Abstract
Background:
Multidrug-resistant Pseudomonas aeruginosa (MDR P. aeruginosa) is known as a serious threat to human health worldwide. Limited information is available concerning the prevalence of MDR P. aeruginosa in Iran. The aim of the present study was to investigate the relative frequency of MDR P. aeruginosa in different parts of Iran.
Materials and Methods:
Using appropriate keywords and well-known English and Persian database, available data about MDR P. aeruginosa in Iran were retrieved. After applying predefined criteria, relevant studies were selected.
Results:
By using random-effect models, the pooled incidence of MDR P. aeruginosa was estimated 58% (95% confidence interval [CI]; 0.54–0.61). The highest and lowest prevalence of MDR P. aeruginosa were observed in Tehran (100%) (95% CI; 0.94–1.00) and Zahedan (16%) (95% CI; 0.10–0.24), respectively. The highest resistance rate was against ceftazidime (50%) (95% CI; 0.46–0.54) and amikacin (50%) (95% CI; 0.46–0.54).
Conclusion:
Our findings are of concern since they demonstrate the high prevalence rate of MDR P. aeruginosa in the majority of Iranian hospitals.
Keywords: Pseudomonas aeruginosa, antibiotic resistance, multidrug-resistant Pseudomonas aeruginosa
INTRODUCTION
Multidrug-resistant Pseudomonas aeruginosa (MDR P. aeruginosa) due to simultaneous resistance against different class of antibiotics is of paramount importance to health-care settings worldwide.[1,2] Treatment outcomes of patients infected with MDR P. aeruginosa owing to limited available antibiotics are considered to be a serious threat to health-care providers.[1,2] In fact, infection caused by MDR P. aeruginosa has several negative impacts on patient outcomes, including higher mortality, an increase in the length of hospital stay, and considerable increase in hospital costs.[3]
Although different definition of MDR isolates is applied in literatures, MDR P. aeruginosa is known as an isolate resistant against antibiotics belonged to at least three different classes, especially aminoglycosides, carbapenems, and fluoroquinolones.[4]
Antibiotic-resistant determinants are often spread through mobile genetic elements such as plasmid and integron. Integrons are genetic structures capable of capturing genes, consisting of conserved segments and a variable region between the conserved segments.[5,6]
Effective antibiotic treatment is dependent on antibiotic resistance pattern; therefore, in this study, we investigated the prevalence of MDR P. aeruginosa in different parts of Iran. As a secondary aim, we estimate the prevalence of resistance against other antibiotics which are widely used to treat P. aeruginosa infections.
MATERIALS AND METHODS
We searched international databases (ISI web of science, Scopus, PubMed, and Google Scholar) as well as two national scientific search engines including Magiran (www.magiran.com) and Iranian Scientific Information database (www.sid.ir), without limitation, by using English and Persian keywords. To find relevant articles, following keywords were used, “Pseudomonas aeruginosa,” “multidrug-resistant P. aeruginosa,” “imipenem-resistant P. aeruginosa,” “metallo-beta-lactamase-producing P. aeruginosa,” and “Iran”. Finally, to find additional data, reference lists of obtained papers were manually searched. The search was restricted to original research or brief reports with full text available, describing the prevalence of MDR P. aeruginosa. All steps were performed by two authors, independently.
Inclusion and exclusion criteria
After evaluation of abstract and full text, the study was included if first, the clinical specimens were taken from patients referring to Iranian hospitals; second, standard antibiotic susceptibility testing methods according to Clinical Laboratory Standard Institute guidelines were applied; third, MDR-P. aeruginosa was defined as isolate resistant against antibiotics belonged to at least three different classes, especially aminoglycosides, carbapenems, and fluoroquinolones. Papers were also excluded if first, investigation published in language other than Persian or English; second, study designed other than cross-sectional; third, duplicate studies or duplicate specimens; fourth, poor materials and methods, especially regarding antibiotic concentration and producer company; fifth, P. aeruginosa isolated from environmental and non-clinical samples; and sixth, based on applied criteria, the quality of study was recognized as ineligible.
Quality assessment
The quality of papers was evaluated using checklist provided by Joanna Briggs Institute.[7] In this checklist in order to assess the quality of the study, following items are checked: sample size, research objectives, statistical analysis, sample collection, and appropriate materials and methods. One score was assigned to each parameter and study was included if at least seven scores achieved.
Data extraction
According to inclusion and exclusion criteria, all collected data from the selected studies were tabulated as follows: (1)First author, (2) publication date, (3) enrollment time, (4) province of study, (5) prevalence of MDR P. aeruginosa, and (6) prevalence of resistance against antibiotics. Two authors extracted data from involved studies independently. Inconsistency between the reviewers was resolved through discussion.
Statistical analysis
The numbers of total participants and the numbers of participants with MDR P. aeruginosa were used to estimate the logit event rate and its corresponding standard as effect size for meta-analysis.[8] The logit event rates were turned back to event rate for illustrating the meta-analysis results. The random-effect model which takes the between-study heterogeneity into account was used to derive the summary effects. Between-study heterogeneities were assessed using Cochran's Q-test and I-squared (I2) test.[9] In order to explore the extent to which the overall calculations might depend on a specific study, sensitivity of study was performed. Publication bias was checked by Egger's regression asymmetry test and Begg's adjusted rank correlation test.[8,9,10] Statistical analyses were done using the STATA software package version 11.2 (STATA Corp, College Station, TX, USA).
RESULTS
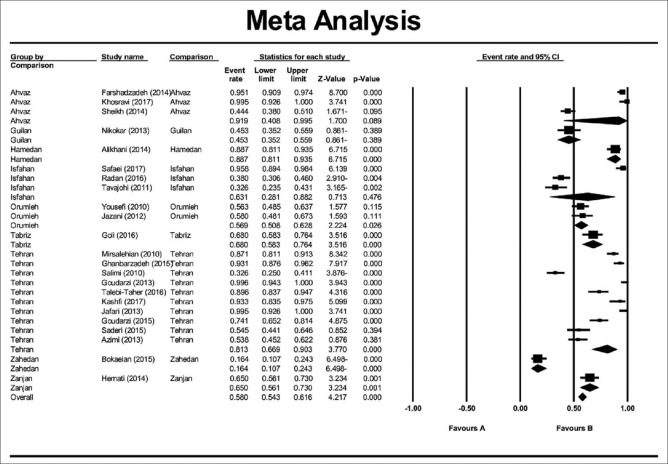
In this study, a total of 4854 articles were found through database search [Figure 1]. In first step, 2359 articles were excluded due to duplication. In the secondary screening and after abstract evaluation, 2145 of publications were excluded. Finally, 350 articles were retained for detailed full-text evaluation. According to quality assessment criteria and inclusion/exclusion criteria, a total of 23 articles with full text reporting the prevalence of MDR P. aeruginosa were recruited for the systematic review and meta-analysis[11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33] [Table 1]. In total, 10 studies from Tehran, 3 studies from Isfahan, 3 studies from Ahvaz, 2 studies from Orumieh, 1 study from Zahedan, 1 study from Zanjan, 1 study from Tabriz, 1 study from Guilan, and 1 study from Hamedan were involved.[11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33] Figure 2 shows the distribution of MDR P. aeruginosa in different parts of Iran. By using random-effect models, the pooled prevalence of MDR P. aeruginosa was estimated to be 58% (95% confidence interval [CI]; 0.54–0.61). However, an evident heterogeneity of MDR P. aeruginosa-relative frequency was seen (Cochrane Q test, Q statistic = 463.38, P < 0.001, I2 = 95.25) [Figure 3]. The highest and lowest prevalence of MDR P. aeruginosa were observed in Tehran (100%) (95% CI; 0.94–1.00) and Zahedan (16%) (95% CI; 0.10–0.24), respectively. We also checked the prevalence rate of resistance against ceftazidime, imipenem, meropenem, aztreonam, amikacin, gentamycin, ciprofloxacin, and piperacillin/tazobactam [Table 2]. The highest resistance rate was against ceftazidime (50%) (95% CI; 0.46–0.54) and amikacin (50%) (95% CI; 0.46–0.54) followed by piperacillin/tazobactam (49%) (95% CI; 0.44–0.54) and the lowest rate was against imipenem (31%) (95% CI; 0.27–0.35) [Table 2]. There was an asymmetry in Begg's funnel plot when depicting the effect sizes (logit event rate for MDR resistance) against their standard error [Figure 4]. The Begg's and Egger's test also confirmed an asymmetry (Begg's test, P = 0.008, Egger's test, P = 0.002). We explored the magnitude of the bias using trim and till analysis. Two studies could be added using trim and fill analysis; however, the overall prevalence was not changed notably after filling the two studies (event rate = 58%, 95% CI: 56–60). The funnel plot showing the observed studies as well as studies filled after the trim and fill analysis is provided in Figure 4.
Figure 1.
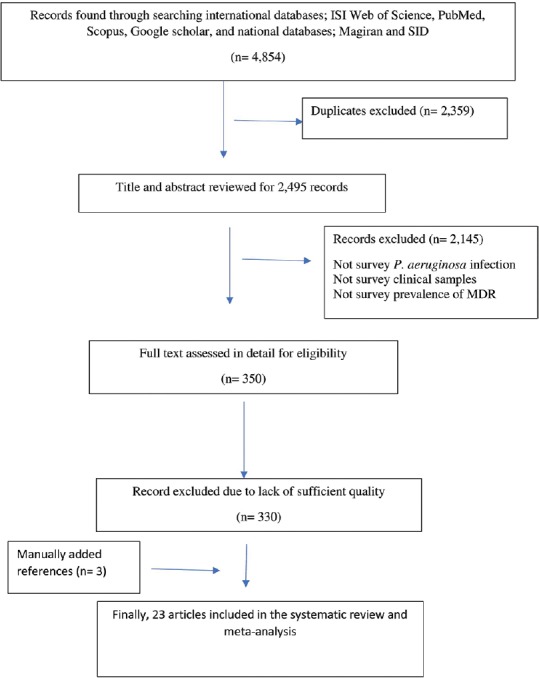
Schematic flow diagram for literature review and study selection
Table 1.
Characteristics of studies involved in the systematic review and meta-analysis
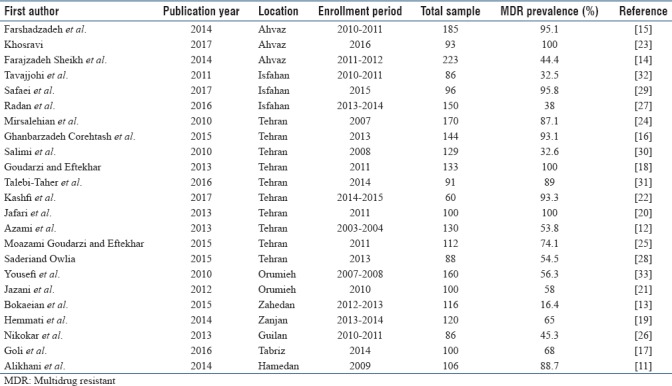
Figure 2.
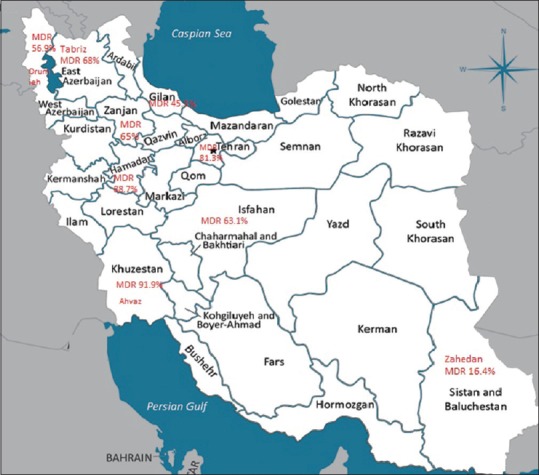
Distribution of multidrug-resistant Pseudomonas aeruginosa in different parts of Iran
Figure 3.
Meta-analysis examining the overall prevalence of multidrug-resistant Pseudomonas aeruginosa on studies conducted in Iran. The analysis revealed that the overall prevalence was about 58%
Table 2.
Antibiotic resistance patterns of Pseudomonas aeruginosa in different provinces of Iran
Figure 4.
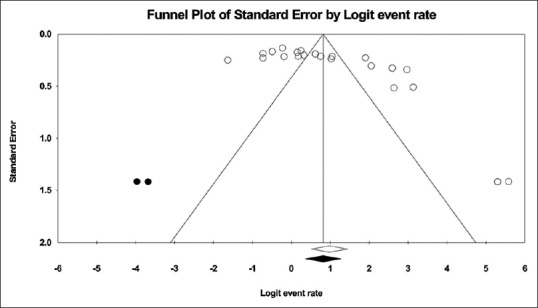
Funnel plot with pseudo 95% confidence interval demonstrating the effect sizes derived from each study (logit event rate) against their corresponding standard errors
DISCUSSION
Appropriate selection of antibiotics is dependent on antibiotic resistance profile and active surveillance of changing trends in resistance patterns; therefore, we conducted this study to estimate the prevalence and distribution of MDR P. aeruginosa in different parts of Iran, using data provided by published papers.
The prevalence of P. aeruginos a infection in different parts of Iran is high.[34] Our findings revealed that the prevalence of MDR P. aeruginosa was 58% and is varied in different provinces of Iran, with highest and lowest rates observed in Tehran (100%) (95% CI; 0.94–1.00) and Zahedan (16%) (95% CI; 0.10–0.24), respectively [Table 1].
The study by Gill et al. between 2014 and 2015 on MDR P. aeruginosa rates of patients admitted to Intensive Care Unit showed similar percentage of resistance, with 50% of all isolates being MDR.[35] In addition, the finding of Khan et al. demonstrated that the prevalence of MDR P. aeruginosa in different hospitals of Karachi, Pakistan, is lower than our findings, with 30% of isolates being MDR.[36] A comprehensive study conducted at 28 hospitals in Thailand from 2000 to 2005 revealed that the prevalence of MDR P. aeruginosa was 20%–30%,[37] which is lower than our findings.
Comprehensive antibiotic resistance surveillance in European countries demonstrated that the percentages of MDR P. aeruginosa isolates in thirty participated countries ranged from 0% (Estonia and Iceland) to 49.4% (Romania).[38] Sixteen countries (Germany, Bulgaria, Austria, Lithuania, Malta, Ireland, Luxembourg, Finland, Cyprus, Sweden, Norway, United Kingdom, Netherlands, Denmark, Iceland, and Estonia) reported resistance percentages below 10%, 11 reported 10%–25% (including Belgium, Slovenia, Portugal, Spain, France, Poland, Croatia, Hungary, Czech Republic, Latvia, and Italy), and the remaining three (Slovakia, Greece, and Romania) reported MDR percentages above 25%.[38]
Unfortunately, despite the existence of several reports on antibiotic resistance patterns on P. aeruginosa isolated from clinical samples in Iran, there is not a comprehensive study on the prevalence of MDR P. aeruginosa in Iranian hospitals; hence, we tried to do a comprehensive study across Iran.
Based on our data, resistance to ceftazidime (50%) is higher than the percentage reported from Iceland (0%), United Kingdom (3.7%), and Sweden (6.8%).[38] Furthermore, our study revealed that compared with most European countries, resistance to other antibiotics such as imipenem, meropenem, ciprofloxacin, piperacillin/tazobactam, amikacin, gentamycin, and aztreonam is high [Table 2]. For example, Europe antimicrobial resistance surveillance in 2013 reported the percentage of fluoroquinolones-resistant isolates ranged from 0% (Iceland) to 53.1% (Slovakia). At the same time, the percentage of aminoglycosides-resistant isolates ranged from 0% (Iceland and Malta) to 51.2% (Romania). Carbapenem-resistant isolates of P. aeruginosa in Denmark was 2.9%, which is significantly lower than our results.[38]
The emergence and dissemination of MDR P. aeruginosa is of paramount concern because these isolates are simultaneously resistant against multiple antibiotics; therefore, limited choices such as colistin and polymyxin B remain available to treat patients infected by these isolates.
This study faces some limitations that should be considered; first, due to restricted access to some data provided by theses, in-press articles, or nonopen access articles, some data might have been missed; second, for some parts of country, the relevant data were unavailable; hence, this study could not completely represent the status of prevalence rate for Iran.
CONCLUSION
P. aeruginosa is one of the most important pathogens in Iranian hospitals. Our findings are of concern since they demonstrate the high prevalence rate of MDR P. aeruginosa in the majority of Iranian hospitals. Indiscriminate use of antibiotics has resulted in the development of multidrug-resistant P. aeruginosa infections, which is a serious threat to health of patients. To prevent further dissemination of these isolates, appropriate infection control practices must be implemented.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
Acknowledgment
The authors confirm that their work is not supported in any way by the Iranian military nor the Iranian government's nuclear program.
REFERENCES
- 1.Hirsch EB, Tam VH. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev Pharmacoecon Outcomes Res. 2010;10:441–51. doi: 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaez H, Salehi-Abargouei A, Khademi F. Systematic review and meta-analysis of imipenem-resistant Pseudomonas aeruginosa prevalence in Iran. Germs. 2017;7:86–97. doi: 10.18683/germs.2017.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nakamura A, Miyake K, Misawa S, Kuno Y, Horii T, Kondo S, et al. Meropenem as predictive risk factor for isolation of multidrug-resistant Pseudomonas aeruginosa. J Hosp Infect. 2013;83:153–5. doi: 10.1016/j.jhin.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 5.El Zowalaty ME, Al Thani AA, Webster TJ, El Zowalaty AE, Schweizer HP, Nasrallah GK, et al. Pseudomonas aeruginosa: Arsenal of resistance mechanisms, decades of changing resistance profiles, and future antimicrobial therapies. Future Microbiol. 2015;10:1683–706. doi: 10.2217/fmb.15.48. [DOI] [PubMed] [Google Scholar]
- 6.Chen J, Su Z, Liu Y, Wang S, Dai X, Li Y, et al. Identification and characterization of class 1 integrons among Pseudomonas aeruginosa isolates from patients in Zhenjiang, China. Int J Infect Dis. 2009;13:717–21. doi: 10.1016/j.ijid.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 7.Munn Z, Moola S, Lisy K, Riitano D, Tufanaru C. Methodological guidance for systematic reviews of observational epidemiological studies reporting prevalence and cumulative incidence data. Int J Evid Based Healthc. 2015;13:147–53. doi: 10.1097/XEB.0000000000000054. [DOI] [PubMed] [Google Scholar]
- 8.Egger M, Davey-Smith G, Altman D. Systematic Reviews in Health Care: Meta-Analysis in Context. 2nd ed. London: BMJ Books; 2001. [Google Scholar]
- 9.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 10.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–34. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alikhani MY, Karimi Tabar Z, Mihani F, Kalantar E, Karami P, Sadeghi M, et al. Antimicrobial resistance patterns and prevalence of blaPER-1 and blaVEB-1 genes among ESBL-producing Pseudomonas aeruginosa isolates in West of Iran. Jundishapur J Microbiol. 2014;7:e8888. doi: 10.5812/jjm.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azami S, Ali A, Asgarani E. Association between metallo-β-lactamases and integrons with multi- drug resistance in Pseudomonas aeruginosa isolates. J Med Microbiol Infect Dis. 2013;1:46–51. [Google Scholar]
- 13.Bokaeian M, Shahraki Zahedani S, Soltanian Bajgiran M, Ansari Moghaddam A. Frequency of PER, VEB, SHV, TEM and CTX-M genes in resistant strains of Pseudomonas aeruginosa producing extended spectrum β-lactamases. Jundishapur J Microbiol. 2015;8:e13783. doi: 10.5812/jjm.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farajzadeh Sheikh A, Rostami S, Jolodar A, Tabatabaiefar MA, Khorvash F, Saki A, et al. Detection of metallo-beta lactamases among carbapenem-resistant Pseudomonas aeruginosa. Jundishapur J Microbiol. 2014;7:e12289. doi: 10.5812/jjm.12289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farshadzadeh Z, Khosravi AD, Alavi SM, Parhizgari N, Hoveizavi H. Spread of extended-spectrum β-lactamase genes of blaOXA-10, blaPER-1 and blaCTX-M in Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2014;40:1575–80. doi: 10.1016/j.burns.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 16.Ghanbarzadeh Corehtash Z, Khorshidi A, Firoozeh F, Akbari H, Mahmoudi Aznaveh A. Biofilm formation and virulence factors among Pseudomonas aeruginosa isolated from burn patients. Jundishapur J Microbiol. 2015;8:e22345. doi: 10.5812/jjm.22345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goli HR, Nahaei MR, Ahangarzadeh Rezaee M, Hasani A, Samadi Kafil H, Aghazadeh M, et al. Emergence of colistin resistant Pseudomonas aeruginosa at Tabriz hospitals, Iran. Iran J Microbiol. 2016;8:62–9. [PMC free article] [PubMed] [Google Scholar]
- 18.Goudarzi S, Eftekhar F. Assessment of carbapenem susceptibility and multidrug-resistance in Pseudomonas aeruginosa burn isolates in Tehran. Jundishapur J Microbiol. 2013;6:162–5. [Google Scholar]
- 19.Hemmati F, Sourouri R, Haghi F, Zeighami H. Determination of antibiotic resistance profile and frequency of metallo-beta-lactamases in Pseudomonas aeruginosa isolates. J Zanjan Univ Med Sci Health Serv. 2014;22:85–77. [Google Scholar]
- 20.Jafari M, Fallah F, Shams Borhan R, Navidinia M, Karimi A, Rafiei Tabatabaei S, et al. The first report of CMY, aac (6′)-Ib and 16S rRNA methylase genes among Pseudomonas aeruginosa isolates from Iran. Arch Pediatr Infect Dis. 2013;2:109–12. [Google Scholar]
- 21.Jazani N, Zahedi A, Garebagi N. Phenotypic detection of metallo-β-lactamase producing Pseudomonas aeruginosa isolated from Urmia hospitals. Afr J Microbiol Res. 2012;6:1387–92. [Google Scholar]
- 22.Kashfi M, Hashemi A, Sadredin Amin M, Tarashi S, Taki E. The prevalence of aminoglycoside-modifying enzyme genes among Pseudomonas aeruginosa strains isolated from burn patients. Arch Clin Infect Dis. 2017;12:1–5. [Google Scholar]
- 23.Khosravi AD, Motahar M, Abbasi Montazeri E. The frequency of class1 and 2 integrons in Pseudomonas aeruginosa strains isolated from burn patients in a burn center of Ahvaz, Iran. PLoS ONE. 2017;12:e0183061. doi: 10.1371/journal.pone.0183061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mirsalehian A, Feizabadi M, Nakhjavani FA, Jabalameli F, Goli H, Kalantari N, et al. Detection of VEB-1, OXA-10 and PER-1 genotypes in extended-spectrum beta-lactamase-producing Pseudomonas aeruginosa strains isolated from burn patients. Burns. 2010;36:70–4. doi: 10.1016/j.burns.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 25.Moazami Goudarzi S, Eftekhar F. Multidrug resistance and integron carriage in clinical isolates of Pseudomonas aeruginosa in Tehran, Iran. Turk J Med Sci. 2015;45:789–93. doi: 10.3906/sag-1408-120. [DOI] [PubMed] [Google Scholar]
- 26.Nikokar I, Tishayar A, Flakiyan Z, Alijani K, Rehana-Banisaeed S, Hossinpour M, et al. Antibiotic resistance and frequency of class 1 integrons among Pseudomonas aeruginosa, isolated from burn patients in Guilan, Iran. Iran J Microbiol. 2013;5:36–41. [PMC free article] [PubMed] [Google Scholar]
- 27.Radan M, Moniri R, Khorshidi A, Gilasi H, Norouzi Z, Beigi F, et al. Emerging carbapenem-resistant Pseudomonas aeruginosa isolates carrying bla IMP among burn patients in Isfahan, Iran. Arch Trauma Res. 2016;5:e33664. doi: 10.5812/atr.33664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saderi H, Owlia P. Detection of multidrug resistant (MDR) and extremely drug resistant (XDR) P. aeruginosa isolated from patients in Tehran, Iran. Iran J Pathol. 2015;10:265–71. [PMC free article] [PubMed] [Google Scholar]
- 29.Safaei HG, Moghim S, Isfahani BN, Fazeli H, Poursina F, Yadegari S, et al. Distribution of the strains of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa isolates from burn patients. Adv Biomed Res. 2017;6:74. doi: 10.4103/abr.abr_239_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Salimi H, Yakhchali B, Owlia P, Lari A. Molecular epidemiology and drug susceptibility of Pseudomonas aeruginosa strains isolated from burn patients. Lab Med. 2010;41:540–4. [Google Scholar]
- 31.Talebi-Taher M, Majidpour, Gholami A, Rasouli-Kouhi S, Adabi M. Role of efflux pump inhibitor in decreasing antibiotic cross-resistance of Pseudomonas aeruginosa in a burn hospital in Iran. J Infect Dev Ctries. 2016;10:600–4. doi: 10.3855/jidc.7619. [DOI] [PubMed] [Google Scholar]
- 32.Tavajjohi Z, Moniri R, Khorshidi A. Detection and characterization of multidrug resistance and extended-spectrum-beta-lactamase-producing (ESBL) Pseudomonas aeruginosa isolates in teaching hospital. Afr J Microbiol Res. 2011;5:3223–8. [Google Scholar]
- 33.Yousefi S, Nahaei M, Farajnia S, Ghojazadeh M, Akhi M, Sharifi Y, et al. Class 1 integron and imipenem resistance in clinical isolates of Pseudomonas aeruginosa: Prevalence and antibiotic susceptibility. Iran J Microbiol. 2010;2:115–21. [PMC free article] [PubMed] [Google Scholar]
- 34.Vaez H, Faghri J, Nasr Esfahani B, Moghim S, Fazeli H, Sedighi M, et al. Antibiotic resistance patterns and genetic diversity in clinical isolates of Pseudomonas aeruginosa isolated from patients of a referral hospital, Isfahan, Iran. Jundishapur J Microbiol. 2015;8:e20130. doi: 10.5812/jjm.20130v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gill JS, Arora S, Khanna SP, Kumar KH. Prevalence of multidrug-resistant, extensively drug-resistant, and pandrug-resistant Pseudomonas aeruginosa from a tertiary level Intensive Care Unit. J Glob Infect Dis. 2016;8:155–9. doi: 10.4103/0974-777X.192962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Khan F, Khan A, Kazmi SU. Prevalence and susceptibility pattern of multi drug resistant clinical isolates of Pseudomonas aeruginosa in Karachi. Pak J Med Sci. 2014;30:951–4. doi: 10.12669/pjms.305.5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suwantarat N, Carroll KC. Epidemiology and molecular characterization of multidrug-resistant Gram-negative bacteria in Southeast Asia. Antimicrob Resist Infect Control. 2016;5:15. doi: 10.1186/s13756-016-0115-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Annual Report of the European Antimicrobial Resistance Surveillance Network (EARS-Net) Stockholm: European Centre for Disease Prevention and Control; 2014. European Centre for Disease Prevention and Control. Antimicrobialresistance Surveillance in Europe 2013. [Google Scholar]