Abstract

In the many scientific endeavors that are driven by organic chemistry, unambiguous identification of small molecules is of paramount importance. Over the past 50 years, NMR and other powerful spectroscopic techniques have been developed to address this challenge. While almost all of these techniques rely on inference of connectivity, the unambiguous determination of a small molecule’s structure requires X-ray and/or neutron diffraction studies. In practice, however, X-ray crystallography is rarely applied in routine organic chemistry due to intrinsic limitations of both the analytes and the technique. Here we report the use of the electron cryo-microscopy (cryoEM) method microcrystal electron diffraction (MicroED) to provide routine and unambiguous structural determination of small organic molecules. From simple powders, with minimal sample preparation, we could collect high-quality MicroED data from nanocrystals (∼100 nm, ∼10–15 g) resulting in atomic resolution (<1 Å) crystal structures in minutes.
Short abstract
From powder to structure: The cryo-electron microscopy (cryoEM) method micro-electron diffraction (MicroED) is described as a new technique for routine structural analysis providing high-resolution crystal structures from seemingly amorphous powders.
The history of organic chemistry closely parallels the development of new methods for structural characterization. The earliest studies were driven by melting point determination, and over the past 175 years more complex methods for interrogation of structure have been developed. Techniques such as polarimetry,1 UV–vis,2 and infrared spectroscopy,3 coupled with electron paramagnetic resonance,4 vibrational circular dichroism,5 circular dichroism,6 and mass spectrometry7 have been commonly employed over the years, dramatically expanding our ability to assign structures. In the past 50 years, however, the explosion of NMR spectroscopy8 and the accompanying abundance of individual NMR experiments have produced a wealth of detailed structural information for organic chemists. Indeed, NMR is a mainstay in chemistry and the most predominant method employed in both routine synthetic chemistry experiments and in advanced structural elucidation of complex small molecules. In the current state of the art, only single crystal X-ray diffraction holds a higher place in terms of precision, producing unequivocal structural information about the position, orientation, connectivity, and placement of individual atoms and bonds within a given molecule.
For decades, small molecule X-ray analysis has been the definitive tool for structural analysis.9 This technique, however, is not without limitations. The process is considered by many an art, where the production of high-quality crystals suitable for X-ray diffraction requires uncodified “tricks of the trade” as well as a certain amount of luck! Additionally, even after a substance has been successfully crystallized, there is no guarantee that the particular crystal form will be amenable to X-ray diffraction. Since crystal growth is both a slow and arduous process, X-ray diffraction has not been an effective tool for rapid, on-the-fly structural determination of small molecules. For this reason, X-ray diffraction is generally not implemented as a routine analytical tool for the practicing organic chemist, despite the fact that the structural data provided are far superior to any other characterization method to date.
Results and Discussion
Herein, we employ the recently developed electron cryo-microscopy (CryoEM) method microcrystal electron diffraction (MicroED)10 to address the long-standing need for fast and reliable structure determination in organic chemistry. Recently, electron diffraction was used to solve the structure of a methylene blue derivative, although no scope studies were undertaken to allow the reader to assess the applicability of the methodology.11 Moreover, a specialized detector was used for their experiments, limiting the broad adaptability of their approach to the wider synthetic community.11 We demonstrate that with minimal sample preparation and experiment time, simple powders and seemingly amorphous materials (in some cases, solids isolated via silica gel chromatography and rotary evaporation) could be directly used in MicroED studies, leading to rapid, high-quality structural elucidation of several classes of complex molecules with atomic resolution, in many cases better than 1 Å. Moreover, we utilize a commercially available microscope that is already in use at universities around the world. MicroED has the potential to dramatically accelerate and impact the fields of synthetic chemistry, natural product chemistry, drug discovery, and many others by delivering rapid, high-resolution atomic structures of complex, small molecules with minimal sample preparation or formal crystallization procedures.
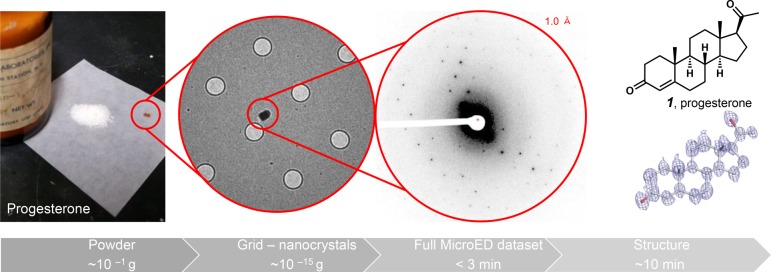
The applicability of MicroED was initially tested on the naturally occurring steroid progesterone (1) as a model system (Figure 1). The sample was obtained as a powder from chemical supplier Preparations Laboratories Inc. (we estimate the bottle to be more than 20 years old). Small quantities of the seemingly amorphous solid were transferred directly from the bottle onto glass cover slides and ground between another slide to produce a fine powder. The powder was then deposited on a holey carbon–copper grid, cooled to liquid nitrogen temperatures, and transferred to a cryoelectron microscope operating at an acceleration voltage of 200 kV (Thermo Fisher Talos Arctica). An overview of the preparation is shown in Figure 1. Upon imaging, thousands of nanocrystals were easily discernible on the grid surface providing ample nanocrystals to investigate for diffraction. Typically, for samples such as these, the vast majority of nanocrystals diffracted to ∼1 Å resolution or better (Figure 1). Through continuous rotation of the sample stage, 140 degrees of diffraction data could be collected from a single nanocrystal,12 while the improved autoloader and piezo stage of the Talos Arctica allowed us to travel through the zero degree point without introducing errors in crystal position. Typically, the stage was rotated at approximately 0.6 deg/s, and an entire data set was collected in less than 3 min as a movie (Video 1) using a bottom mount CetaD CMOS detector (Thermo Fisher) fitted with a thick scintillator for diffraction studies. Software adapted from previous studies13 was used to convert the diffraction movie frames into SMV format for expeditious processing in the readily available XDS software package commonly used for X-ray crystallography.13 By collecting data from just a single nanocrystal, the structure of steroid 1 was resolved to an impressive 1 Å resolution. The entire process, from bottle to structure, was easily accomplished in less than 30 min.
Figure 1.
Process of applying MicroED to small molecule structural analysis. Here commercial progesterone (1) was analyzed, and an atomic resolution structure was determined at 1 Å resolution. Grid holes are 1 μm in diameter.
Encouraged by these results, we wanted to investigate a wide range of natural products to fully explore the scope and applicability of this powerful structural determination method for small molecules (Figure 2A). The Talos Arctica was particularly amenable to our studies as it is capable of storing up to 12 different grids at once, providing effortless swapping of sample grids for rapid investigation of multiple compounds. Once a reliable sample prep routine had been established, over-the-counter medications were purchased from local pharmacies for investigation. Tablets of CVS-branded acetaminophen and Kroger-branded ibuprofen were crushed using a mortar and pestle, and the ground powder was placed on electron microscopy grids as described above. Despite the heterogeneity of such pharmaceuticals, which typically include a multitude of coatings, binders, and other formulation agents, we were astonished to obtain such clearly resolved atomic resolution structures of both acetaminophen (2) and ibuprofen (3) in rapid succession. Just as impressively, structures of the sodium channel blocker carbamazepine (4) and the macrocyclic polypeptide antibiotic thiostrepton (5) were also determined from seemingly amorphous powders used as received from Sigma-Aldrich. We went on to further study several commercially available natural products and derivatives. Once again, compounds were used as received, without any crystallization, to yield atomic resolution structures of biotin (6), ethisterone (7), cinchonine (8), and brucine (9). Of the 11 different commercial bioactives examined, all 11 yielded processable MicroED data. Of those 11 compounds, 10 were amenable to rapid structure determination by direct methods,14 while one was determined by molecular replacement.15 As mentioned previously, all structures were obtained without any crystallization attempts or chemical modifications to compounds examined. While these pharmaceutical and commercial natural products were likely recrystallized for purification purposes by the manufacturer, the powders examined by MicroED possessed nanocrystals a billionth the size (∼100 nm) of crystals typically needed for X-ray crystallography. This was powder to structure.
Figure 2.
Different types of small molecules solved by MicroED. (A) Several pharmaceuticals, vitamins, commercial natural products, and synthetic samples resolved through MicroED. (B) Example of an amorphous film utilized in this study leading to 1 Å resolution data. (C) Protons could be observed for several compounds through MicroED. Green density are Fo – Fc maps showing positive density belonging to hydrogen atoms of the molecule.
Next we decided to investigate compounds that were never crystallized, but instead were purified by flash column chromatography. Since silica gel chromatography is the most common method of purification in early stage research for complex molecules in drug discovery, natural product isolation efforts, and synthesis efforts in general, we were interested to see whether solid samples prepared in such a way would be amenable to analysis by MicroED. Four compounds, purified by chromatographic methods, were collected from our laboratories, and samples of these seemingly amorphous solids were analyzed. Here, two of four compounds diffracted, yielding atomic resolution structures at or below 1 Å (10 and 11, Figure 2A,B). While the success rate for these compounds was 50%, it is worthy to note that no crystallization procedures were employed in the isolation of these materials. Notably, (+)-limaspermidine (10), an alkaloid natural product synthesized by our laboratories,16 was resolved from a residue of only milligram quantities of material following flash chromatography and rotary evaporation from a scintillation vial (Figure 2B). Furthermore, while it is extremely challenging to observe protons in X-ray structures, electrons interact with matter more strongly than X-rays and are affected by charge, making them relatively common to observe in MicroED data.17−21 For all structures resolved from our samples, at least some, if not most, protons could be observed on the molecule. In particular, the density maps obtained for limaspermidine (10) and carbamazepine (4) after refinement showed protons associated with almost all atoms in these molecules (Figure 2C).
Astounded by the ease with which such high-quality data were obtained and the apparent generality of MicroED to small molecules, we undertook studies to examine heterogeneous samples (mixtures of compounds). In the case of heterogeneous samples, single crystal X-ray diffraction precludes the study of mixtures, and NMR is poorly suited for this task. For this experiment, mixtures of four compounds (4, 6, 8 and 9, cf. Figure 3) were crushed together and deposited on a holey copper–carbon. Several crystal forms belonging to the different compounds in the mixture were visually identified on the grids (Figure 3). MicroED data were collected from several nanocrystals, and the identity of each species was resolved within minutes by confirmation of unit cell parameters. After compound identification, atomic resolution structures could be rapidly determined for all small molecules present in the mixture (Figure 3). (Note: no unexpected or unusually high safety hazards were encountered.)
Figure 3.
Identification of compounds from heterogeneous mixtures. An EM grid was prepared as above with biotin, brucine, carbamazepine, and cinchonine powders mixed together. All four compounds identified by unit cell parameters using MicroED data from within the same grid square. All structures were solved to ∼1 Å resolution. Grid holes are 2 μm in diameter.
The results described here introduce a powerful new characterization tool into the organic chemist’s toolbox. While MicroED was initially developed for structure determination of biological materials such as proteins in a frozen, hydrated state,22,23 we demonstrate that cross pollination of macromolecular structural methods of CryoEM are powerful tools for chemical synthesis, drug characterization, and drug discovery. Prior to this work, MicroED has allowed for the structural characterization of several proteins from crystals which had generally been unsuitable for X-ray crystallography due to their small size or morphologies.22−24 Despite this success, the MicroED method has largely gone unnoticed in the small molecule communities. On the basis of our findings, we anticipate that MicroED will be enthusiastically received by many types of small molecule chemists in both academia and industry. Here we have shown that a variety of seemingly amorphous solid materials can lead to rapid atomic resolution structure determination by MicroED with little or no additional sample preparation or crystallization. The fact that a solid film in a flask, following solvent removal from a flash chromatography purification, can lead to an atomic resolution molecular structure, is evidence that MicroED will likely have a profound effect on the structural characterization work-flow of organic chemists. Although the past 50 years have seen huge advances in the state of the art, no completely new techniques have been introduced that alter the routine structural interrogation of organic substances. NMR,8 IR,3 UV–vis,2 and X-ray diffraction9 have been routinely in place since the 1960s and are still utilized today as the most common methods for structure determination in chemistry. We believe that electron diffraction is potentially the next big advance in the field and are enthusiastic about the prospects of expanding its utility as a routine analytical technique for chemists.25,26
Post Preprint Addendum and Background
MicroED was developed for the determination of atomic resolution protein structures from submicron thick, frozen-hydrated crystals that are typically too small for X-ray diffraction.22,23 In MicroED, crystals are illuminated by an extremely low dose (typically <0.01 e–/Å2/s) electron beam, while the crystals are continuously rotated and diffraction is collected on a fast camera as a movie.13 MicroED data are then processed using broadly available software for X-ray crystallography without the need for specialized software for structure analysis and refinement.12
Other electron diffraction methods include automated diffraction tomography27 (ADT) and rotation electron diffraction28 (RED). These methods differ from MicroED in the sampling of diffraction space, through tilt series and/or precession. The broad applicability of MicroED has been demonstrated since its conception10,12 as structures of large globular proteins,23 small proteins,22 peptides,19 membrane proteins,29 and inorganic compounds18 have been successfully determined. In many of these examples, hydrogens were observed and reported with the first example in 201519 followed by an independent report from Palatinus and co workers two years later.30 Most recently, the structure of a small organic molecule, carbamezapine, was determined by MicroED to sub-Angstrom resolution.24
Acknowledgments
We thank Profesor Doug Rees (Caltech), Professor Bil Clemons (Caltech), and Dr. Michael Sawaya (UCLA) for useful discussions. We thank Byungkuk Yoo and Michael Takase (Caltech) for technical assistance with data analysis. We thank Beau Pritchett and Hendrik Klare for providing synthetic samples.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acscentsci.8b00760.
Accession Codes
Data and materials availability: MicroED density maps have been deposited to the PDB (thiostrepton, 6MXF), EMDB (EMD-9282, EMD-9284, EMD-9285, EMD-9286, EMD-9287, EMD-9288, EMD-9289, EMD-9290, EMD-9291, EMD-9292), and CCDC (1876036, 1876037, 1876038, 1876039, 1876040, 1876041, 1876042, 1876043, 1876044, 1876045).
Author Contributions
# C.G.J. and M.W.M. contributed equally. C.G.J. performed experiments, developed the sample preparation techniques, refined structural data, prepared figures, and assisted with manuscript preparation. M.W.M performed experiments, developed the sample preparation techniques, collected data, refined structural data, prepared figures, and assisted with manuscript preparation. T.F. performed experiments. J.H. wrote the software for image conversion, participated in data analysis, refinement, and structure determination. J.A.R. performed experiments, developed the sample preparation techniques, and assisted with manuscript preparation. B.M.S. conceived of the project, designed experiments, and assisted with manuscript preparation. H.M.N. conceived of the project, designed experiments, performed experiments, developed the sample preparation techniques, and assisted with manuscript and figure preparation. T.G. performed experiments, collected data, developed the sample prep techniques, developed microscope data collection parameters, provided the microscope and expertise, and assisted in manuscript and figure preparation.
C.G.J. would like to acknowledge the National Science Foundation for a predoctoral fellowship (DGE-1650604). B.M.S. acknowledges the NIH-NIGMS for generous funding (R01GM080269). J.A.R. is supported by DOE Grant DE-FC02-02ER63421, NIH-NIGMS Grant R35GM128867, and as a Beckman Young Investigator, a Searle Scholar, and a Pew Scholar. H.M.N. thanks The Packard Foundation, The Sloan Foundation, Pew Charitable Trusts, and the NIH-NIGMS (R35 GM128936) for generous funding. The Gonen laboratory is supported by funds from the Howard Hughes Medical Institute.
The authors declare no competing financial interest.
Supplementary Material
References
- Schreier P.; Bernreuther A.; Huffer M.. Analysis of Chiral Organic Molecules: Methodology and Applications; Walter de Gruyter: Berlin, 2011. [Google Scholar]
- Scott A. I.Interpretation of the Ultraviolet Spectra of Natural Products: International Series of Monographs on Organic Chemistry; Elsevier: Amsterdam, 2013; Vol. 7. [Google Scholar]
- Coates J.Interpretation of infrared spectra, a practical approach. In Encyclopedia of Analytical Chemistry: Applications, Theory and Instrumentation; Meyers R. A., Ed.; Wiley: New York, 2000; Vol. 12, pp 10815–10837. [Google Scholar]
- Dougherty D. A. Spin control in organic molecules. Acc. Chem. Res. 1991, 24, 88–94. 10.1021/ar00003a005. [DOI] [Google Scholar]
- Stephens P. J.; Devlin F. J.; Pan J. J. The determination of the absolute configurations of chiral molecules using vibrational circular dichroism (VCD) spectroscopy. Chirality 2008, 20, 643–663. 10.1002/chir.20477. [DOI] [PubMed] [Google Scholar]
- Berova N.; Bari L. D.; Pescitelli G. Application of electronic circular dichroism in configurational and conformational analysis of organic compounds. Chem. Soc. Rev. 2007, 36, 914–931. 10.1039/b515476f. [DOI] [PubMed] [Google Scholar]
- De Hoffmann E.Mass spectrometry. In Kirk Othmer Encyclopedia of Chemical Technology; John Wiley & Sons: New York, 2000. [Google Scholar]
- Günther H.NMR Spectroscopy: Basic Principles, Concepts and Applications in Chemistry; John Wiley & Sons: New York, 2013. [Google Scholar]
- Dunitz J. D.X-ray Analysis and the Structure of Organic Molecules; Verlag Helvetica Chimica Acta: Zürich, 1995. [Google Scholar]
- Shi D.; Nannenga B. L.; Iadanza M. G.; Gonen T. Three-dimensional electron crystallography of protein microcrystals. eLife 2013, 2, e01345. 10.7554/eLife.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruene T.; Wennmacher J. T. C.; Zaubitzer C.; Holstein J. J.; Heidler J.; Fecteau-Lefebvre A.; De Carlo S.; Müller E.; Goldie K. N.; Regeni I.; Li T.; Santiso-Quinones G.; Steinfeld G.; Handschin S.; van Genderen E.; van Bokhoven J. A.; Clever G. H.; Pantelic R. Rapid structure determination of microcrystalline molecular compounds using electron diffraction. Angew. Chem., Int. Ed. 2018, 10.1002/anie.201811318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannenga B. L.; Shi D.; Leslie A. G. W.; Gonen T. High-resolution structure determination by continuous-rotation data collection in MicroED. Nat. Methods 2014, 11, 927–930. 10.1038/nmeth.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattne J.; Reyes F. E.; Nannenga B. L.; Shi D.; de la Cruz M. J.; Leslie A. G. W.; Gonen T. MicroED data collection and processing. Acta Crystallogr., Sect. A: Found. Adv. 2015, 71, 353–360. 10.1107/S2053273315010669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldrick G. SHELXT - Integrated space-group and crystal-structure determination. Acta Crystallogr., Sect. A: Found. Adv. 2015, 71, 3–8. 10.1107/S2053273314026370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin A.; Teplyakov A. MOLREP : an Automated Program for Molecular Replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. 10.1107/S0021889897006766. [DOI] [Google Scholar]
- Pritchett B. P.; Donckele E. J.; Stoltz B. M. Enantioselective Catalysis Coupled with Stereodivergent Cyclization Strategies Enables Rapid Syntheses of (+)-Limaspermidine and (+)-Kopsihainanine A. Angew. Chem., Int. Ed. 2017, 56, 12624–12627. 10.1002/anie.201707304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q. Rev. Biophys. 1995, 28, 171–193. 10.1017/S003358350000305X. [DOI] [PubMed] [Google Scholar]
- Vergara S.; Lukes D. A.; Martynowycz M. W.; Santiago U.; Plascencia-Villa G.; Weiss S. C.; de la Cruz M. J.; Black D. M.; Alvarez M. M.; López-Lozano X.; Barnes C. O.; Lin G.; Weissker H.-C.; Whetten R. L.; Gonen T.; Yacaman M. J.; Calero G. MicroED Structure of Au146(p-MBA)57 at Subatomic Resolution Reveals a Twinned FCC Cluster. J. Phys. Chem. Lett. 2017, 8, 5523–5530. 10.1021/acs.jpclett.7b02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J. A.; Ivanova M. I.; Sawaya M. R.; Cascio D.; Reyes F. E.; Shi D.; Sangwan S.; Guenther E. L.; Johnson L. M.; Zhang M.; Jiang L.; Arbing M. A.; Nannenga B. L.; Hattne J.; Whitelegge J.; Brewster A. S.; Messerschmidt M.; Boutet S.; Sauter N. K.; Gonen T.; Eisenberg D. S. Structure of the toxic core of alpha-synuclein from invisible crystals. Nature 2015, 525, 486–490. 10.1038/nature15368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya M. R.; Rodriguez J.; Cascio D.; Collazo M. J.; Shi D.; Reyes F. E.; Hattne J.; Gonen T.; Eisenberg D. S. Ab initio structure determination from prion nanocrystals at atomic resolution by MicroED. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 11232–11236. 10.1073/pnas.1606287113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattne J.; Shi D.; Glynn C.; Zee C.-T.; Gallagher-Jones M.; Martynowycz M. W.; Rodriguez J. A.; Gonen T. Analysis of Global and Site-Specific Radiation Damage in Cryo-EM. Structure 2018, 26, 759–766. 10.1016/j.str.2018.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz M. J.; Hattne J.; Shi D.; Seidler P.; Rodriguez J.; Reyes F. E.; Sawaya M. R.; Cascio D.; Weiss S. C.; Kim S. K.; Hinck C. S.; Hinck A. P.; Calero G.; Eisenberg D.; Gonen T. Atomic resolution structures from fragmented protein crystals by the cryoEM method MicroED. Nat. Methods 2017, 14, 399–402. 10.1038/nmeth.4178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannenga B. L.; Shi D.; Hattne J.; Reyes F. E.; Gonen T. Structure of catalase determined by MicroED. eLife 2014, 3, e03600. 10.7554/eLife.03600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher-Jones M.; Glynn C.; Boyer D. R.; Martynowycz M. W.; Hernandez E.; Miao J.; Zee C.-T.; Novikova I. V.; Goldschmidt L.; McFarlane H. T.; Helguera G. F.; Evans J. E.; Sawaya M. R.; Cascio D.; Eisenberg D. S.; Gonen T.; Rodriguez J. A. Sub-ångström cryo-EM structure of a prion protofibril reveals a polar clasp. Nat. Struct. Mol. Biol. 2018, 25, 131–134. 10.1038/s41594-017-0018-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martynowycz M. W.; Gonen T. From electron crystallography of 2D crystals to MicroED of 3D crystals. Curr. Opin. Colloid Interface Sci. 2018, 34, 9–16. 10.1016/j.cocis.2018.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nannenga B. L.; Gonen T. Protein structure determination by MicroED. Curr. Opin. Struct. Biol. 2014, 27, 24–31. 10.1016/j.sbi.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb U.; Mugnaioli E.; Gorelik T. E. Automated electron diffraction tomography – a new tool for nano crystal structure analysis. Cryst. Res. Technol. 2011, 46, 542–554. 10.1002/crat.201100036. [DOI] [Google Scholar]
- Wan W.; Sun J.; Su J.; Hovmoller S.; Zou X. Three-dimensional rotation electron diffraction: software RED for automated data collection and data processing. J. Appl. Crystallogr. 2013, 46, 1863–1873. 10.1107/S0021889813027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S.; Gonen T. MicroED structure of the NaK ion channel reveals a Na+ partition process into the selectivity filter. Commun. Biol. 2018, 38, 1–36. 10.1038/s42003-018-0040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palatinus L.; Brázda P.; Boullay P.; Perez O.; Klementová M.; Petit S.; Eigner V.; Zaarour M.; Mintova S. Hydrogen positions in single nanocrystals revealed by electron diffraction. Science 2017, 355, 166–169. 10.1126/science.aak9652. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.