, molecular epidemiology study of EGFR mutations in Asian patients with advanced non‐small‐cell lung cancer of adenocarcinoma histology (PIONEER). J Thorac Oncol 2014; 9: 154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Russell PA, Barnett SA, Walkiewicz M et al Correlation of mutation status and survival with predominant histologic subtype according to the new IASLC/ATS/ERS lung adenocarcinoma classification in stage III (N2) patients. J Thorac Oncol 2013; 8: 461–8. [DOI] [PubMed] [Google Scholar]
- 12. Girard N, Sima CS, Jackman DM et al Nomogram to predict the presence of EGFR activating mutation in lung adenocarcinoma. Eur Respir J 2012; 39: 366–72. [DOI] [PubMed] [Google Scholar]
- 13. Sharma SV, Bell DW, Settleman J, Haber DA. Epidermal growth factor receptor mutations in lung cancer. Nat Rev Cancer 2007; 7: 169–81. [DOI] [PubMed] [Google Scholar]
- 14. Xue C, Hu Z, Jiang W et al National survey of the medical treatment status for non‐small cell lung cancer (NSCLC) in China. Lung Cancer 2012; 77: 371–5. [DOI] [PubMed] [Google Scholar]
- 15. Caicedo C, Garcia‐Velloso MJ, Lozano MD et al Role of [18F]FDG PET in prediction of KRAS and EGFR mutation status in patients with advanced non‐small‐cell lung cancer. Eur J Nucl Med Mol Imaging 2014; 41: 2058–65. [DOI] [PubMed] [Google Scholar]
- 16. Liu Y, Kim J, Balagurunathan Y, Li Q et al Radiomic features are associated with EGFR mutation status in lung adenocarcinomas. Clin Lung Cancer 2016; 17: 441–8.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang L, Chen B, Liu X, Song J, Fang M et al Quantitative biomarkers for prediction of epidermal growth factor receptor mutation in non‐small cell lung cancer. Transl Oncol 2018; 1: 94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lv Z, Fan J, Xu J et al Value of 18F‐FDG PET/CT for predicting EGFR mutations and positive ALK expression in patients with non‐small cell lung cancer: A retrospective analysis of 849 Chinese patients. Eur J Nucl Med Mol Imaging 2018; 45: 735–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xu J, Jin B, Chu T et al EGFR tyrosine kinase inhibitor (TKI) in patients with advanced non‐small cell lung cancer (NSCLC) harboring uncommon EGFR mutations: A real‐world study in China. Lung Cancer 2016; 96: 87–92. [DOI] [PubMed] [Google Scholar]
- 20. Pi C, Xu CR, Zhang MF et al EGFR mutations in early‐stage and advanced‐stage lung adenocarcinoma: Analysis based on large‐scale data from China. Thorac Cancer 2018; 9: 814–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sun Y, Yin X, Wen MM et al EGFR mutations subset in Chinese lung squamous cell carcinoma patients. Mol Med Rep 2018; 17: 7575–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Quan X, Gao H, Wang Z et al Epidermal growth factor receptor somatic mutation analysis in 354 Chinese patients with non‐small cell lung cancer. Oncol Lett 2018; 15: 2131–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhou Q, Song Y, Zhang X et al A multicenter survey of first‐line treatment patterns and gene aberration test status of patients with unresectable stage IIIB/IV nonsquamous non‐small cell lung cancer in China (CTONG 1506). BMC Cancer 2017; 17: 462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang Z, Yang S, Lu H. Preoperative serum carcinoembryonic antigen levels are associated with histologic subtype, EGFR mutations, and ALK fusion in patients with completely resected lung adenocarcinoma. Onco Targets Ther 2017; 10: 3345–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao Y, Song P, Li H, Jia H, Zhang B. Elevated serum CEA levels are associated with the explosive progression of lung adenocarcinoma harboring EGFR mutations. BMC Cancer 2017; 17 (1): 484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cho A, Hur J, Moon YW et al Correlation between EGFR gene mutation, cytologic tumor markers, 18F‐FDG uptake in non‐small cell lung cancer. BMC Cancer 2016; 16: 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tanaka K, Hata A, Kaji R et al Cytokeratin 19 fragment predicts the efficacy of epidermal growth factor receptor‐tyrosine kinase inhibitor in non‐small‐cell lung cancer harboring EGFR mutation. J Thorac Oncol 2013; 8: 892–8. [DOI] [PubMed] [Google Scholar]
- 28. NCCN Clinical Practice Guidelines in Oncology (NCCN Guidelines®) : Non‐small cell lung cancer. Version 4. http://nccn.org 2018. Retrieved from https://www.nccn.org/patients/guidelines/cancers.aspx#nsclc [accessed 30 May 2018]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Proportion of patients with EGFR del19/L858R mutations in groups divided by different variables.
Figure S1. Receiver operating characteristic curves of the candidate variables. AUC, area under curve; CEA, carcinoembryonic antigen.


