Abstract
Oceanic harmful algal blooms of Pseudo-nitzschia diatoms produce the potent mammalian neurotoxin domoic acid (DA). Despite decades of research, the molecular basis for its biosynthesis is not known. By using growth conditions known to induce DA production in Pseudo-nitzschia multiseries, we implemented transcriptome sequencing in order to identify DA biosynthesis genes that colocalize in a genomic four-gene cluster. We biochemically investigated the recombinant DA biosynthetic enzymes and linked their mechanisms to the construction of DA’s diagnostic pyrrolidine skeleton, establishing a model for DA biosynthesis. Knowledge of the genetic basis for toxin production provides an orthogonal approach to bloom monitoring and enables study of environmental factors that drive oceanic DA production.
Oceanic harmful algal blooms (HABs) are intensifying in frequency and severity in association with climate change (1–3). This phenomenon was exemplified in the summer of 2015 when the North American Pacific coast experienced the largest HAB ever recorded, spanning the Aleutian Islands of Alaska to the Baja peninsula of Mexico (fig. S1) (4). This bloom caused widespread ecological and economic devastation, resulting in the deaths of marine mammals and the closure of beaches and fisheries. The dominant algal species in the 2015 HAB were pennate diatoms of the globally distributed genus Pseudo-nitzschia, which are often associated with high production of the excitatory glutamate receptor agonist domoic acid (DA; 1). Mammalian consumption of DA-contaminated shellfish exerts its toxicity at the AMPA and kainate ionotropic glutamate receptors of the central nervous system (5, 6). In humans, a high, single-exposure dose of DA can cause amnesic shellfish poisoning (ASP), which involves symptoms of amnesia, seizures, coma, and in extreme cases, death. Even chronic, low-level consumption of DA may lead to kidney damage, cognitive deficit, and impairment of fetal development, making DA outbreaks an important human health problem (7–10). Similar neurotoxic symptoms have been observed in birds and marine mammals such as sea lions, which suffer spatial memory impairment linked to DA consumption, likely leading to increases in sea lion strandings (11).
Although abiotic (12) and biotic (13) factors have been shown to affect toxicity in culture, the biological and physicochemical mechanisms underlying DA production in Pseudo-nitzschia are unclear. Moreover, not all Pseudo-nitzschia blooms produce DA (12). An understanding of the genetic basis of DA biosynthesis in diatoms would facilitate determination of the cellular pathway that controls oceanic DA production and thereby di-atom toxicity.
Stable isotope experiments (14, 15) suggest that DA is composed of glutamic acid (Glu) and geranyl pyrophosphate (GPP) building blocks. No pathway that involves these starting materials to construct the characteristic pyrrolidine ring of DA has been previously characterized, and thus, our ability to predict candidate bio-synthetic genes using traditional bioinformatic methods was limited. However, we postulated that a redox enzyme, such as a cytochrome P450 (CYP450), generates the 7′-carboxylic acid of DA through three successive oxidations, a reaction common in all branches of life (Fig. 1A) (16).
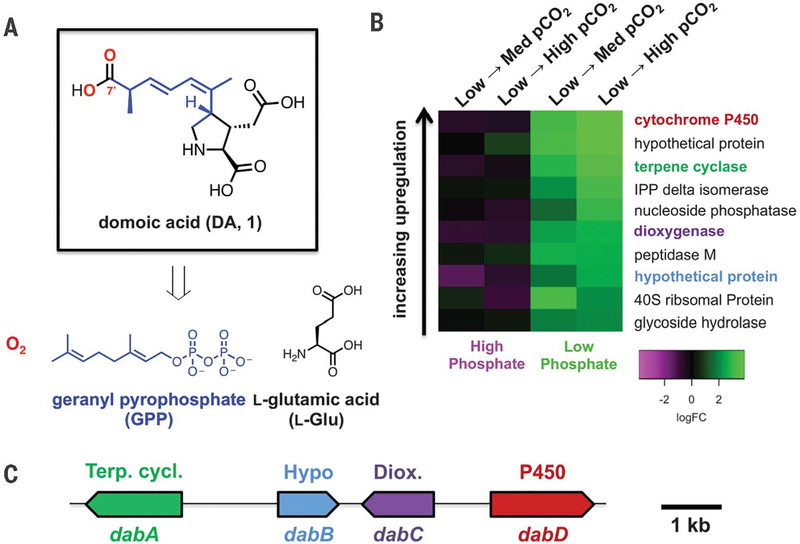
Fig. 1. Identification of domoic acid biosynthetic genes from transcriptomic and genomic data.
(A) Structure of DA (1) and its proposed biosynthetic building blocks (14, 15). (B) Ten most up-regulated P. multiseries transcripts under previously reported DA-induction conditions (17). Differential expression changes are each shown with reference to the Pco2 conditions listed across the top of the heatmap, with phosphate concentrations held constant. (C) DA biosynthesis (dab) gene cluster in the P. multiseries genome.
To identify putative DA biosynthetic genes, we examined patterns of transcriptional activity under previously established conditions—phosphate limitation and elevated CO2 (17)—that stimulate DA production. We created a differential expression dataset for nearly 20,000 Pseudo-nitzschia multiseries RNA transcripts (table S1). A small fraction (~500; 2.5%) showed consistent up-regulation under phosphate limitation (fig. S2 and table S2), and further refinement of this subset to include genes that were also up-regulated with increasing partial pressure of CO2 (PCO2) highlighted only 43 (0.22%) transcripts (table S3). A CYP450 gene showed the highest fold change of all analyzed transcripts under the high PCO2 and low-phosphate DA-inducing condition (Fig. 1B). This transcript was the only annotated CYP450 gene out of 20 total within the P. multiseries genome (220 Mbp; Joint Genome Institute accession no. PRJNA32659) that showed increased transcription (fig. S3 and table S4). When mapped to the public P. multiseries genome, this CYP450 was localized to a compact genomic island spanning ~8 kb that possesses three other similarly up-regulated genes that are indicative of canonical gene clustering more typically observed in bacterial- and fungal-specialized metabolism (Fig. 1C) (18). Genomic organization in diatoms is not generally typified by clustering of metabolic genes (19, 20). However, clusters of transcriptionally coregulated genes, sensitive to Fe and Si limitation, have been reported (21, 22). The annotated gene functions suggest involvement in terpenoid and redox biochemistry, which are two predicted hallmarks for DA biosynthesis. These gene candidates for DA biosynthetic (Dab) enzymes were annotated as dabA (terpene cyclase), dabB (hypothetical protein), dabC [α-ketoglutarate (αKG)–dependent dioxygenase], and dabD (CYP450) (Fig. 1C). We independently sequenced this gene cluster from an environmental isolate of P. multiseries in order to validate its conservation (GenBank accession no. MH202990).
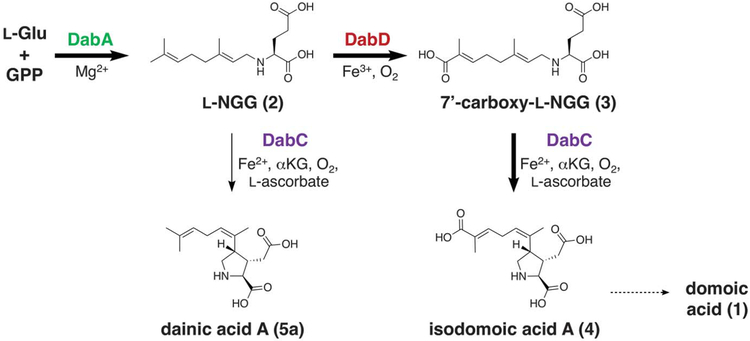
Despite the rarity of the pyrrolidine kainoid skeleton in nature, the putative dab gene functions enabled us to initially map our genes onto the suggested biosynthetic pathway (14, 15). We hypothesized that DabA catalyzes the first committed step of DA biosynthesis: N-prenylation of L-Glu with GPP to form N-geranyl-L-glutamic acid (L-NGG; 2). DabC and DabD would perform subsequent oxidative reactions (Fig. 2).
Fig. 2.
Domoic acid (1) biosynthetic pathway based on dab gene annotations and in vitro enzyme activities.
We began in vitro validation of the dab genes with dabA, which contains a chloroplast transit peptide sequence and an intron but low similarity to any characterized protein in the National Center for Biotechnology Information (NCBI) database. Structural prediction by using Phyre2 (23) suggested that DabA may possess a terpene cyclase–like fold, and phylogenetic analysis intimated an evolutionary history related to red algal and bacterial genes (fig. S4). Among diatoms, dabA appears restricted to Pseudo-nitzschia spp. We expressed recombinant His6-DabA without the N-terminal transit peptide in Escherichia coli and purified the enzyme using Ni2+ affinity and size-exclusion chromatography (fig. S5). In vitro, DabA catalyzes the N-geranylation of L-Glu to form L-NGG (2) in a Mg+2-dependent manner (Fig. 3A). DabA interrogation with structurally similar substrates shows modest promiscuity toward prenyl pyrophosphates but high specificity for Glu (fig. S6). Recently, L-NGG was isolated in low abundance from the DA-containing red alga Chondria armata and shown through labeling experiments in P. multiseries to be a precursor to DA (24). These observations further implicate dabA as a gene that encodes a major step in DA biosynthesis.
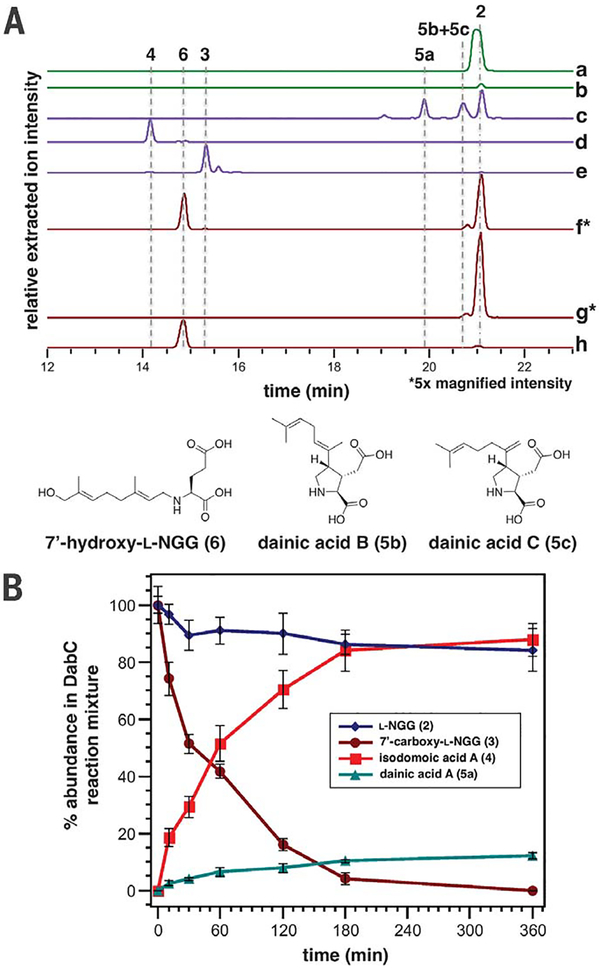
Fig. 3. In vitro characterization of Dab enzymes.
(A) Relative intensities of negative ionization–extracted ion chromatogram liquid chromatography–mass spectrometry traces [(282.1711, 298.1660, 312.1453, 280.1554, 310.1296) ± 0.01 mass/charge ratio] for in vitro Dab reactions. a, DabA, L-Glu (500 μM), GPP (500 μM), MgCl2 (10 mM); b, DabA, L-Glu(500 μM), GPP (500 μM), without MgCl2; c, DabC, L-NGG (2; 500 μM), FeSO4, αKG, L-ascorbicacid; d, DabC, 7′-carboxy-L-NGG (3; 500 μM), FeSO4, αKG, L-ascorbic acid; e, DabC, 7′-carboxy-L-NGG (3; 500 μM), FeSO4, αKG, L-ascorbic acid, EDTA (1.0 mM); f, DabD and PmCPR1 containing S. cerevisiae microsomes, L-NGG (2; 500 μM), NADPH; g, empty vector S. cerevisiae micro-somes, L-NGG (2; 500 μM), NADPH; andh, synthetic 7′-hydroxy-L-NGG (6) standard. Additional enzyme and cosubstrate concentrations can be found in the supplementary materials. Asterisks indicate that relative extracted ion intensities for traces f and g have been 5× magnified. (B) Comparison of in vitro DabC substrate consumption of L-NGG (2; 1.0 mM) and 7′-carboxy-L-NGG (3; 1.0 mM) and relative formation of respective dainic acid A (5a) and isodomoic acid A (4) products (n = 3 replicate DabC experiments).
To investigate the subsequent transformations of L-NGG, we individually interrogated the activity of the DabC and DabD oxygenases toward this substrate. In the presence of Fe2+, L-ascorbic acid, and cosubstrate aKG, recombinant DabC purified from E. coli cyclized L-NGG to form three pyrrolidine ring–containing molecules: 7′-methylisodomoic acids A, B, and C, termed dainic acids A, B, and C (5a to 5c), respectively (Fig. 3A and figs. S7 and S8), of which 5a and 5b had been recently isolated from C. armata (24). DabC, however, showed minimal cyclization of other similarly N-prenylated glutamic acids (fig. S7). Enzymatic synthesis of the dainic acids was made more amenable through a one-pot coupled assay using L-Glu, GPP, DabA, DabC, and their requisite cofactors and cosubstrates (fig. S9). Despite DabC generating much of the structural diversity observed within the kainoid metabolites (25), the slow rate of L-NGG consumption and failure to go to completion led us to suspect that this enzyme may instead act on an oxidized substrate, placing DabD oxidation ahead of DabC cyclization in the DA biosynthetic pathway. Incubation of L-NGG with Saccharomyces cerevisiae micro-somes that possess coexpressed transmembrane proteins DabD (fig. S10) and P. multiseries CYP450 reductase (PmCPR1) generated small but reproducible quantities of 7′-hydroxy-L-NGG (6) and 7′-carboxy-L-NGG (3) (Fig. 3A and fig. S11), which was validated through comparison with synthetic standards. Conversely, the dainic acid isomers were not substrates of the DabD P450 (fig. S11). 7′-carboxy-L-NGG incubation with DabC showed rapid cyclization to the P. multiseries natural product isodomoic acid A (4) (Fig. 3A), which did not occur in the presence of metal chelator ethylenediaminetetraacetic acid (EDTA). When comparing both DabC in vitro substrates for physiological relevance, 7′-carboxy-L-NGG was consumed at a much faster rate than L-NGG, further suggesting that 7′-carboxy-L-NGG is an on-pathway intermediate (Fig. 3B and fig. S12). Our cumulative in vitro biochemical results imply a DA biosynthetic pathway that begins with the DabA-catalyzed geranylation of L-Glu to yield L-NGG, likely in the chloroplast. DabD then performs three successive oxidation reactions at the 7′-methyl of L-NGG to produce 7′-carboxy-L-NGG, which is then cyclized by DabC to generate the naturally occurring isodomoic acid A (Fig. 2). A putative isomerase likely converts isodomoic acid A to DA. We biochemically interrogated the coclustered dabB gene product but did not observe isomerase activity (fig. S13). Further examination of additionally up-regulated transcripts did not suggest an obvious candidate gene (fig. S14). We are actively investigating this final isomerization reaction to complete the pathway to DA.
In addition to Pseudo-nitzschia spp., the kainoid structure has been observed in other diatom, red macroalgal, and fungal compounds (fig. S15) (12, 26, 27). In silico application of the dab genes as a kainoid ring biosynthetic query identified dabA and dabC homologs in several red algal transcriptomes (28, 29), including the known kainic acid producer Palmaria palmata (fig. S16). Like DA, kainic acid (26) is a structurally related glutamate agonist constructed from a shorter terpene substrate without the need of the DabD P450 oxidation.
With the establishment of the DabACD-dependent biosynthetic pathway to isodomoic acid A in P. multiseries, we next linked this discovery to the marine environment. We identified dab genes with the same genetic organization and high sequence identity in the genome of the known DA-producing Pseudo-nitzschia multistriata (fig. S17) (30). Moreover, of the eight publicly available Pseudo-nitzschia transcriptomes, only the highly toxic DA-producing species Pseudo-nitzschia australis expressed the dab genes (figs. S18 and S19) (31). No dab homologs were found in any other sequenced microalgal genera. By virtue of its limited distribution, dabA thus presents an opportunity for genetic monitoring of the DA-producing capabilities of Pseudo-nitzschia blooms orthogonal to currently established mass spectrometry–based and enzyme-linked immunosorbent assay–based identification approaches. We anticipate that knowledge of the dab genes will allow for greater understanding of the basis and diversity of Pseudo-nitzschia toxicity, the physiological function of DA, and the environmental conditions that promote HAB formation so that we may better anticipate risk of exposure to this toxic marine natural product.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge B. Duggan, A. Mrse, and Y. Su (all University of California, San Diego) for assistance and maintenance of nuclear magnetic resonance and high-resolution mass spectrometry machinery; W. Fenical (Scripps Institution of Oceanography) for helpful discussion and feedback; and M. Kahru (Scripps Institution of Oceanography) for providing processed satellite images.
Funding:
This work was supported by grants from the National Science Foundation (NSF OCE-1313747 to B.S.M., NSF-ANT-1043671 to A.E.A., and OCE 1538525 and OCE 1638804 to D.A.H.), the National Institute of Environmental Health Sciences (NIEHS P01-ES021921 to B.S.M.), the U.S. Department of Energy Genomics Science program (DE-SC0008593 and DE-SC0018344 to A.E.A.), the Gordon and Betty Moore Foundation (GBMF3828 to A.E.A. and GBMF4960 to G.J.S.), and the Czech Science Foundation (18–13458S to M.O.) and funding from the National Institutes of Health (Chemical Biology Interfaces–University of California, San Diego, training grant 5T32GM112584 to J.K.B.), the Dickinson-McCrink Fellowship (J.K.B.), the Natural Sciences and Engineering Research Council of Canada (NSERC-PDF to S.M.K.M.), and the Simons Foundation Fellowship of the Life Sciences Research Foundation (J.R.C.).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: Transcriptome sequences are deposited in NCBI’s Sequence Read Archive, BioProjectIDs SAMN08773590-SAMN08773622. The sequence of the dab cluster as amplified from P. multiseries 15901C3 is deposited in GenBank, accession no. MH202990. Data used to construct fig. S1 are courtesy of the NASA Ocean Biology Processing Group (OBPG) and are available from https://oceancolor.gsfc.nasa.gov. All other data needed to evaluate the paper are present in the supplementary materials.
REFERENCES AND NOTES
- 1.Wells ML et al. , Harmful Algae 49, 68–93 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McKibben SM et al. , Proc. Natl. Acad. Sci. U.S.A. 114, 239–244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gobler CJ et al. , Proc. Natl. Acad. Sci. U.S.A. 114, 4975–4980 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCabe RM et al. , Geophys. Res. Lett 43, 10366–10376 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stewart GR, Zorumski CF, Price MT, Olney JW, Exp. Neurol 110, 127–138 (1990). [DOI] [PubMed] [Google Scholar]
- 6.Larm JA, Beart PM, Cheung NS, Neurochem. Int 31, 677–682 (1997). [DOI] [PubMed] [Google Scholar]
- 7.Funk JA et al. , J. Am. Soc. Nephrol 25, 1187–1197 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre KA et al. , Harmful Algae 64, 20–29 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramsdell JS, Zabka TS, Mar. Drugs 6, 262–290 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grattan LM et al. , Toxins (Basel) 10, 103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cook PF et al. , Science 350, 1545–1547 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Lelong A, Hégaret H, Soudant P, Bates SS, Phycologia 51, 168–216 (2012). [Google Scholar]
- 13.Sison-Mangus MP, Jiang S, Tran KN, Kudela RM, ISME J 8, 63–76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramsey UP, Douglas DJ, Walter JA, Wright JL, Nat. Toxins 6, 137–146 (1998). [PubMed] [Google Scholar]
- 15.Savage TJ, Smith GJ, Clark AT, Saucedo PN, Toxicon 59, 25–33 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Zhang X, Li S, Nat. Prod. Rep 34, 1061–1089 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Sun J et al. , Limnol. Oceanogr 56, 829–840 (2011). [Google Scholar]
- 18.Medema MH et al. , Nat. Chem. Biol 11, 625–631 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armbrust EV et al. , Science 306, 79–86 (2004). [DOI] [PubMed] [Google Scholar]
- 20.Bowler C et al. , Nature 456, 239–244 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Allen AE et al. , Proc. Natl. Acad. Sci. U.S.A. 105, 10438–10443 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sapriel G et al. , PLOS ONE 4, e7458 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelley LA, Mezulis S, Yates CM, Wass MN,Sternberg MJE, Nat. Protoc 10, 845–858 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maeno Y et al. , Sci. Rep 8, 356 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clayden J, Read B, Hebditch KR, Tetrahedron 61, 5713–5724 (2005). [Google Scholar]
- 26.Nitta I, Watase H, Tomiie Y, Nature 181, 761–762 (1958). [DOI] [PubMed] [Google Scholar]
- 27.Konno K, Shirahama H, Matsumoto T, Tetrahedron Lett. 24, 939–942 (1983). [Google Scholar]
- 28.Saunders GW, Jackson C, Salomaki ED, Mol. Phylogenet. Evol 119, 151–159 (2018). [DOI] [PubMed] [Google Scholar]
- 29.Matasci N et al. , Gigascience 3, 17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basu S et al. , New Phytol. 215, 140–156 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keeling PJ et al. , PLOS Biol. 12, e1001889 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.