Abstract
A nuclear magnetic resonance- (NMR-) based metabolomics method was used to identify differential metabolites of methanol extracts obtained from six parts of Peganum harmala L. (P. harmala), namely, the root, stem, leaf, flower, testa, and seed. Two multivariate statistical analysis methods, principal component analysis (PCA) and partial least squares-discriminant analysis (PLS-DA), were combined to clearly distinguish among the P. harmala samples from the six different parts. Eleven differential components were screened by the PLS-DA loading plot, and the relative contents were calculated by univariate analysis of variance. Chemometric results showed significant differences in the metabolites of the different parts of P. harmala. The seeds contained large amounts of harmaline, harmine, and vasicine compared to other organs. The acetic acid, proline, lysine, and sucrose contents of the roots were significantly higher than those of the other parts. In the testa, the vasicine, asparagine, choline, and 4-hydroxyisoleucine contents were clearly dominant. The obtained data revealed the distribution characteristics of the metabolomes of the different P. harmala parts and provided fundamental knowledge for the rational development of its medicinal parts.
1. Introduction
P. harmala is the only salt-tolerant perennial herb in the Peganum genus of the family Zygophyllaceae [1]. It has been used as a popular traditional ethnodrug for a long time due to its phytotherapeutic value [2]. Traditionally, the seeds have been used to relieve pain, to promote blood circulation, and to treat rheumatism and illnesses such as cough and asthma [3, 4]. The whole plant has been used for pain relief and as a rheumatism treatment [5]. P. harmala extract is a rich source of bioactive substances [6], including large amounts of primary and secondary metabolites [7]. The whole plant and its seeds contain several kinds of alkaloids [8]. The alkaloid content of the seeds accounts for 3.92–7% of the dry weight [9]. The main alkaloid components are derivatives of quinazoline and β-carboline [10], which have exhibited anticancer and antibacterial activities in pharmacological studies [4, 10]. Because these metabolites located in different parts of the plant can vary greatly in type and quantity, the resulting pharmacological activities and antibacterial effects are significantly different [11]. Previous studies mainly focused on the extraction and activities of alkaloid compounds in P. harmala [12]. Only few alkaloids (β-carbolines and quinazoline derivatives) isolated from the different organs in P. harmala have been investigated [4, 13, 14], and no comprehensive studies of the metabolome of each of its parts have been performed.
Plant metabolomics is a method for investigating the dynamic changes in small-molecule metabolites or components in plants, and it has played an increasingly important role in explaining plant growth and reproduction [15, 16]. However, it is challenging to use metabolomics to comprehensively annotate metabolites and analyse the physiological and ecological roles of metabolomes [17]. As a rule, the structure of a compound is determined by spectral methods after being isolated and purified by various chromatographic techniques [13]. This process is time-consuming and laborious. At present, nuclear magnetic resonance (NMR) metabolomics technology offers a fast and sensitive method to detect distinctive signals and resolve the structures of compounds in a mixture by comparing with available data [18]. This technique has been widely applied in pharmacology, pharmacodynamics, and pharmacokinetics studies because of its advantages, including simple preparation, its unbiased nature, and its ability to qualitatively and quantitatively detect multiple metabolic components simultaneously [19].
In the current study, an NMR-based plant metabolomics method was used to analyse fingerprint spectra of methanol extracts obtained from different parts of P. harmala. A multivariate unsupervised analysis method, namely, principal component analysis (PCA), was employed to distinguish between the metabolomes of six different P. harmala parts. To enlarge the difference found in the PCA model and detect influential variables, partial least squares-discriminant analysis (PLS-DA), a supervised pattern recognition method, was used to recognize the characteristic differential metabolites among the groups classified according to different organs of plants. The relative contents of the major metabolites were analysed to help in explaining their ecological significance. This investigation of the characteristic components of the metabolomes of different P. harmala organs provides a molecular-level understanding of the distribution pattern of the metabolomes in this plant, which is expected to facilitate the rational development of medicinal plant resources.
2. Materials and Methods
NaH2PO4 and Na2HPO4 were used to prepare the buffer (pH = 6). Methanol and NaN3 were used to inhibit the activity of the decomposing enzymes in the samples (analytical pure, Beijing Chemical Plant). A 1 mmol/L trimethylsilylpropanoic acid (TSP) solution was prepared using D2O (99.9% deuterated, Cambridge Isotope Laboratories, USA). The deuterated 3-(trimethylsilyl)propionate sodium (TSP, 99% purity, J&K Scientific Co., Ltd.) solution was used as the internal standard. Milli-Q deionized water was used in the experiments. The P. harmala plant samples were collected in the Changji prefecture of Xinjiang, China. The flowers, stems, roots, testas, seeds, and leaves of the samples were collected, dried in air, ground, dried to a constant weight, and stored in a desiccator until use.
Sample preparation: first, 100 mL of methanol was added to a precisely weighed 2.0000 g crushed and dried plant sample three times for ultrasonic extraction. After centrifugation for 10 minutes, the supernatants were combined and subjected to rotary evaporation. The sample was freeze-dried to obtain the extract. Then, 0.2 mL of the deuterated TSP solution (1 mmol), 0.3 mL of a phosphate buffer (pH = 6.0), and 0.2 mL of NaN3 (10 mmol) solution were added to 10 mg of the extract, which was precisely weighed and mixed well. After sonication and centrifugation, 0.6 mL of the supernatant was pipetted into a 5 mm tube for NMR analysis. Five parallel preparations were performed for each sample.
NMR experiments: 1H-NMR measurements were performed with a 500 MHz NMR spectrometer at 298K. 1H and 13C nuclear resonance frequencies were 500 and 125 MHz, respectively. For the one-dimensional hydrogen spectra, the noesygppr1d pulse sequence with suppressed water peaks was used with the following parameters: a water peak suppression power of 41 dB (Bruker nomenclature), pulse delay time of 2 s, scan number of 128, spectrum width of 12 ppm, pulse time of 9.8 µs, sampling time of 2.72 s, relaxation time of 2.0 s, sampled data point number of 32,768, and free-induction decay (FID) resolution of 0.18 Hz. The FID was processed with an exponential window function with a widening factor of 0.3 Hz. The baseline adjustment and phase correction were all performed manually. The methanol-extracted, water-soluble metabolites were determined by NMR using the presaturation technique to suppress the water peaks; TSP was used as the internal standard, and deuterated water was used to lock the field. The 1H-NMR spectra were heavily superimposed, and it was difficult to identify many of the signals. Two-dimensional nuclear magnetic resonance experiments, including correlated spectroscopy (COSY), heteronuclear single quantum coherence (HSQC) spectroscopy, and heteronuclear multiple bond correlation (HMBC) spectroscopy, enabled the facile identification of the metabolites from the collected signals of the methanol extracts of the different P. harmala plant parts. For the COSY spectra, the spectral width was 5000 Hz, the number of sampling points was 400 (F1) × 4096 (F2), and the number of time increments was 24. For the HSQC spectra, the 1H and 13C spectral widths were 5000 Hz and 22638 Hz, respectively; the number of sampling points was 256 (F1) × 2048 (F2), and the number of time increments was 60. For the HMBC spectra, the 1H and 13C spectral widths were 5000 Hz and 30184 Hz, respectively; the number of sampling points was 256 (F1) × 4096 (F2), and the number of time increments was 112.
NMR data analysis: all the 1H-NMR spectra were processed using the TopSpin 3.2 software (Bruker Biospin). The baseline and phase were calibrated manually. After the chemical shifts were calibrated using TSP (δ H = 0.00), the spectra were imported into the MestReNova software (version 8.0.1, Mestrelab Research, Santiago de Compostela, Spain) for data processing. After calibrating the phase and adjusting the baseline, the spectrum was integrated in the range of δ H 0.5–10 ppm with an integration interval of 0.02 ppm. However, the spectrum was not integrated in the range of δ H 4.71–5.05 ppm (residual water peak). The sum of the integral areas of the spectrum was normalized to generate an Excel data file, which was imported into MATLAB (Umetrics, Umea, Sweden). PCA and PLS-DA were performed after mean centre processing, and the differential metabolites were screened using a variable importance factor (VIP) of >1 in the loading model. The effectiveness of the PLS-DA model was validated by a permutation test. The relative contents of some metabolites were analysed by analysis of variance (ANOVA) and t-tests. Duncan's new multiple range test was used to correct the p value [20].
3. Results and Discussion
3.1. 1H-NMR Fingerprint Peak Assignment for the Methanol Extracts of the Different Parts of P. harmala
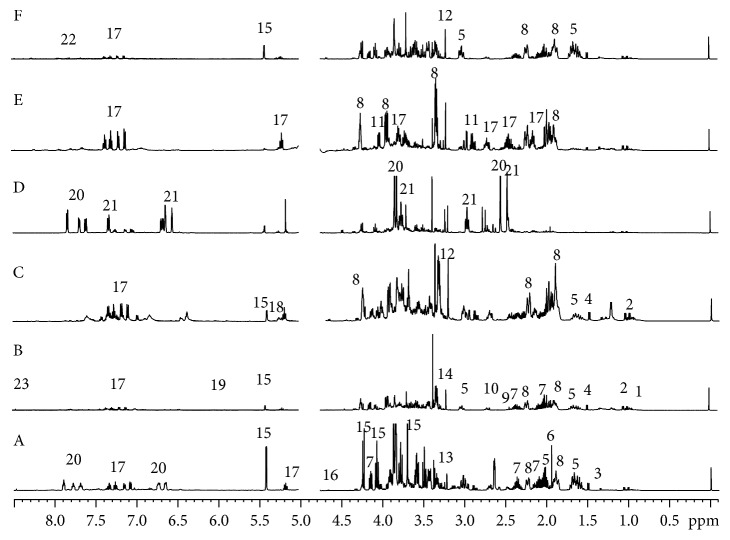
The fingerprint spectra of the polar extracts of P. harmala revealed the presence of 23 metabolites, 19 primary and 4 secondary (Figure 1, Table 1). The 19 primary metabolites were three carbohydrate compounds, five organic acids, eight amino acids, and the last three were compounds of other types. The four secondary metabolites were vasicine, vasicinone, harmine, and harmaline. These twenty-three compounds were identified in the methanol extracts of P. harmala by comparing the chemical shifts and coupling splitting values in the nuclear magnetic spectra and the relevant information from the two-dimensional NMR spectra to related literature data [21] and the standard spectra of amino acids, organic acids, and sugar compounds in the Human Metabolome Database [22] (HMDB) (http://www.hmdb.ca/). The 1H-NMR spectra of methanol extracts from different organs of P. harmala and the characteristic signals of the identified metabolites are shown in Figure 1. The main amino acids identified in the organic acid and amino acid region (δ H 0.5–3.0 ppm) were isoleucine, valine, threonine, alanine, proline, lysine, 4-hydroxyisoleucine, and asparagine. The main organic acids identified in this region included acetic acid, succinic acid, and malic acid. The signals of the sugar compounds in the range of δ H 3.0–6.0 ppm overlapped considerably. However, diagnostic anomeric proton signal of sucrose and glucose could be easily identified. Meanwhile, the characteristic signals of nitrogen-containing metabolites, such as choline, phosphorylcholine, and betaine, were clearly observed in this region, as shown in Table 1.
Figure 1.
Typical 500 MHz 1H-NMR spectra of the metabolites of the different parts of P. harmala: (A) root, (B) leaf, (C) flower, (D) seed, (E) testa, and (F) stem. Key metabolites: 1, leucine; 2, valine; 3, threonine; 4, alanine; 5, lysine; 6, acetic acid; 7, proline; 8, 4-hydroxyisoleucine; 9, succinic acid; 10, malic acid; 11, asparagine; 12, choline; 13, phosphorylcholine; 14, betaine; 15, sucrose; 16, β-glucose; 17, vasicine; 18, α-glucose; 19, maleic acid; 20, harmine; 21, harmaline; 22, vasicinone; 23, formic acid.
Table 1.
1H-NMR assignments of major metabolites in P. harmala extracts.
| No. | Metabolite | δ H (multiplicity, J) | Sample |
|---|---|---|---|
| 1 | Isoleucine | 1.02 (d, 7.06), 0.95 (t, 7.15) | Bb |
| 2 | Valine | 1.05 (d, 7.0), 0.99 (d, 7.0) | Bb |
| 3 | Threonine | 4.25 (m), 1.33 (d, 6.55) | Ab |
| 4 | Alanine | 3.57 (m), 1.48 (d, 7.3) | Bb |
| 5 | Lysine | 3.6 (m), 1.65, 1.89 (m), 2.25 (m), 3.02, 3.4 (m) | Fb |
| 6 | Acetic acid | 1.92 (s) | Ab |
| 7 | Proline | 3.30–3.35 (m), 2.31–2.37 (m), 1.95–2.00 (m), 4.12 (dd, 8.63, 6.56) | Ab |
| 8 | 4-Hydroxyisoleucine | 4.25 (m), 2.167 (m), 3.835 (m), 2.22, 1.98 (m), 1.88 (m) | Ea |
| 9 | Succinic acid | 2.43 (s), 2.43 (s) | Fb |
| 10 | Malic acid | 2.71 (dd, 2.9, 15.62), 4.31 (dd, 3.1, 10.2) | Fb |
| 11 | Asparagine | 4.03 (dd, 4.4, 7.25), 2.94 (m), 2.84 (m) | Eb |
| 12 | Choline | 4.07, 3.21 (s) | Cb |
| 13 | Phosphorylcholine | 3.23 (s) | Ab |
| 14 | Betaine | 3.27 (s) | Bb |
| 15 | Sucrose | 3.69 (s), 4.22 (d, 8.65), 4.06 (t, 8.89), 3.91 (m), 3.87 (d, 3.15), 5.42 (d, 3.8), 3.59 (m), 3.78 (t, 9.48), 3.49 (t, 9.28), 3.91(m) | Ab |
| 16 | β-Glucose | 4.65 (d, 8.0) | Ab |
| 17 | Vasicine | 3.77, 3.69 (m),2.15, 2.71 (m), 5.22 (t, 8.80), 7.11 (m), 7.36 (m), 7.28 (m), 7.19 (d, 7.5) | Ea |
| 18 | α-Glucose | 5.24 (d, 3.83) | Cb |
| 19 | Maleic acid | 6.01 (s) | Bb |
| 20 | Harmine | 6.64 (d, 1.9), 6.69 (dd, 8.75, 2.25), 7.84 (d, 5.64), 2.55 (s), 7.69 (d, 5.42), 7.61 (d, 8.59), 3.84 (s) | Da |
| 21 | Harmaline | 6.46 (dd, 8.85, 1.94), 6.37 (d, 1.66), 7.0 (d, 8.64), 2.47 (s), 2.96(t, 8.83), 3.22(t, 8.83), 3.81(s) | Da |
| 22 | Vasicinone | 4.11, 3.91 (m), 1.99, 2.45 (m), 4.99 (t, 6), 7.71 (m), 7.93 (m), 7.64 (m), 8.25 (d, 6) | Fa |
| 23 | Formic acid | 8.46 (s) | Ab |
Note. “a” was determined by the 1H-NMR, COSY, HSQC, and HMBC spectra. “b” was determined by the 1H-NMR and HSQC spectra. s, singlet; d, doublet; t, triplet; m, multiplet; A, root; B, leaf; C, flower; D, seed; E, testa; F, stem.
In the aromatic region, vasicine, vasicinone, harmaline, and harmine were identified in the methanol extracts of P. harmala. Moreover, a comparison with the literature [23–25] and analysis of the two-dimensional COSY, HMBC, and HSQC spectra further confirmed the existence of these compounds, and the peak assignments are shown in Table 1. In addition to alkaloids described above, signal related to formic acid was also identified in the aromatic region of the spectra.
3.2. Multivariate Analysis of the 1H-NMR Data
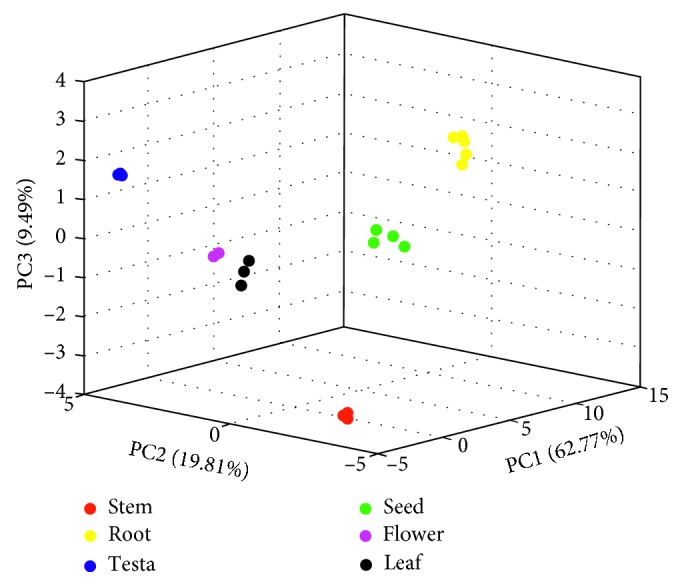
The 1H-NMR fingerprint spectra of the methanol extracts of the six parts of P. harmala are shown in Figure 1. A visual observation of the spectra revealed that the presence of many types of metabolites in stems, leaves, roots, and flowers, especially amino acids, with relatively high contents, whereas the metabolite levels in the testas and seeds were low. To further determine the potential differential metabolites, a multivariate statistical analysis (PCA) of the nuclear magnetic data of the polar metabolites of the six different P. harmala parts was performed, and the results are shown in Figure 2.
Figure 2.
PCA score plot of the NMR spectra of the methanol extracts of the six different P. harmala parts (“red,” “yellow,” “blue,” “green,” “magenta,” and “black” dots represent the stem, root, testa, seed, flower, and leaf of the plant, respectively).
PCA is the most commonly performed unsupervised pattern recognition method in metabolomics research. The latent variable information can be determined from massive data sets to reduce the data dimensionality [26]. The sample classification information can be obtained from the score plot. In the current work, PCA was undertaken on the obtained data, and then satisfied results were generated. As shown in the PCA score plot in Figure 2, the first three principal components (PC1, PC2, and PC3) accounted for 92.07% of the original variable information (PC1: 62.77%, PC2: 19.81%, and PC3: 9.49%). The metabolites of the six P. harmala parts were significantly different, and the separation trend was obvious. Along the first ordination axis, the seeds and roots were clearly distinguished from the other plant parts and exhibited a positive correlation. In the PC2 direction of the score plot, the stems and roots were clearly distinguished from the extracts of the other plant parts, exhibiting a negative correlation.
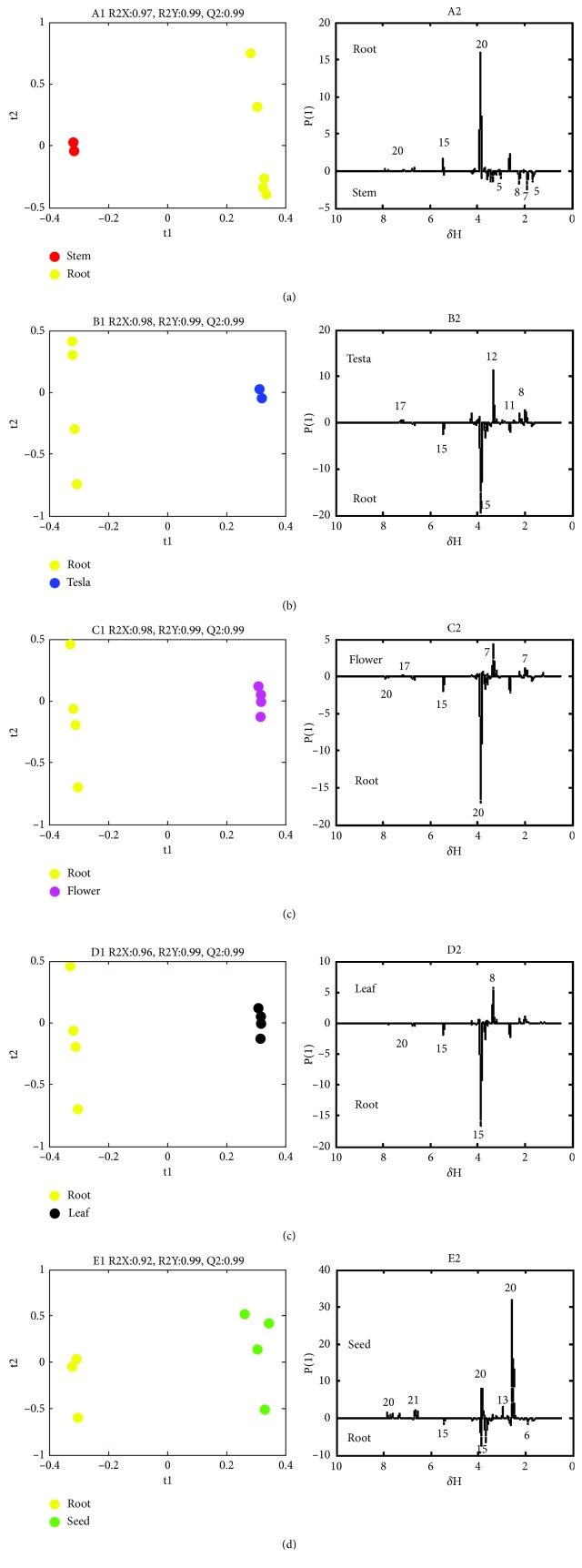
To identify the differential metabolites of the six parts, the root sample was used as the control and the samples of the other parts were compared to it and sorted to identify the metabolites that contribute the most to the group classification. PLS-DA was applied to the 1H-NMR data of the stem, testa, flower, leaf, and seed to obtain score and loading plots (Figure 3). The score plot of A1, B1, C1, D1, and E1 in Figure 3 shows that the root was completely separated from the other parts. The parameters R2X, R2Y, and Q2 of the five models are shown in Figure 3 and were all greater than 0.9, indicating that the models had strong predictive power [27, 28]. Permutation tests were performed to PLS-DA models to verify that these parameters generated by the established models were not overfitted. Therefore, the models were validated, showing that the results were reliable. The loading plot can be used to determine the variables contributing to the classification, and depending on the levels of their contributions, the variables that can be considered as potential biomarkers can be discovered. Based on the A2, B2, C2, D2, and E2 groups shown in the PLS-DA loading plot, the significant differential metabolites included acetic acid (6), asparagine (11), lysine (5), proline (7), choline (12), phosphocholine (13), 4-hydroxyisoleucine (8), sucrose (15), vasicine (17), harmine (20), and harmaline (21).
Figure 3.
PLS-DA score plot (A1-E1) and linear loading plot (A2-E2) of the methanol extracts of the different P. harmala parts (“red,” “yellow,” “blue,” “green,” “magenta,” and “black” dots represent the methanol extracts of the stem, root, testa, seed, flower, and leaf of the plant, respectively; t1 and t2 are the scoring points for the first and second principal components, respectively; δ H is the 1H-NMR chemical shift; P[1] is the loading point of the first principal component. The numbers represent different metabolites, as shown in Table 2).
3.3. Screening and Univariate Analysis of the Potential Characteristic Metabolites
The data with VIP>1 in the PLS-DA loading plot of the five methanol extracts of P. harmala were analysed to obtain the most significant metabolites for their classification, which included acetic acid, asparagine, choline, harmine, harmaline, 4-hydroxyisoleucine, phosphocholine, proline, lysine, sucrose, and vasicine. Specifically, to obtain the relative contents of the metabolites, which were subjected to univariate analysis of variance, their characteristic peaks were compared with the peak area of the internal standard. The calculated relative contents of the metabolites in the methanol extracts of the different P. harmala parts are shown in Table 2. All the data were analysed by ANOVA and t-tests. Differences were considered to be statistically significant at p < 0.05. The data in Table 2 show that the acetic acid, proline, lysine, sucrose, and vasicine contents of the P. harmala roots were significantly higher than those of the other parts. In addition to vasicine, the asparagine, choline, and 4-hydroxyisoleucine contents in the testas were clearly dominant. The seeds contained large amounts of harmaline, harmine, and phosphocholine. However, the differences in the metabolomes of the P. harmala stems, leaves, and flowers were not significant, which is consistent with the results of the multivariate analysis.
Table 2.
Relative quantification of principal metabolites in different parts of P harmala (mg/g).
| Compounds | Stem | Root | Testa | Seed | Flower | Leaf |
|---|---|---|---|---|---|---|
| Proline | 37.88 ± 18.95bc | 86.53 ± 10.27a | 26.70 ± 2.14c | — | 55.10 ± 7.88b | 55.93 ± 2.39b |
| Lysine | 153.63 ± 5.41b | 180.02 ± 12.46a | — | — | 95.87 ± 12.23c | 68.85 ± 4.08d |
| Asparagine | 17.77 ± 1.75c | 17.96 ± 3.32c | 123.84 ± 12.11a | 29.37 ± 29.68c | 76.03 ± 9.36b | 32.03 ± 4.48c |
| 4-Hydroxyisoleucine | 204.85 ± 9.81b | 189.29 ± 16.19b | 364.89 ± 50.07a | — | 195.70 ± 15.17b | 160.35 ± 8.92b |
| Acetic acid | 1.85 ± 0.17c | 7.77 ± 0.98a | 4.39 ± 0.85b | — | 4.18 ± 0.54b | 2.68 ± 0.09c |
| Sucrose | 113.024 ± 8.45b | 339.19 ± 35.69a | 13.29 ± 4.92d | 63.28 ± 2.0c | 60.08 ± 5.91c | 40.63 ± 1.63c |
| Vasicine | 17.19 ± 9.8b | 64.31 ± 8.44ab | 124.13 ± 71.58a | — | 68.36 ± 6.69ab | 26.69 ± 2.56b |
| Harmine | — | 19.27 ± 1.68b | — | 40.77 ± 5.98a | — | — |
| Harmaline | — | — | — | 58.46 ± 9.61 | — | — |
| Choline | 4.24 ± 0.17c | 2.67 ± 0.18d | 8.59 ± 0.96a | 2.31 ± 1.45d | 6.12 ± 0.51b | 3.31 ± 0.19cd |
| Phosphorylcholine | 0.55 ± 0.13d | 2.92 ± 0.20b | 3.56 ± 0.52b | 5.50 ± 1.33a | 0.97 ± 0.14cd | 1.99 ± 0.13bc |
Note. The numbers are the contents of the compounds in the methanol extracts of the different P. harmala parts (mean value ± standard deviation). “—” indicates that the compound was not detected. Duncan's new multiple range test was used to calculate the p value (when p < 0.05, different letters within same row represent subsets with significant differences; the order of the letters indicates the range of sample concentrations from large to small).
3.4. Biological Significance of the Characteristic Metabolites
The results showed that the sucrose, proline, acetic acid, betaine, and lysine contents of the P. harmala roots were much higher than those of the other parts. Of these metabolites, sucrose, proline, betaine, and acetic acid are osmotic regulators in plants, controlling osmotic adjustment [29]. The roots of a plant are the vegetative organ responsible for the physiological functions of nutrient absorption, production, and transport [30]. Sucrose is also used as an energy carrier in plants and is the main source of carbon and energy for plant growth and development. P. harmala roots are well suited to performing the main nutrient functions for plant growth and development and to contributing to plant metabolism, and sucrose provides energy for the growth, development, and reproduction of the plant [31]. The proline and betaine contents can increase under abiotic stress conditions, such as exposure to drought, high salinity, high temperatures, and heavy metals [32]. Besides to serve as valuable sources of nitrogen and carbon, proline and betaine prevent plant damage caused by osmotic stress and act as free radical scavengers [33–35]. It is generally believed that proline is mostly synthesized in the roots and most of the product is transported to the above-ground parts. The acidic substances in the roots can enhance the effectiveness of the nutrients in the rhizosphere soil, promote the acidification, chelation, ion exchange, and reduction in the insoluble nutrients in the plant, and participate in circulation and energy flow of the nutrient elements through the plant [36].
The experimental results showed that the alkaloid contents of the P. harmala seeds, including the harmine, harmaline, and phosphocholine contents, were most abundant compared to other parts [37]. Alkaloids are secondary metabolites that are generated when plants resist invasion from the environment and defend against external factors. They do not directly participate in plant growth, development, and reproduction, but they can improve the adaptability of the plant to adverse environments. The seed of P. harmala is the reproductive organ and is rich in alkaloids. To germinate and grow in an arid environment, the plant must strongly compete for water, nutrients, and space. The alkaloids in P. harmala seeds must be washed out by rainwater before germination. When these compounds enter the soil environment, they might inhibit the germination of other plant seeds and the growth of their seedlings through allelopathy [38]. The results demonstrate a higher concentration of phosphocholine in P. harmala seeds than other organs. The phosphate in the plant fluid exists mainly in the form of phosphocholine, a phosphorus carrier that accounts for 5–20% of the total phosphorus in plants, and is a key component of the plant cell membrane [39].
The data show an increase in the level of vasicine, asparagine, choline, and 4-hydroxyisoleucine contents in testa than the other groups. The testa is a protective layer on the outside of the plant seed that protects the seed embryo from mechanical damage and prevents pests, disease invasion, and water loss [40]. Its main physiological role is to control the germination of the seed by enhancing its dormancy, limit unfavourable biochemical activities during seed storage, and protect the seed and root from animal foraging and microbiological and viral invasion [41]. Harmaline and choline in the P. harmala testa might play a role in these defensive functions. 4-Hydroxyisoleucine is a unique nonprotein amino acid in plants that exhibits physiological and chemical activities that are responsible for glucose and lipid metabolism [42]. The amino acid asparagine is a soluble form of nitrogen that is predominantly absorbed directly by the plant roots. These compounds in the testa of P. harmala are closely related to its self-protection capabilities [42].
4. Conclusions
In this study, NMR was used to analyse methanol extracts of six parts of P. harmala and the plant metabolomics technique aimed at investigating the characteristic chemical composition of these extracts. The results showed that the differential metabolites in the different P. harmala parts included sucrose, harmine, proline, lysine, betaine, acetic acid, harmaline, vasicine, choline, 4-hydroxyisoleucine, and asparagine. From the quantitative point of view, the relative contents of these 11 metabolites in the methanol extracts of the six P. harmala parts studied were determined. The results revealed the significant differences in the types of metabolites in the P. harmala roots, seeds, and testas. The NMR-based plant metabolomics method established in this research can provide a detailed metabolomic profile of biological samples from different plant parts and allow the holistic analysis of metabolome. Resulting in improved understanding of P. harmala metabolism, the results can provide important data to aid in plant ecophysiological studies and facilitate the development of P. harmala medicinal resources.
Acknowledgments
We wish to thank Professor Jin Li, School of Life Sciences of Xinjiang Normal University, for sample identification. This work was supported by the National Natural Science Foundation of China (Nos. 21165081 and 21465023) and the National Key Research and Development Program (No. 2016YFC0500805).
Contributor Information
Zhufeng Geng, Email: gengzhufeng@bnu.edu.cn.
Zhiwei Deng, Email: dengzw@bnu.edu.cn.
Data Availability
The Excel data used to support the findings of this study are included within the supplementary information file.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Supplementary Materials
Supplemental Table: the 1H-NMR raw data of the different parts of P. harmala methanol extract.
References
- 1.Ahmed M. Z., Khan M. A. Tolerance and recovery responses of playa halophytes to light, salinity and temperature stresses during seed germination. Flora-Morphology, Distribution, Functional Ecology of Plants. 2010;205(11):764–771. doi: 10.1016/j.flora.2009.10.003. [DOI] [Google Scholar]
- 2.Aziz M. A., Khan A. H., Adnan M., Izatullah I. Traditional uses of medicinal plants reported by the indigenous communities and local herbal practitioners of Bajaur Agency, federally administrated tribal areas, Pakistan. Journal of Ethnopharmacology. 2017;198:268–281. doi: 10.1016/j.jep.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 3.Niroumand M. C., Farzaei M. H., Amin G. Medicinal properties of Peganum harmala L. in traditional Iranian medicine and modern phytotherapy: a review. Journal of Traditional Chinese Medicine. 2015;35(1):104–109. doi: 10.1016/s0254-6272(15)30016-9. [DOI] [PubMed] [Google Scholar]
- 4.Li S., Wang K., Gong C., et al. Cytotoxic quinazoline alkaloids from the seeds of Peganum harmala . Bioorganic and Medicinal Chemistry Letters. 2018;28(2):103–106. doi: 10.1016/j.bmcl.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Bournine L., Bensalem S., Fatmi S., et al. Evaluation of the cytotoxic and cytostatic activities of alkaloid extracts from different parts of Peganum harmala L. (Zygophyllaceae) European Journal of Integrative Medicine. 2017;9:91–96. doi: 10.1016/j.eujim.2016.10.002. [DOI] [Google Scholar]
- 6.Khlifi D., Sghaier R. M., Amouri S., et al. Composition and anti-oxidant, anti-cancer and anti-inflammatory activities of Artemisia herba-alba, Ruta chalpensis L. and Peganum harmala L. Food and Chemical Toxicology. 2013;55:202–208. doi: 10.1016/j.fct.2013.01.004. [DOI] [PubMed] [Google Scholar]
- 7.Li S., Cheng X., Wang C. A review on traditional uses, phytochemistry, pharmacology, pharmacokinetics and toxicology of the genus Peganum. Journal of Ethnopharmacology. 2017;203:127–162. doi: 10.1016/j.jep.2017.03.049. [DOI] [PubMed] [Google Scholar]
- 8.Moloudizargari M., Mikaili P., Aghajanshakeri S., Asghari M., Shayegh J. Pharmacological and therapeutic effects of Peganum harmala and its main alkaloids. Pharmacognosy Reviews. 2013;7(14):p. 199. doi: 10.4103/0973-7847.120524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Apostolico I., Aliberti L., Caputo L., et al. Chemical composition, antibacterial and phytotoxic activities of Peganum harmala seed essential oils from five different localities in Northern Africa. Molecules. 2016;21(9):p. 1235. doi: 10.3390/molecules21091235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daoud A., Song J., Xiao F., Shang J. B-9-3, a novel β-carboline derivative exhibits anti-cancer activity via induction of apoptosis and inhibition of cell migration in vitro. European Journal of Pharmacology. 2014;724:219–230. doi: 10.1016/j.ejphar.2013.12.038. [DOI] [PubMed] [Google Scholar]
- 11.Asgarpanah J., Ramezanloo F. Chemistry, pharmacology and medicinal properties of Peganum harmala L. African Journal of Pharmacy and Pharmacology. 2012;6(22):1573–1580. doi: 10.5897/ajpp11.876. [DOI] [Google Scholar]
- 12.Lamchouri F., Zemzami M., Jossang A., et al. Cytotoxicity of alkaloids isolated from Peganum harmala seeds. Pakistan Journal of Pharmaceutical Sciences. 2013;26(4):699–706. [PubMed] [Google Scholar]
- 13.Ma X., Liu D., Tang H., et al. Purification and characterization of a novel antifungal protein with antiproliferation and anti-HIV-1 reverse transcriptase activities from Peganum harmala seeds. Acta Biochimica et Biophysica Sinica. 2013;45(2):87–94. doi: 10.1093/abbs/gms094. [DOI] [PubMed] [Google Scholar]
- 14.Mahmoudian M. H. J. A. Toxicity of Peganum harmala: review and a case report. Iranian Journal of Pharmacology and Therapeutics. 2002;1:1–4. [Google Scholar]
- 15.Kim H. K., Choi Y. H., Verpoorte R. NMR-based metabolomic analysis of plants. Nature Protocols. 2010;5(3):536–549. doi: 10.1038/nprot.2009.237. [DOI] [PubMed] [Google Scholar]
- 16.Hall R. D. Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytologist. 2006;169(3):453–468. doi: 10.1111/j.1469-8137.2005.01632.x. [DOI] [PubMed] [Google Scholar]
- 17.Jones O. A. H., Maguire M. L., Griffin J. L., et al. Metabolomics and its use in ecology. Austral Ecology. 2013;38(6):713–720. doi: 10.1111/aec.12019. [DOI] [Google Scholar]
- 18.Fan T. W. M., Lane A. N. Applications of NMR spectroscopy to systems biochemistry. Progress in Nuclear Magnetic Resonance Spectroscopy. 2016;92-93:18–53. doi: 10.1016/j.pnmrs.2016.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Markley J. L., Bruschweiler R., Edison A. S., et al. The future of NMR-based metabolomics. Current Opinion in Biotechnology. 2017;43:34–40. doi: 10.1016/j.copbio.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duncan D. B. Multiple range and multiple F tests. Biometrics. 1955;11(1):1–42. doi: 10.2307/3001478. [DOI] [Google Scholar]
- 21.Li Y., He Q., Geng Z., et al. NMR-based metabolomic profiling of Peganum harmala L. reveals dynamic variations between different growth stages. Royal Society Open Science. 2018;5(7) doi: 10.1098/rsos.171722.171722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wishart D. S., Tzur D., Knox C., et al. HMDB: the human metabolome Database. Nucleic Acids Research. 2007;35:521–526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaw B. S., Bai Y., Puar M. S., Dubose K. K., William Pelletier S. 1H- and 13C-NMR assignments for some pyrroloe[2,lb] quinazoline alkaloids of adhatoda vaszca. Journal of Natural Products. 1994;57(7):953–962. doi: 10.1021/np50109a012. [DOI] [Google Scholar]
- 24.Zheng X., Zhang Z., Chou G., et al. Acetylcholinesterase inhibitive activity-guided isolation of two new alkaloids from seeds of Peganum nigellastrum bunge by an in vitro TLC- bioautographic assay. Archives of Pharmacal Research. 2009;32(9):1245–1251. doi: 10.1007/s12272-009-1910-x. [DOI] [PubMed] [Google Scholar]
- 25.Mhaske S. B., Argade N. P. Concise and efficient synthesis of bioactive natural products pegamine, deoxyvasicinone, and (−)-Vasicinone. Journal of Organic Chemistry. 2001;66(26):9038–9040. doi: 10.1021/jo010727l. [DOI] [PubMed] [Google Scholar]
- 26.Maulidiani M., Mediani A., Abas F., et al. 1 H NMR and antioxidant profiles of polar and non-polar extracts of persimmon ( Diospyros kaki L.) – metabolomics study based on cultivars and origins. Talanta. 2018;184:277–286. doi: 10.1016/j.talanta.2018.02.084. [DOI] [PubMed] [Google Scholar]
- 27.Basoglu A., Sen I., Meoni G., Tenori L., Naseri A. NMR-based plasma metabolomics at set intervals in newborn dairy calves with severe sepsis. Mediators of Inflammation. 2018;2018:12. doi: 10.1155/2018/8016510.8016510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim S., Shin B., Lim D. K., et al. Expeditious discrimination of four species of the Panax genus using direct infusion-MS/MS combined with multivariate statistical analysis. Journal of Chromatography B. 2015;1002:329–336. doi: 10.1016/j.jchromb.2015.08.034. [DOI] [PubMed] [Google Scholar]
- 29.Parvaiz Ahmad M. R. W. Physiological Mechanisms and Adaptation Strategies in Plants Under Changing Environment. New York, NY, USA: Springer; 2014. [Google Scholar]
- 30.Iversen C. M. Using root form to improve our understanding of root function. New Phytologist. 2014;203(3):707–709. doi: 10.1111/nph.12902. [DOI] [PubMed] [Google Scholar]
- 31.Hu L., Chen L., Liu L., et al. Metabolic acclimation of source and sink tissues to salinity stress in bermudagrass (Cynodon dactylon) Physiologia Plantarum. 2015;155(2):166–179. doi: 10.1111/ppl.12312. [DOI] [PubMed] [Google Scholar]
- 32.Li Z., Duan X., Min X., Zhou Z. Methylglyoxal as a novel signal molecule induces the salt tolerance of wheat by regulating the glyoxalase system, the antioxidant system, and osmolytes. Protoplasma. 2017;254(5):1995–2006. doi: 10.1007/s00709-017-1094-z. [DOI] [PubMed] [Google Scholar]
- 33.BRIENS M., LARHER F. Osmoregulation in halophytic higher-plants-a comparative-study of soluble carbohydrates, polyols, betaines and free proline. Plant, Cell and Environment. 1982;5(4):287–292. doi: 10.1111/1365-3040.ep11572682. [DOI] [Google Scholar]
- 34.Li Z., Duan X., Min X., Zhou Z. Methylglyoxal as a novel signal molecule induces the salt tolerance of wheat by regulating the glyoxalase system, the antioxidant system, and osmolytes. Protoplasma. 2017;254(5):1995–2006. doi: 10.1007/s00709-017-1094-z. [DOI] [PubMed] [Google Scholar]
- 35.Ashraf M., Foolad M. R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environmental and Experimental Botany. 2007;59(2):206–216. doi: 10.1016/j.envexpbot.2005.12.006. [DOI] [Google Scholar]
- 36.Jia W., Davies W. J. Modification of leaf apoplastic pH in relation to stomatal sensitivity to root-sourced abscisic acid signals. Plant Physiology. 2006;143(1):68–77. doi: 10.1104/pp.106.089110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Herraiz T., Guillén H., Arán V. J., Salgado A. Identification, occurrence and activity of quinazoline alkaloids in Peganum harmala . Food and Chemical Toxicology. 2017;103:261–269. doi: 10.1016/j.fct.2017.03.010. [DOI] [PubMed] [Google Scholar]
- 38.Shao H., Huang X., Zhang Y., Zhang C. Main alkaloids of Peganum harmala L. And their different effects on dicot and monocot crops. Molecules. 2013;18(3):2623–2634. doi: 10.3390/molecules18032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi H., Chen T., Murata N. Transformation with a gene for choline oxidase enhances the cold tolerance of Arabidopsis during germination and early growth. Plant, Cell and Environment. 1998;21:232–239. [Google Scholar]
- 40.Steinbrecher T., Leubner-Metzger G. The biomechanics of seed germination. Journal of Experimental Botany. 2016;68,(4):765–783. doi: 10.1093/jxb/erw428. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Z., Bao W., Wu N. Dormancy and germination in rosa multibracteata hemsl. & E. H. Wilson. Scientia Horticulturae. 2009;119(4):434–441. doi: 10.1016/j.scienta.2008.08.017. [DOI] [Google Scholar]
- 42.Zandi P., Basu S. K., Khatibani L. B., et al. Fenugreek (Trigonella foenum-graecum L.) seed: a review of physiological and biochemical properties and their genetic improvement. Acta Physiologiae Plantarum. 2015;37(1) doi: 10.1007/s11738-014-1714-6. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table: the 1H-NMR raw data of the different parts of P. harmala methanol extract.
Data Availability Statement
The Excel data used to support the findings of this study are included within the supplementary information file.