Abstract
The sheep rumen submodel MollyRum14 was evaluated on its methane and VFA predictions against data from respiration-chamber trials conducted with sheep fed perennial ryegrass, white clover, chicory, forage rape, turnip (leafy and bulb varieties), swedes, kale, or forage radish. We assessed the model’s response to substrate degradation rate (settings that affect the rate of cellulose and hemicellulose digestion) and to fermentation stoichiometry (settings that alter nonglucogenic to glucogenic short-chain fatty acid ratios). Model predictions were evaluated against data for methane production (pCH4: g/d), methane yield (yCH4: g/kg DMI), and acetate to propionate ratio (A:P). The predictive ability of the model for both pCH4 and yCH4 was superior for perennial ryegrass than for other forages. Except for swedes and chicory, predictions for yCH4 were correctly ranked across the forages evaluated. Except for forage rape, robust predictions were obtained for all forages using fast degradation kinetics and a predominantly acetogenic stoichiometry. Model predictions for forage rape were enhanced using slow degradation kinetics and a predominantly propionic stoichiometry. These results indicate that MollyRum14 is suitable to predict methane emissions from sheep fed a variety of fresh forages including annual fodder crops. However, a clear understanding of degradation rates and stoichiometries is needed to enhance the utility of the model as a predictive tool. This would allow continuous adjustment of digestion rates and stoichiometries to be potentially tailored to individual forage species.
Keywords: digestion, methane, modeling, rumen, sheep
INTRODUCTION
Ruminant livestock significantly contribute to methane production on a global scale (Steinfeld et al., 2006). Mathematical modeling has potential for investigating methane mitigation strategies, but requires robust models. Alemu et al. (2011) and Morvay et al. (2011) reported that fermentation stoichiometries used in a range of models need improvement for reliably predicting methane emissions. Also, van Lingen et al. (2016) suggested that detailed mechanistic consideration is required to explain rumen fermentation stoichiometries.
We previously modified the rumen submodel of Baldwin’s (1995) Molly95 dairy cow model to produce a new sheep rumen model (MollyRum14). Relative to the original model, MollyRum14 included a rumen hydrogen (H2) pool as a dynamic variable regulating the estimation of methane formation and a correction of the Molly95 equation for microbial H2 utilization, but the fermentation stoichiometry (FS) and fiber digestion were left unchanged (Vetharaniam et al., 2015).
Using data from studies in which sheep were fed fresh perennial ryegrass (Lolium perenne), MollyRum14 predicted methane production better than Molly95 (Vetharaniam et al., 2015), with relative prediction errors of 4.4% and 17.7%, respectively. Although New Zealand’s sheep sector relies heavily on perennial ryegrass and white clover (Trifolium repens) pastures, other forage crops such as chicory (Cichorium intybus) and a variety of Brassica species have grown in importance. In sheep-feeding trials, some Brassica species were effective in reducing CH4 (g/g DMI) compared with perennial ryegrass (Pacheco et al., 2014). One explanation for this effect was that sheep fed forage rape (Brassica napus L.) had fermentation patterns similar to those in animals fed grain diets (i.e., lower ruminal pH and acetate to propionate ratio) (Sun et al., 2015). MollyRum14, like its predecessor Molly95, has descriptive specifications that categorize the diet being eaten, and these are used within the model to set degradation rates and fermentation stoichiometries. Our objective was to test how well the current specifications in MollyRum14 actually performed on a range of fresh forages.
This study evaluated the ability of MollyRum14 to predict methane emissions from respiration-chamber trials with sheep fed a variety of fresh temperate forages. We also tested the effect on the prediction of changing the descriptive feed categories which regulate fiber degradation rates and FS.
MATERIALS AND METHODS
Parameters to Identify the Nature of Feeds
Diets in MollyRum14, like Molly95, are nominally specified as legume, grass, or corn silage, which are then used to set rate constants in fiber degradation functions. For these categories (legume, grass, or corn silage), the degradation rate constants are, respectively 6.0, 9.0, or 9.0 d−1 (kg fibrolytic microbes)−1 for cellulose, and 6.0, 7.0, or 9.0 d−1 (kg fibrolytic microbes)−1 for hemicellulose. To avoid confusion when testing the categorization of forages, we will henceforth refer to these specifications as SLOW, moderate (MOD), and FAST degradation rates (Table 1). Although MollyRum14 was formulated to allow modeling of diets containing a mixture of forage types, Molly95 does not have that capability. Thus, we have not considered diets with mixed forage types, and only monocultures are considered here.
Table 1.
Parameters used in MollyRum14 to characterize the chemical composition of forages (as sole feed sources)
| Parameter | Interpretation |
|---|---|
| FdAc | Acetate |
| FdAi | Insoluble ash |
| FdAs | Soluble ash |
| FdBu | Butyrate |
| FdCe | Cellulose |
| Fdfat | Fat added to diet |
| FdHc | Hemicellulose |
| FdLa | Lactate |
| FdLg | Lignin |
| FdLi | Lipids |
| FdNn | NPN |
| FdOa | Organic acids |
| FdPe | Pectin |
| FdPi | Insoluble protein |
| FdPs | Soluble protein |
| FdSc | Soluble carbohydrate |
| FdSt | Starch |
| FdUr | Urea |
| PSF | Small particle fraction |
| Stsol | Starch soluble fraction |
| FS = 0, FS = 0.5, FS = 1 | Fermentation stoichiometry that alters nonglucogenic to glucogenic short-chain fatty acid ratios |
| SLOW, MOD, FAST | Substrate (cellulose and hemicellulose) degradation rate categorized as slow, moderate, or fast |
| WaterConc | Fraction of water in diet |
A prefix Fd indicates a fraction in feed expressed as kg/kg DM.
The model Molly95 has 3 Boolean input parameters (exactly one of which is set to 1) that nominally specify the type of diet and are used to set the FS parameters for short-chain fatty acids (SCFA) formation. FORSET = 1 indicates that the diet is largely forage and sets a stoichiometry that favors mainly acetogenesis. CONSET = 1 indicates that the diet is largely concentrate and sets a stoichiometry that favors mainly propionogenesis. MIXSET = 1 indicates a diet that is half forage, half concentrate and sets a stoichiometry that is intermediate the FORSET and CONSET stoichiometries. The presence of a greater proportion of concentrates in the feed would be associated with an increase in the production of propionate at the expense of acetate (Janssen, 2010). To allow us to test different stoichiometries against different feed types without causing confusion, in MollyRum14, we have replaced these Boolean parameters with a single FS parameter that linearly interpolates between these stoichiometry options: FS = 0, 0.5, and 1.0 corresponds, respectively, to true values for, FORSET = 1, MIXSET, and CONSET.
Parameters Used to Identify Chemical Composition of Feeds
In addition to the categorical parameters discussed above, 21 parameters are used in MollyRum14 to define the chemical composition of feed and water content (or DM) of the feed (Table 1). Any combination of feeds/feed types fed to an animal is represented by this set of parameters. We assumed that this characterization is sufficient to capture the essential differences in diets and thus focused on model responses to different chemical compositions of diets and to assumptions regarding the forage and feed types. These descriptive variables are included in Table 1.
Database Assembly
Data on forage composition were collated from a number of trials in which methane emissions were measured from sheep housed in respiration chambers (Sun et al., 2011; Sun et al., 2012b; Sun et al., 2013; Sun et al., 2016), together with information on mean dry matter intake (DMI), methane production (pCH4; g/d), methane yield (yCH4; g/kg DMI), and the ratio of ruminal acetate to propionate molar concentrations (A:P). The dataset comprised 43 experimental means, each one representing the chemical composition of a forage fed to a group of sheep (Table 2). Each experimental mean represented between 4 and 24 individual sheep. The dataset included treatment means from experiments in which sheep were fed 100% of the diet as 1 of 9 different fresh forages. These included perennial ryegrass (L. perenne L.; n = 14 means), forage rape (B. napus ssp. biennis; n = 15), white clover (T. repens L.; n = 4), bulb turnip (Brassica rapa ssp. rapa; n = 3), leafy turnip (Brassica campestris ssp. rapifera; n = 2), chicory (C. intybus L.; n = 2), swedes (B. napus ssp. napobrassica; n = 1), forage radish (Raphanus raphanistrum; n = 1), and kale (Brassica oleracea ssp. acephala; n = 1). The data used (Table 2) did not include data from ryegrass-fed sheep that were previously used for calibration and validation of the model (Vetharaniam et al., 2015). To complete the MollyRum14 input requirements in Table 1, estimation of missing composition data was required. Total ash was calculated by subtracting percentage organic matter from 100. If cellulose or lignin values were missing for a trial, the values were calculated by assuming that ADF was the sum of cellulose and lignin. Soluble protein, insoluble protein, and nonprotein nitrogen were calculated from crude protein, following the default proportions for ryegrass-white clover pastures currently used in the model (54.7%, 42.0%, and 3.3%, respectively: Beukes et al., 2014). Water concentration was calculated as the reciprocal of DM content.
Table 2.
Feed composition (g/kg DM, unless stated otherwise), methane emissions (methane production: pCH4 in g CH4/d), and dry matter intake (DMI) used to generate predictions of pCH4, ruminal pH, and acetate to propionate molar ratios using MollyRum14
| Forage | DM (% of FW) | OM | CP | HWSC | Pectin | RFC | aNDF | ADF | Hcell | Cell | RFC:SC | EE | DMI | pCH4 (g/d) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chicory | 11.9 | 80.4 | 11.4 | 15.3 | 7.5 | 22.8 | 23.9 | 18.8 | 5.1 | 10.6 | 1.48 | 3.7 | 666 | 15.1 |
| Chicory | 11.9 | 80.4 | 11.4 | 15.3 | 7.5 | 22.8 | 23.9 | 18.8 | 5.1 | 10.6 | 1.48 | 3.7 | 1114 | 23.8 |
| Ryegrass | 16.5 | 87.3 | 19.7 | 11.4 | 1.0 | 12.4 | 42.3 | 21.8 | 20.4 | 19.1 | 0.31 | 4.1 | 685 | 17.5 |
| Ryegrass | 16.5 | 87.3 | 19.7 | 11.4 | 1.0 | 12.4 | 42.3 | 21.8 | 20.4 | 19.1 | 0.31 | 4.1 | 1053 | 22.7 |
| Bulb turnip | 10.0 | 85.1 | 13.0 | 23.8 | 9.4 | 33.2 | 24.0 | 18.0 | 5.9 | 11.7 | 1.88 | 1.7 | 874 | 18.0 |
| Kale | 13.9 | 86.1 | 16.7 | 17.3 | 8.0 | 25.4 | 20.1 | 12.9 | 7.1 | 7.3 | 1.76 | 3.4 | 866 | 17.0 |
| Forage rape | 11.7 | 86.0 | 19.3 | 19.6 | 8.9 | 28.5 | 23.4 | 16.3 | 7.1 | 10.0 | 1.67 | 3.4 | 899 | 14.7 |
| Swedes | 9.4 | 90.8 | 16.2 | 30.1 | 6.9 | 37.0 | 17.6 | 12.1 | 5.5 | 6.9 | 3.11 | 1.1 | 792 | 13.3 |
| Forage rape | 13.1 | 85.2 | 21.5 | 14.2 | 7.6 | 21.8 | 20.9 | 16.1 | 4.8 | 12.4 | 1.31 | 3.4 | 862 | 11.8 |
| Forage rape | 14.2 | 91.7 | 15.8 | 24.0 | 7.5 | 31.5 | 17.0 | 12.3 | 4.7 | 8.6 | 2.38 | 3.3 | 896 | 16.0 |
| Forage rape | 13.1 | 90.2 | 18.3 | 20.4 | 7.7 | 28.1 | 17.5 | 13.0 | 4.5 | 7.7 | 2.3 | 3.0 | 1173 | 13.2 |
| Forage rape | 11.6 | 90.6 | 16.7 | 24.1 | 7.2 | 31.3 | 18.2 | 13.0 | 5.2 | 8.4 | 2.32 | 2.4 | 1293 | 17.6 |
| Ryegrass | 12.5 | 86.9 | 22.8 | 7.4 | 1.2 | 8.6 | 51.9 | 27.0 | 24.9 | 25.0 | 0.17 | 3.7 | 791 | 18.4 |
| Forage rape | 9.7 | 86.6 | 32.7 | 15.0 | 8.7 | 23.7 | 18.3 | 13.3 | 4.9 | 7.4 | 2.01 | 2.9 | 978 | 8.1 |
| Forage rape | 8.4 | 87.1 | 26.1 | 16.7 | 9.8 | 26.5 | 22.7 | 17.5 | 5.2 | 9.4 | 1.84 | 3.3 | 1454 | 20.8 |
| Forage rape | 10.3 | 87.1 | 26.1 | 16.7 | 9.8 | 26.5 | 22.7 | 17.5 | 5.2 | 9.4 | 1.84 | 3.3 | 1792 | 24.5 |
| Forage rape | 11.1 | 88.1 | 20.2 | 22.8 | 8.7 | 31.4 | 22.5 | 17.0 | 5.5 | 7.8 | 2.39 | 2.9 | 1670 | 23.1 |
| Forage rape | 9.8 | 88.1 | 20.2 | 22.8 | 8.7 | 31.4 | 22.5 | 17.0 | 5.5 | 7.8 | 2.39 | 2.9 | 1629 | 22.7 |
| Forage rape | 10.1 | 88.1 | 20.2 | 22.8 | 8.7 | 31.4 | 22.5 | 17.0 | 5.5 | 7.8 | 2.39 | 2.9 | 1516 | 20.2 |
| Forage rape | 11.6 | 89.1 | 17.1 | 24.1 | 8.3 | 32.4 | 24.0 | 17.0 | 7.0 | 9.6 | 1.99 | 2.7 | 1830 | 25.1 |
| Forage rape | 11.3 | 89.1 | 17.1 | 24.1 | 8.3 | 32.4 | 24.0 | 17.0 | 7.0 | 9.6 | 1.99 | 2.7 | 1824 | 26.9 |
| Bulb turnip | 9.8 | 89.3 | 11.5 | 30.7 | 6.7 | 37.3 | 20.3 | 14.5 | 5.9 | 7.5 | 2.79 | 1.1 | 1042 | 17.3 |
| Leafy turnip | 10.1 | 80.6 | 22.6 | 12.2 | 8.8 | 21.0 | 24.5 | 17.6 | 6.9 | 8.2 | 1.42 | 2.6 | 908 | 12.5 |
| Radish | 9.1 | 81.3 | 23.9 | 10.4 | 8.4 | 18.9 | 24.8 | 18.5 | 6.3 | 8.3 | 1.34 | 2.9 | 658 | 11.0 |
| Forage rape | 13.2 | 85.8 | 17.6 | 14.7 | 10.3 | 25.0 | 27.2 | 20.0 | 7.2 | 7.8 | 1.69 | 3.5 | 948 | 17.2 |
| Bulb turnip | 10.9 | 86.3 | 11.5 | 25.1 | 7.6 | 32.7 | 21.3 | 15.8 | 5.6 | 4.6 | 3.43 | 1.5 | 914 | 22.0 |
| Leafy turnip | 11.9 | 82.8 | 18.7 | 14.3 | 9.0 | 23.3 | 22.6 | 14.8 | 7.8 | 6.7 | 1.81 | 1.8 | 965 | 17.6 |
| Forage rape | 18.9 | 90.0 | 9.5 | 21.0 | 10.3 | 31.3 | 26.2 | 16.1 | 10.1 | 6.4 | 2.18 | 2.1 | 779 | 14.2 |
| Ryegrass | 14.8 | 84.2 | 18.1 | 8.3 | 0.9 | 9.2 | 46.4 | 24.2 | 22.2 | 21.5 | 0.21 | 4.3 | 792 | 15.4 |
| Ryegrass | 19.8 | 90.1 | 16.0 | 12.3 | 1.1 | 13.4 | 44.5 | 23.1 | 21.4 | 21.4 | 0.31 | 3.6 | 929 | 21.2 |
| White clover | 16.6 | 90.5 | 25.5 | 9.8 | 6.5 | 16.3 | 27.6 | 18.3 | 9.3 | 9.8 | 0.85 | 2.4 | 930 | 18.4 |
| Ryegrass | 17.2 | 86.4 | 19.2 | 11.4 | 0.7 | 12.1 | 44.4 | 23.0 | 21.4 | 19.2 | 0.3 | 3.1 | 1100 | 24.5 |
| White clover | 16.2 | 91.1 | 21.4 | 12.5 | 6.5 | 19.0 | 26.9 | 18.3 | 8.6 | 9.0 | 1.04 | 2.2 | 470 | 12.6 |
| Ryegrass | 16.3 | 90.3 | 12.5 | 16.4 | 0.7 | 17.1 | 41.5 | 22.0 | 19.5 | 20.2 | 0.43 | 2.8 | 480 | 12.2 |
| White clover | 16.2 | 91.1 | 21.4 | 12.5 | 6.5 | 19.0 | 26.9 | 18.3 | 8.6 | 9.0 | 1.04 | 2.2 | 1160 | 27.1 |
| Ryegrass | 16.3 | 90.3 | 12.5 | 16.4 | 0.7 | 17.1 | 41.5 | 22.0 | 19.5 | 20.2 | 0.43 | 2.8 | 1190 | 25.9 |
| White clover | 16.0 | 90.3 | 22.0 | 10.2 | 6.1 | 16.9 | 29.6 | 20.3 | 9.3 | 11.6 | 0.57 | 2.3 | 1140 | 25.7 |
| Ryegrass | 18.4 | 89.8 | 11.7 | 12.3 | 0.7 | 13.3 | 46.6 | 23.7 | 22.9 | 21.6 | 0.29 | 2.7 | 1120 | 24.5 |
| Ryegrass | 24.0 | 88.8 | 10.2 | 10.4 | 0.8 | 11.4 | 55.4 | 28.3 | 27.1 | 23.7 | 0.21 | 2.7 | 490 | 13.1 |
| Ryegrass | 24.0 | 88.8 | 10.2 | 10.4 | 0.8 | 11.4 | 55.4 | 28.3 | 27.1 | 23.7 | 0.21 | 2.7 | 720 | 19.5 |
| Ryegrass | 24.0 | 88.8 | 10.2 | 10.4 | 0.8 | 11.4 | 55.4 | 28.3 | 27.1 | 23.7 | 0.21 | 2.7 | 920 | 23.2 |
| Ryegrass | 24.0 | 88.8 | 10.2 | 10.4 | 0.8 | 11.4 | 55.4 | 28.3 | 27.1 | 23.7 | 0.21 | 2.7 | 1070 | 27.1 |
| Ryegrass | 24.0 | 88.8 | 10.2 | 10.4 | 0.8 | 11.4 | 55.4 | 28.3 | 27.1 | 23.7 | 0.21 | 2.7 | 1340 | 31.9 |
DM = dry matter; OM = organic matter; CP = crude protein; aNDF = neutral detergent fiber with a heat stable amylase and inclusive of residual ash, ADF = acid detergent fiber; HSWC = hot water soluble carbohydrates; RFC = readily fermentable carbohydrates (HWSC plus pectin); Hcell = hemicellulose; Cell = cellulose; RFC:SC = ratio of RFC to structural carbohydrates; EE = ether extract.
Simulations in MollyRum14
Nine combinations of parameter sets are obtained by combining the 3 degradation rate settings (SLOW, MOD, and FAST) with the 3 stoichiometry settings (0, 0.5, and 1). For each trial, simulations were run using each of the 9 combinations and predictions were compared with observed data, in order to assess the standard diet descriptor options in the model against the diverse range of pasture species and fodder crops chosen.
Comparisons between observed and predicted values of A:P and ruminal pH were made from a subset of experiments that measured VFA concentrations (n = 20, including representation of the 9 forages studied) and ruminal pH (n = 10: comprising data from bulb and leafy turnips, radish, and rape). Reductions in yCH4 have been associated with lower ruminal pH when forage rape is fed to sheep, compared with ryegrass diets (Sun et al., 2015). Thus, it would be expected that a mechanistic model should represent not only the changes in methane emissions, but also the correct trend in A:P and pH values for these forages.
Evaluation of Model Performance
The performance of the model was evaluated using several parameters derived from the comparison of observed and predicted values. The association between observed and predicted values was measured by the coefficient of determination (R2), which indicates the model precision by quantifying how much of the variance in observed values is predicted by the model. Also, the concordance correlation coefficient (CCC; Lin, 1989) was estimated to assess the agreement between the observed and predicted values (i.e., how does the regression line between observed and predicted values deviate from the line of unity [observed = predicted]). Values of CCC closer to 1 indicate a more accurate prediction.
The prediction error was assessed in terms of the root-mean-squared prediction error (RMSPE), which represents the mean difference between observed and predicted values. To allow comparisons of models, RMSPE is generally expressed as a percentage of the observed mean (relative prediction error, RPE). An RPE value of less than 10% indicates that the model predictions are good, a value between 10% and 20% suggests a reasonable prediction, and a value greater than 20% is indicative of poor prediction (Bryant and Lopez Villalobos, 2007). The RMSPE was partitioned to assess systematic biases (mean or slope) in the prediction error (Bibby and Toutenburg, 1977). Ideally, a robust model should have small biases, with most of the errors in prediction being random. All the above model evaluations were conducted using the Model Evaluation System v. 3.1.15 (Texas A&M University). Finally, the ratio of RMSPE to standard deviation of observed values (RSR) was calculated to assess the error associated with the model predictions relative to the inherent variation in observed values. With this statistic, values closer to zero are considered better than large positive values (Moriasi et al., 2007). Values of <0.5 and <0.75 for RSR are indicative of a very good and good model performance, respectively, whereas values >1 suggest that the model performance is poor.
RESULTS AND DISCUSSION
Methane Predictions—All Forages Tested
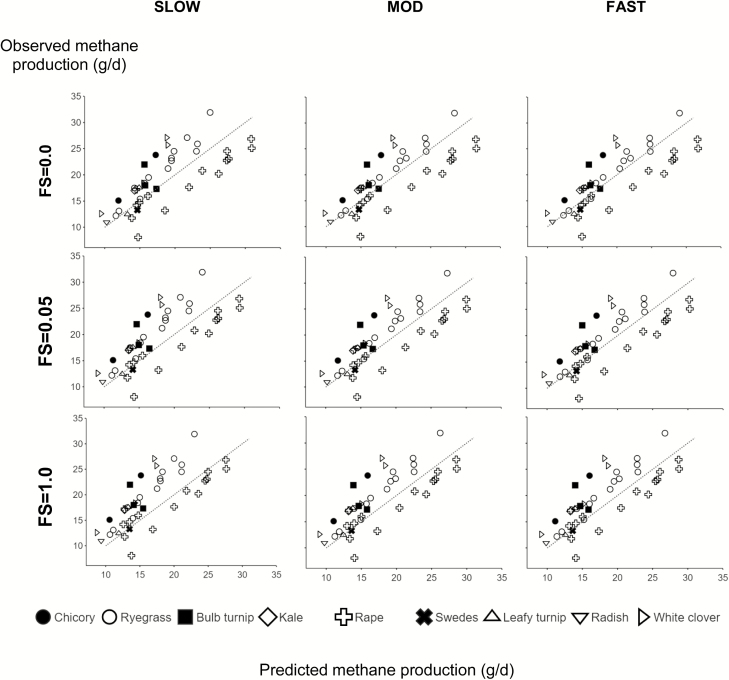
The model predictions of pCH4 across the different forages in the database can be considered adequate, with R2 and CCC values of about 0.6 and 0.75, respectively (Figure 1). The error in the prediction is in accordance with the inherent variation in observed values (RSR about 0.7). The best predictions were obtained when the degradation rate of forages was set as MOD or FAST, whereas categorizing forages as SLOW provided slightly poorer results for predictions of methane production (Table 3). This is demonstrated by the smaller RPE and RSR values, and greater R2 and CCC values, for forages set as MOD or FAST degradation rate compared with setting the degradation rate as SLOW (Table 3).
Figure 1.
Comparison of MollyRum14 model predictions for methane production (g CH4/d) with data from respiration-chamber trials, and the effect of substrate degradation rate (SLOW, MOD, or FAST) and FS. FS = 0 (favors a more acetogenic fermentation), FS = 0.5 (intermediate), and FS = 1 (favors a more propionogenic fermentation) used in the model calculations. Dotted line is x = y.
Table 3.
Measures of predictive ability of MollyRum14 for methane production (g CH4/d) from sheep fed varying forages (sole diet), and the effect of substrate degradation rate (SLOW, MOD or FAST) and fermentation stoichiometry (FS = 0 favors a more acetogenic fermentation, FS = 0.5 intermediate, FS = 1.0 favors a more propionogenic fermentation)
| Variable | Substrate degradation rate | FAST | |||||||
|---|---|---|---|---|---|---|---|---|---|
| MOD | |||||||||
| SLOW | FS | ||||||||
| 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | |
| Mean observed | 19.2 | 19.2 | 19.2 | 19.2 | 19.2 | 19.2 | 19.2 | 19.2 | 19.2 |
| Mean predicted | 18.3 | 17.5 | 16.6 | 19.0 | 18.2 | 17.4 | 19.2 | 18.4 | 17.6 |
| Mean bias | −0.9 | −1.7 | −2.6 | −0.1 | −0.9 | −1.7 | 0.1 | −0.7 | −1.5 |
| RPE1 | 20.5 | 21.4 | 23.3 | 18.8 | 18.9 | 20.1 | 18.5 | 18.5 | 19.5 |
| Error decomposition: | |||||||||
| % bias | 4.9 | 17.0 | 32.7 | 0.1 | 6.5 | 20.1 | <0.1 | 4.3 | 17.0 |
| % slope | 12.4 | 7.4 | 3.6 | 15.0 | 10.2 | 5.7 | 15.5 | 10.8 | 6.3 |
| % random | 82.7 | 75.5 | 63.7 | 84.9 | 83.4 | 74.2 | 84.4 | 84.9 | 76.7 |
| R 22 | 0.56 | 0.56 | 0.56 | 0.62 | 0.62 | 0.62 | 0.64 | 0.63 | 0.63 |
| CCC3 | 0.74 | 0.71 | 0.67 | 0.79 | 0.78 | 0.74 | 0.80 | 0.79 | 0.76 |
| RSR4 | 0.72 | 0.75 | 0.82 | 0.66 | 0.67 | 0.70 | 0.65 | 0.65 | 0.69 |
N = 43.
1Relative prediction error = (Root mean square prediction error: RMSPE)/mean observed) × 100. An RPE value of <10% indicates good predictive ability, between 10% and 20% reasonable predictive ability and >20% poor predictive ability.
2 R 2 of the regression predicted vs. observed.
3Lin’s concordance correlation coefficient (closer to 1 is better).
4Ratio of RMSPE to standard deviation of observed values: <0.5, very good prediction; 0.5–0.75, good prediction; 0.75–1, moderate prediction; and >1.0, model needs improvement.
Across all feed categories, the assumption of FS = 0 provided better predictions than the assumption of FS = 1 for the whole dataset, whereas FS = 0.5 provided results that were intermediate. Increasing the FS score in the feed resulted in an increase in mean bias and reduced both the slope bias and error due to random variation (Table 3). Considering all trials together, there was a better prediction for experiments with lower methane emissions compared with experiments with higher methane emissions, and this effect was exacerbated by increasing the assumed FS score (Figure 1).
Despite the low R2 and CCC values for yCH reported as part of the original calibration of the model (Vetharaniam et al., 2015), the predictions for yCH4 obtained across the whole database suggest that the predictive ability of MollyRum14 is moderately good (R2 and CCC values of about 0.60 and 0.65, respectively), provided that the categories for substrate degradation rate and FS are chosen judiciously (Table 4). In the original calibration, the performance of the model was assessed using individual sheep data, which adds to the variability in observed values and is likely to be the cause of the modest performance previously reported in our publication of MollyRum14. In this study, we have used experimental means, which could contribute to the better than expected performance of the model for predicting yCH4.
Table 4.
Measures of predictive ability of MollyRum14 for methane yield (g CH4/kg DMI) from sheep fed varying forages (sole diet), and the effect of substrate degradation rate (SLOW, MOD, or FAST) and fermentation stoichiometry (FS = 0 favors a more acetogenic fermentation, FS = 0.5 intermediate, FS = 1.0 favors a more propionogenic fermentation)
| Variable | SLOW | Substrate degradation rate | FAST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MOD | |||||||||
| FS | |||||||||
| 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | |
| Mean observed | 19.3 | 19.3 | 19.3 | 19.3 | 19.3 | 19.3 | 19.3 | 19.3 | 19.3 |
| Mean predicted | 18.0 | 17.2 | 16.3 | 18.7 | 17.9 | 17.2 | 18.9 | 18.1 | 17.4 |
| Mean bias | −1.3 | −2.1 | −3.0 | −0.6 | −1.4 | −2.1 | −0.4 | −1.15 | −1.9 |
| RPE1 | 19.2 | 21.2 | 23.9 | 16.8 | 18.2 | 20.3 | 16.3 | 17.5 | 19.6 |
| Error decomposition: | |||||||||
| % bias | 13.1 | 27.3 | 41.4 | 3.3 | 14.9 | 29.6 | 1.5 | 11.6 | 26.4 |
| % slope | 9.8 | 8.0 | 6.0 | 7.5 | 6.6 | 5.1 | 6.2 | 5.8 | 4.6 |
| % random | 77.1 | 64.6 | 52.5 | 89.2 | 78.5 | 65.2 | 92.4 | 82.6 | 69.0 |
| R 22 | 0.54 | 0.52 | 0.51 | 0.59 | 0.58 | 0.56 | 0.60 | 0.59 | 0.57 |
| CCC3 | 0.55 | 0.49 | 0.42 | 0.66 | 0.62 | 0.56 | 0.69 | 0.65 | 0.59 |
| RSR4 | 0.77 | 0.85 | 0.95 | 0.67 | 0.73 | 0.81 | 0.65 | 0.70 | 0.78 |
N = 43.
1RPE: Relative prediction error = (Root mean square prediction error RMSPE/mean observed) × 100. An RPE value of <10% indicates good predictive ability, between 10% and 20% reasonable predictive ability, and >20% poor predictive ability.
2 R 2 of the regression predicted vs. observed.
3Lin’s concordance correlation coefficient (closer to 1 is better).
4Ratio of RMSPE to standard deviation of observed values: <0.5, very good prediction; 0.5–0.75, good prediction; 0.75–1, moderate prediction; and >1.0, model needs improvement.
Although the R2 and CCC values in Table 4 seem modest, it is important to note that yCH4 predictions have proven to be more challenging than predicting pCH4 from sheep. This was evidenced by Hammond et al. (2009) and Zhao et al. (2016), who developed empirical models based on single and multiple regression approaches to predict pCH4 and yCH4 from sheep fed ryegrass pastures in respiration chamber studies. The models reported by Hammond et al. (2009) explained up to 80% of the variation in pCH4, but only a maximum of 20% of the variance in yCH4 using feed composition as predictors. Similarly, Zhao et al. (2016) reported R2 values between 0.09 and 0.93 for pCH4 (i.e., 9% to 93% of variance explained) depending on complexity of the model and predictor variables, whereas for yCH4, the same authors reported R2 values between 0.28 and 0.62. It is worth noting that the use of R2 as a measure of model performance by both Hammond et al. (2009) and Zhao et al. (2016) can only describe model precision, whereas in this study, both accuracy and precision are assessed for MollyRum14. The R2 values reported in these 2 reports can be expected to be greater than those reported herein (Table 3) because empirical model development is inherently about explaining as much of the variation in the dataset. Furthermore, in both reports, the evaluation of the models was against the data used for model parameterization, which would be expected to yield a large R2. In contrast, in our study, the model accuracy (CCC) and precision (R2) could be interpreted as an indication of how well the mechanisms controlling methane production in the sheep rumen are represented by the model. Therefore, the R2 and CCC values of about 0.6 presented in Table 4 are encouraging and suggest that MollyRum14 has potential as a tool to generate better predictions of yCH4 from forage diets fed to sheep than empirical models using forage composition as predictors (Hammond et al., 2009). Also, MollyRum14 yCH4 predictions are comparable or better than those from empirical models that include chemical composition, feeding levels, and diet digestibility as predictors (Zhao et al., 2016).
Predictive models, both empirical and mechanistic, for pCH4 and yCH4 have been extensively reported for other ruminants, such as beef and dairy cattle (e.g., Kebreab et al. (2008), Ellis et al. (2010), Bannink et al. (2011), Moraes et al. (2013), and Charmley et al. (2016)). The performance of MollyRum14 is similar to, or better than, the empirical models evaluated by Kebreab et al. (2006) for dairy cattle diets (e.g., RPE ranging from 23.8% to 33.3% and CCC from 0.44 to 0.71). Moraes et al. (2013) reported the use of Bayesian approaches to develop improved models to predict methane emissions from different cattle classes (lactating, nonlactating, heifers, and steers). The Bayesian-based models reported by Moraes et al. (2013) predicted pCH4 with an RPE between 14% and 19%, depending on the cattle classes, compared with 19% and 30% for regression-based models. Depending on the choice of FS and degradation rate, the prediction performance of MollyRum14 is within the performance range of the improved models of Moraes et al. (2013). Although similar in predictive performance to MollyRum14, the Bayesian-based models reported by Moraes et al. (2013) are better suited for improvement of national inventories, because of their empirical nature. In contrast, MollyRum14 has utility as a tool for exploration of methane mitigation mechanisms. Kebreab et al. (2008) evaluated the Molly95 (the “parent” model of MollyRum14) and reported RPE and CCC for pCH4 predictions for dairy cattle that were 32% and 0.50, respectively. For beef cattle in feedlots, the same authors reported RPE and CCC values of 22% and 0.69, respectively. Therefore, we can conclude that MollyRum14 extends the repertoire of predictive mechanistic models for species other than cattle.
Individual Forages Tested
The effects of substrate degradation rate and FS were assessed for prediction from forage rape and perennial ryegrass rations, the forages with the largest number of records in the database (Table 5). This assessment was performed by comparing the RPE values generated for each combination of substrate degradation rate and FS assumption for these 2 forages. This analysis suggested that the best predictions of pCH4 and yCH4 for forage rape are obtained when the SLOW degradation rate was selected. It also indicated that using a greater FS score (i.e., towards a propionogenic profile) in the simulations improves the predictions for forage rape, which is consistent with observations that fermentation profiles and microbial communities in the rumen of forage rape-fed sheep resemble those from animals fed grain-based concentrates (Sun et al., 2015). Interestingly, the best predictions for both pCH4 and yCH4 for rations of perennial ryegrass were obtained when the FAST degradation rate was selected. Using a greater FS worsened the predictions for perennial ryegrass.
Table 5.
Selected measures of predictive ability of MollyRum14 for methane production (g CH4/d) and methane yield (g CH4/kg DMI) from sheep fed forage rape (n = 15 experimental means) or perennial ryegrass (n = 14 experimental means) as sole diets
| Variable | SLOW | Substrate degradation rate | FAST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MOD | |||||||||
| FS | |||||||||
| 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | |
| Methane production (g CH4/d) | |||||||||
| Forage rape | |||||||||
| Mean observed | 18.4 | 18.4 | 18.4 | 18.4 | 18.4 | 18.4 | 18.4 | 18.4 | 18.4 |
| Mean predicted | 21.6 | 20.5 | 19.5 | 22.0 | 21.0 | 20.1 | 22.1 | 21.1 | 20.2 |
| RPE1 | 22.2 | 17.8 | 14.3 | 24.1 | 19.8 | 16.0 | 24.6 | 20.2 | 16.3 |
| CCC2 | 0.79 | 0.85 | 0.89 | 0.77 | 0.82 | 0.87 | 0.77 | 0.82 | 0.87 |
| Perennial ryegrass | |||||||||
| Mean observed | 21.2 | 21.2 | 21.2 | 21.2 | 21.2 | 21.2 | 21.2 | 21.2 | 21.2 |
| Mean predicted | 18.3 | 17.5 | 16.8 | 19.6 | 18.9 | 18.2 | 20.0 | 19.3 | 18.5 |
| RPE1 | 16.3 | 19.5 | 22.8 | 9.5 | 12.6 | 15.8 | 8.0 | 11.0 | 14.3 |
| CCC2 | 0.79 | 0.71 | 0.64 | 0.92 | 0.87 | 0.81 | 0.95 | 0.90 | 0.84 |
| Methane yield (g CH4/kg DMI) | |||||||||
| Forage rape | |||||||||
| Mean observed | 14.3 | 14.3 | 14.3 | 14.3 | 14.3 | 14.3 | 14.3 | 14.3 | 14.3 |
| Mean predicted | 16.6 | 15.8 | 15.0 | 16.9 | 16.2 | 15.4 | 17.0 | 16.3 | 15.5 |
| RPE1 | 21.8 | 18.7 | 17.0 | 23.3 | 20.1 | 17.8 | 23.6 | 20.3 | 17.9 |
| CCC2 | 0.22 | 0.23 | 0.20 | 0.21 | 0.22 | 0.21 | 0.21 | 0.22 | 0.22 |
| Perennial ryegrass | |||||||||
| Mean observed | 23.8 | 23.8 | 23.8 | 23.8 | 23.8 | 23.8 | 23.8 | 23.8 | 23.8 |
| Mean predicted | 20.6 | 19.8 | 19.0 | 22.0 | 21.3 | 20.5 | 22.5 | 21.6 | 20.8 |
| RPE1 | 14.7 | 17.7 | 20.9 | 8.8 | 11.6 | 14.7 | 7.4 | 10.1 | 13.2 |
| CCC2 | 0.35 | 0.26 | 0.20 | 0.64 | 0.49 | 0.37 | 0.72 | 0.56 | 0.42 |
Varying substrate degradation rate (SLOW, MOD, or FAST) and fermentation stoichiometry (FS = 0 favors a more acetogenic fermentation, FS = 0.5 intermediate, FS = 1.0 favors a more propionogenic fermentation) were assumed.
1RPE: relative prediction error = [Root mean square prediction error (RMSPE)/mean observed] × 100. An RPE value of <10% indicates good predictive ability, between 10% and 20% reasonable predictive ability, and >20% poor predictive ability.
2CCC: Lin’s concordance correlation coefficient.
According to the RPE for individual feeds, MollyRum14 provides moderate to good pCH4 predictions for ryegrass, forage rape, and both leafy and bulb turnips (Table 6). Although the predictions for swedes, radish, and kale had moderate to small RPE (<13.8%), the small number of observations in our dataset precludes a definite assessment of the performance of the model for these forages. Predictions for chicory and white clover were not satisfactory (RPE > 23%), but the number of observations for chicory is small to make a definitive assessment of the model. As stated above, using a high FS score improved predictions for forage rape, but worsened the predictions for ryegrass. These findings suggest that the adjustment of the FS in the feed seems sensible, at least in terms of the overall change of direction for the prediction.
Table 6.
Allocation of forages in database to the combination of MollyRum14 digestion parameters that generates the best prediction for methane production (g CH4/d) and methane yield (g CH4/kg DMI)
| FS | |||
|---|---|---|---|
| 0.0 | 0.5 | 1.0 | |
| Methane production (g CH4/d) | |||
| SLOW | – | – | Forage rape (14.3) |
| MOD | – | – | – |
| FAST | Bulb turnip (19.0), perennial ryegrass (8.0), white clover (23.6) | – | – |
| Methane yield (g CH4/kg DMI) | |||
| SLOW | – | – | Forage rape (17.0) |
| MOD | – | – | – |
| FAST | Bulb turnip (19.5), perennial ryegrass (7.4), white clover (22.0) | ||
The digestion parameters considered were substrate degradation rate (SLOW, MOD, or FAST) and fermentation stoichiometry (FS = 0 favors a more acetogenic fermentation, FS = 0.5 intermediate, FS = 1.0 favors a more propionogenic fermentation). Assessment of predictions were based on RPE1 values (%) shown in brackets. Only forages with n > 3 are included.
1RPE: relative prediction error = [Root mean square prediction error (RMSPE)/mean observed] × 100. An RPE value of <10% indicates good predictive ability, between 10% and 20% reasonable predictive ability, and >20% poor predictive ability.
For yCH4, selecting a FAST degradation rate and the FS = 0 resulted in the best predictions for most forages. The exceptions to these assumptions were forage rape and swedes, where the lowest RPE values were obtained using FS = 1.0 and SLOW degradation rate (Table 6). Satisfactory predictions of yCH4 (RPE < 10%) were obtained for perennial ryegrass (7.4%), whereas predictions of yCH4 were moderately adequate (10% < RPE < 20%) for leafy turnip and rape (11.8% and 17.0%, respectively).
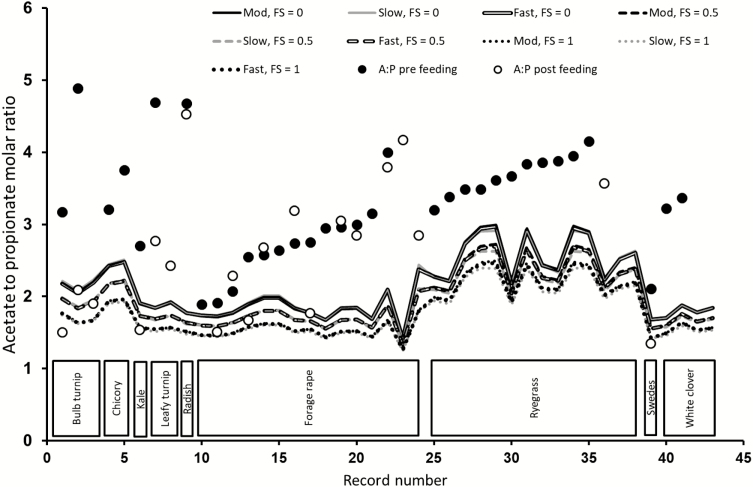
Acetate to Propionate Ratios
Figure 2 provides a comparison of model predictions of A:P ratios and actual values from the simulated trials. The observed and predicted values of A:P ratios are plotted against record number in the horizontal axis, which has been ordered by forage type in the database (Figure 2). It is apparent that the model systematically under-predicted this ratio. Setting the degradation rates of cellulose and hemicellulose of a particular forage to SLOW, MOD, or FAST in the simulations had little effect on the prediction of A:P ratios. This is seen in Figure 2 as the lines for each of the simulations with a different feed category superimpose on each other. Expectedly, using a greater FS score in the simulation resulted in noticeable decreases in the predicted A:P values and exacerbated the underprediction of A:P across the different forages tested in this study. The performance of MollyRum14 in predicting A:P can be considered weak, based on the large mean bias, and the very low R2, consistent with extremely low CCC values when the predictions were compared against the observed postfeeding A:P values (Table 7).
Figure 2.
Comparison of observed A:P ratios (prefeeding and postfeeding, solid and open circles, respectively) against MollyRum14 model predictions (lines) for sheep fed varying forages as sole diets, and the effect of substrate degradation rate (SLOW, MOD, or FAST) and FS (FS = 0 favors a more acetogenic fermentation), FS = 0.5 intermediate, and FS = 1 (favors a more propionogenic fermentation) used in the model calculations.
Table 7.
Measures of predictive ability of MollyRum14 for ruminal acetate to propionate (A:P) ratios from sheep fed varying forages (sole diet), and the effect of substrate degradation rate (SLOW, MOD, or FAST) and fermentation stoichiometry (FS = 0 favors a more acetogenic fermentation, FS = 0.5 intermediate, FS = 1.0 favors a more propionogenic fermentation)
| Variable | SLOW | Substrate degradation rate | FAST | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MOD | |||||||||
| FS | |||||||||
| 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | 0.0 | 0.5 | 1.0 | |
| Mean observed | 2.6 | 2.6 | 2.6 | 2.6 | 2.6 | 2.6 | 2.6 | 2.6 | 2.6 |
| Mean predicted | 1.9 | 1.7 | 1.6 | 1.9 | 1.8 | 1.6 | 1.9 | 1.7 | 1.6 |
| Mean bias | −0.6 | −0.8 | −1.0 | −0.7 | −0.8 | −1.0 | −0.7 | −0.8 | −1.0 |
| Relative prediction error1 | 45.2 | 49.0 | 53.4 | 45.1 | 48.7 | 52.7 | 45.3 | 48.7 | 52.8 |
| Error decomposition: | |||||||||
| % bias | 30.4 | 43.2 | 54.0 | 31.3 | 43.0 | 53.1 | 32.1 | 43.5 | 53.3 |
| % slope | 0.8 | 0.5 | 0.7 | 1.2 | 0.9 | 1.1 | 1.2 | 1.0 | 1.2 |
| % random | 60.8 | 52.1 | 44.3 | 61.3 | 53.1 | 45.5 | 61.1 | 53.0 | 45.4 |
| R22 | 0.02 | 0.01 | 0.01 | 0.01 | 0.00 | 0.00 | 0.01 | 0.01 | 0.00 |
| CCC3 | −0.04 | −0.02 | 0.00 | −0.03 | −0.01 | 0.00 | −0.03 | −0.01 | −0.01 |
| RSR4 | 1.2 | 1.3 | 1.5 | 1.2 | 1.3 | 1.4 | 1.2 | 1.3 | 1.4 |
N = 20.
1RPE = (Root mean square prediction error RMSPE/mean observed) × 100 (a smaller value is better).
2 R 2 of the regression predicted vs. observed.
3Lin’s concordance correlation coefficient (closer to 1 is better).
4Ratio of RMSPE to standard deviation of observed values: <0.5, very good prediction; 0.5–0.75, good prediction; 0.75–1, moderate prediction; and >1.0, model needs improvement.
The complexity behind obtaining suitable predictions of A:P is further increased by the fact that VFA concentrations in the rumen (and the ratio between them) are influenced by production and absorption rates, volume, and passage rate of rumen fluid and oscillate diurnally in response to feeding patterns of the sheep (Sun et al., 2012a). The database included only a subset of records in which VFA concentrations were measured. In some of these cases, the VFA were collected prior to feeding, whereas in other cases, the samples were collected 2 h after feeding. In a limited number of cases, both prefeeding and postfeeding samples were available. MollyRum14 assumes steady state conditions, and therefore, in its current form it cannot be used to predict the diurnal oscillations in VFA patterns.
Despite the large under-predictions across the dataset, in many of the cases, the predictions from MollyRum14 fell within the range of the observed prefeeding and postfeeding ratios (e.g., bulb turnip and some of the forage rape data). Further analysis with more data characterizing the diurnal pattern of the VFA proportions would be recommended for a more definite assessment of the ability of MollyRum14 to predict the mean of the VFA patterns. It is encouraging to note that although the values were under-predicted, MollyRum14 predicted that A:P from forage rape would be lower than ryegrass, which is in general agreement with the in vivo measured data.
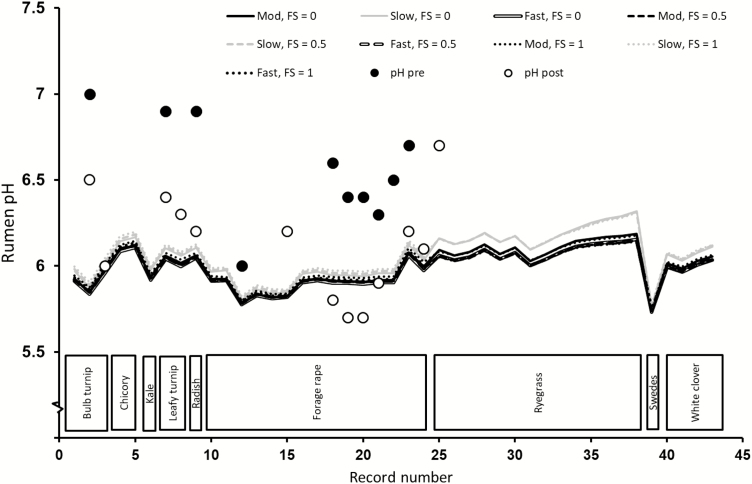
Ruminal pH
The predicted values for different degradation rates and FS in the different forages tested are plotted against the record number in the database ordered by forage type (Figure 3). Similar to VFA profiles, the evaluation of the performance of MollyRum14 predicting ruminal pH is limited by the number of observed values in our dataset. The diurnal variation in ruminal pH (Sun et al., 2012a) further complicates a definite assessment of the pH predictions from the model. The different FS assumed had a minor effect on pH. There were minimal differences in prediction between the MOD and FAST substrate degradation rate categories, since the only difference between these categories was a 22% difference in the degradation rate for hemicellulose. Using the SLOW category resulted in greater pH predicted values, which was more noticeable for ryegrass than for clover. MollyRum14 predicted lower pH values for forage rape than for ryegrass. This pattern is consistent with those observed in trials and has been postulated as a mechanism that partly explained the reductions of methane production and yield from sheep fed the brassica crop (Sun et al., 2015).
Figure 3.
Comparison of observed rumen pH (prefeeding and postfeeding, solid and open circles, respectively) against MollyRum14 model predictions (lines) for sheep fed varying forages as sole diets, and the effect of substrate degradation rate (SLOW, MOD, or FAST) and FS (FS = 0 favors a more acetogenic fermentation), FS = 0.5 intermediate, and FS = 1 (favors a more propiogenic fermentation) used in the model calculations.
Overall Conclusions
MollyRum14 predictions for pCH4 and yCH4 ranged from good to moderate for a variety of fresh forages beyond those used for the calibration of the model. Given the inherent variability and error in the in vivo measurements, the predictions from MollyRum14 adequately describe the fermentation characteristics of forages for sheep. Furthermore, appropriately characterizing forages may increase the effectiveness of MollyRum14 to explore further feeds and feeding scenarios which mitigate methane emissions of sheep. In the case of ryegrass and forage rape, the forages with the largest number of observations, MollyRum14 predictions of A:P and pH seem to follow the behavior of the observed data. This is an important feature as it indicates the value of the model for prediction of other aspects of rumen function that modulate methane emissions.
Across all forages tested, the best predictions of methane production were obtained when the degradation rate of forages was set as MOD or FAST, whereas categorizing forages as SLOW provided slightly poorer results. However, for a given chemical composition of feed, the different substrate degradation rate or FS categories had minor effects on predicted methane emission, despite these governing digestion rates and FS. The apparent lack of sensitivity to these variables may be due to pH having a larger, overriding impact on stoichiometries in MollyRum14 (and Molly95).
Since feed categorisations in both MollyRum14 and Molly95 serve only to modulate (in a prescribed and coarse manner) the FS and degradation rates of cellulose and hemicellulose, it may be worthwhile dispensing with these categories, and supplying stoichiometry and degradation constants as inputs along with feed characteristics. This would allow a more refined control of fermentation characteristics with the possibility of improving model performance.
Footnotes
This study was funded as part of the combined programme of the Pastoral Greenhouse Gas Research Consortium (New Zealand) and the New Zealand Agricultural Greenhouse Gas Research Centre.
LITERATURE CITED
- Alemu A. W., Dijkstra J., Bannink A., France J., and Kebreab E.. 2011. Rumen stoichiometric models and their contribution and challenges in predicting enteric methane production. Anim. Feed Sci. Technol. 166–167:761–778. doi: 10.1016/j.anifeedsci.2011.04.054 [DOI] [Google Scholar]
- Baldwin R. L. 1995. Modeling ruminant digestion and metabolism. Chapman & Hall, London. [DOI] [PubMed] [Google Scholar]
- Bannink A., van Schijndel M. W., and Dijkstra J.. 2011. A model of enteric fermentation in dairy cows to estimate methane emission for the dutch national inventory report using the ipcc tier 3 approach. Anim. Feed Sci. Technol. 166–167:603–618. doi: 10.1016/j.anifeedsci.2011.04.043 [DOI] [Google Scholar]
- Beukes P. C., Gregorini P., Romera A. J., Woodward S. L., Khaembah E. N., Chapman D. F., Nobilly F., Bryant R. H., Edwards G. R., and Clark D. A.. 2014. The potential of diverse pastures to reduce nitrogen leaching on New Zealand dairy farms. Anim. Prod. Sci. 54:1971–1979. doi: 10.1071/AN14563 [DOI] [Google Scholar]
- Bibby J., and Toutenburg H.. 1977. Prediction and improved estimation in linear models. John Wiley & Sons, London, England. [Google Scholar]
- Bryant J. R., and Lopez Villalobos N.. 2007. Choice of the most appropriate models and estimation procedures of lactation curves for grazing dairy cattle. Proc. N. Z. Soc. Anim. Prod. 67:209–214. [Google Scholar]
- Charmley E., Williams S. R. O., Moate P. J., Hegarty R. S., Herd R. M., Oddy V. H., Reyenga P., Staunton K. M., Anderson A., and Hannah M. C.. 2016. A universal equation to predict methane production of forage-fed cattle in Australia. Anim. Prod. Sci. 56:169–180. doi: doi.org/10.1071/AN15365 [Google Scholar]
- Ellis J. L., Bannink A., France J., Kebreab E., and Dijkstra J.. 2010. Evaluation of enteric methane prediction equations for dairy cows used in whole farm models. Glob. Change Biol. 16:3246–3256. doi: 10.1111/j.1365-2486.2010.02188.x [DOI] [Google Scholar]
- Hammond K. J., Muetzel S., Waghorn G. G., Pinares-Patino C. S., Burke J. L., and Hoskin S. O.. 2009. The variation in methane emissions from sheep and cattle is not explained by the chemical composition of ryegrass. Proc. N. Z. Soc. Anim. Prod. 69:174–178. [Google Scholar]
- Janssen P. H. 2010. Influence of hydrogen on rumen methane formation and fermentation balances through microbial growth kinetics and fermentation thermodynamics. Anim. Feed Sci. Technol. 160:1–22. doi: 10.1016/j.anifeedsci.2010.07.002 [DOI] [Google Scholar]
- Kebreab E., France J., McBride B. W., Odongo N., Bannink A., Mills J. A. N., and Dijkstra J.. 2006. Evaluation of models to predict methane emissions from enteric fermentation in North American dairy cattle. In: Kebreab E., Dijkstra J., Gerrits W. J. J., Bannink A. and J. France, editors, Nutrient utilization in farm animals: modelling approaches. CAB Int, Wallingford, UK: p. 299–313. [Google Scholar]
- Kebreab E., K. A. Johnson S. L. Archibeque D. Pape, and Wirth T.. 2008. Model for estimating enteric methane emissions from United States dairy and feedlot cattle. J. Anim. Sci. 86:2738–2748. doi: 10.2527/jas.2008-0960 [DOI] [PubMed] [Google Scholar]
- Lin L. I. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255–268. [PubMed] [Google Scholar]
- Moraes L. E., Strathe A. B., Fadel J. G., Casper D. P., and Kebreab E.. 2013. Prediction of enteric methane emissions from cattle. Glob. Change Biol. 20:2140–2148. doi: 10.1111/gcb.12471 [DOI] [PubMed] [Google Scholar]
- Moriasi D. N., Arnold J. G., Van Liew M. W., Bingner R. L., Harmel R. D., and Veith T. L.. 2007. Model evaluation guidelines for systematic quantification of accuracy in watershed simulations. T. ASABE. 50:885–900. doi: 10.13031/2013.23153 [DOI] [Google Scholar]
- Morvay Y., A. Bannink J. France E. Kebreab, and Dijkstra J.. 2011. Evaluation of models to predict the stoichiometry of volatile fatty acid profiles in rumen fluid of lactating Holstein cows. J. Dairy Sci. 94:3063–3080. doi: 10.3168/jds.2010-3995 [DOI] [PubMed] [Google Scholar]
- Pacheco D., Waghorn G., and Janssen P. H.. 2014. Decreasing methane emissions from ruminants grazing forages: a fit with productive and financial realities?Anim. Prod. Sci. 54:1141–1154. doi: 10.1071/AN14437 [DOI] [Google Scholar]
- Steinfeld H., Gerber P., Wassenaar T., Castel V., Rosales M., and De Haan C.. 2006. Livestock’s long shadow. Environmental issues and options. Food Agric. Organ. United Nations (FAO), Rome. [Google Scholar]
- Sun X., Henderson G., Cox F., Molano G., Harrison S. J., Luo D., Janssen P. H., and Pacheco D.. 2015. Lambs fed fresh winter forage rape (Brassica napus l.) emit less methane than those fed perennial ryegrass (Lolium perenne l.), and possible mechanisms behind the difference. PLoS ONE 10:e0119697. doi: 10.1371/journal.pone.0119697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X. Z., Hoskin S. O., Muetzel S., Molano G., and Clark H.. 2011. Effects of forage chicory (Cichorium intybus) and perennial ryegrass (Lolium perenne) on methane emissions in vitro and from sheep. Anim. Feed Sci. Technol. 166–167:391–397. doi: 10.1016/j.anifeedsci.2011.04.027 [DOI] [Google Scholar]
- Sun X. Z., Pacheco D., Hoskin S. O., and Luo D. W.. 2012a. Rumen fermentation characteristics are influenced by feeding frequency in sheep fed forage chicory and perennial ryegrass at two feeding levels. Proc. N. Z. Soc. Anim. Prod. 72:111–116. [Google Scholar]
- Sun X., Pacheco D., and Luo D.. 2016. Forage brassica: a feed to mitigate enteric methane emissions?Anim. Prod. Sci. 56:451–456. doi: 10.1071/AN15516 [DOI] [Google Scholar]
- Sun X. Z., Pacheco D., Molano G., and Luo D. W.. 2013. Sheep fed fresh forage rape (Brassica napus subsp. Oleifera l.) have lower methane emissions compared with perennial ryegrass (Lolium perenne l.) Advances in Animal Biosciences: Proc. 5th Greenhouse Gases Anim. Agric. Conf. (GGAA 2013) No. 4 Part 2 p 271 Cambridge Univ. Press, Dublin, Ireland. [Google Scholar]
- Sun X. Z., Waghorn G. C., Hoskin S. O., Harrison S. J., Muetzel S. M., and Pacheco D.. 2012b. Methane emissions from sheep fed fresh brassicas (Brassica spp.) compared to perennial ryegrass (Lolium perenne). Anim. Feed Sci. Technol. 176:107–116. doi: 10.1016/j.anifeedsci.2012.07.013 [DOI] [Google Scholar]
- van Lingen H. J., Plugge C. M., Fadel J. G., Kebreab E., Bannink A., and Dijkstra J.. 2016. Thermodynamic driving force of hydrogen on rumen microbial metabolism: a theoretical investigation. PLoS ONE 11:e0161362. doi: 10.1371/journal.pone.0161362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetharaniam I., R. E. Vibart M. D. Hanigan P. H. Janssen M. H. Tavendale, and Pacheco D.. 2015. A modified version of the molly rumen model to quantify methane emissions from sheep. J. Anim. Sci. 93:3551–3563. doi: 10.2527/jas.2015-9037 [DOI] [PubMed] [Google Scholar]
- Zhao Y. G., O’Connell N. E., and Yan T.. 2016. Prediction of enteric methane emissions from sheep offered fresh perennial ryegrass (Lolium perenne) using data measured in indirect open-circuit respiration chambers1. J. Anim. Sci. 94:2425–2435. doi: 10.2527/jas.2016-0334 [DOI] [PubMed] [Google Scholar]