Abstract
Histone deacetylase (HDAC) inhibition attenuates inflammation in rodents and short-chain fatty acids (SCFAs) are effective HDAC inhibitors. Therefore, the objective of this study was to evaluate the role of the SCFAs sodium propionate (SP) and sodium butyrate (SB) as HDAC-dependent regulators of inflammatory gene expression in bovine mammary epithelial cells (MAC-Ts). We postulated that SP and SB would decrease inflammation in MAC-Ts by inhibiting HDAC activity and increasing histone H3 acetylation and consequently decreasing inflammatory gene expression. For this study, MAC-Ts stimulated with lipopolysaccharide (LPS) were used as a model for bovine mammary epithelial cell inflammation. MAC-Ts were cultured in a basal medium. Cell lysates were incubated with SP or SB (0 to 5 mM) for 2 h prior to HDAC substrates incubation for an additional 2 h and HDACs activity was determined. Next, cells were pretreated with SP or SB (0 to 3.0 mM) for 2 h prior to LPS (1 µg/mL) stimulation for an additional 2 h and assessed for histone H3 acetylation. Then, cells were pretreated with SP or SB (1 mM) for 24 h prior to LPS (1 µg/mL) stimulation for an additional 2 h and RNA was isolated for inflammatory gene expression evaluation by PCR array and gene validation was performed using quantitative real-time PCR. One-way ANOVA followed by Tukey post hoc analysis was conducted and statistical significance set at P < 0.05. SP and SB concentration-dependently and selectively inhibited class I HDAC activity, which differed between SCFAs, where SB inhibited (P < 0.05) HDACs 2, 3, and 8, while SP inhibited (P < 0.05) HDACs 2 and 8. Histone H3 acetylation was concentration-dependently increased by SCFAs and likewise the differential regulation of HDAC activity, SCFAs effected differently histone H3 acetylation, where SB increased (P < 0.05) H3K9/14, H3K18 and H3K27 acetylation, while SP increased (P < 0.05) H3K9/14 and H3K18 acetylation. However, SCFAs did not decrease (P > 0.05) overall inflammatory gene expression. Under our experimental conditions, findings suggest that in MAC-Ts, SCFAs regulate epigenetic markers on nucleosomal DNA in addition to regulation of inflammatory gene events independent of HDAC activity. Nevertheless, examination of SCFAs and/or HDACs inhibitors in bovine mammary gland is worth being further investigated to delineate the potential impact of HDAC inhibition and histones hyperacetylation on mammary gland tissue inflammation.
Keywords: histone deacetylase inhibitor, inflammation, short-chain fatty acid
INTRODUCTION
In the 1990s, Huynh et al. (1991) developed an immortalized cell line established from the bovine mammary gland called bovine mammary epithelial cells (MAC-Ts), which have been used as an in vitro model to study mammary gland biology under normal and diseased condition such as inflammation (Silva et al., 2017). Bacterial endotoxin or lipopolysaccharide (LPS) is an outer cell membrane constituent present in Gram-negative bacteria (Baumann and Gauldie, 1994) that exerts a pro-inflammatory effect within the bovine mammary gland (Mattila and Frost, 1989).
Acetylation of lysine residues on the N-terminal tail of histone proteins H2A, H2B, H3, and H4 play a key role in chromatin remodeling, DNA accessibility, and gene expression (Ruijter et al., 2003). Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are enzymes that add or remove acetyl groups from these histone tails, respectively. Under physiological conditions, HATs and HDACs maintain homeostatic balance, which sustains cellular function (Ruijter et al., 2003). However, under metabolic dysfunction or disease, this balance is lost favoring increased HDAC activity that exerts pro-immune and consequently pro-inflammatory actions (Halili et al., 2009).
Zinc-dependent is an HDAC superfamily that is divided into three main classes; I, II (subdivided into IIa and IIb), and IV (Grozinger and Schreiber, 2002). Inhibition of enzymes has been shown efficacious in various inflammatory models including pulmonary hypertension and rheumatoid arthritis (Zhao et al., 2012; Angiolilli et al., 2017) and has been recognized as a possible anti-inflammatory therapeutic (Adcock, 2007; Bush and McKinsey, 2009). Short-chain fatty acids (SCFAs) are end products of ruminal bacteria fermentation and are the major energy source for ruminants (Bergman, 1990). In humans, however, SCFAs have been related to immune regulation (Meijer et al., 2010). Studies have shown that butyrate and propionate can act as HDAC inhibitors (HDACi) exerting effects on the inflammatory process by downregulating the expression of pro-inflammatory genes and consequently suppressing inflammation (Segain et al., 2000; Patnala et al., 2017; Wang et al., 2017).
The role for SCFAs as an HDAC-dependent regulator of inflammatory gene expression on bovine mammary epithelial cells has not yet been examined. To the best of our knowledge, this is the first report to evaluate the effects of SP and SB as HDACi in MAC-Ts. We hypothesized that SP and SB would inhibit inflammation via HDAC-dependent regulation of gene expression. Therefore, we aimed to evaluate the effects of these SCFAs on HDAC activity, histone 3 (H3) acetylation, and inflammatory gene expression.
MATERIAL AND METHODS
Cells and Cell Culture
For this study an immortalized cell line developed by Huynh et al. (1991) from the bovine mammary gland named bovine MAC-Ts was used, tissue dissociation, bovine mammary epithelial cells isolation, and preparation of Type 1 collagen were performed according to Collier et al. (2006). The cells were kindly donated by Dr. Laura Hernandez from University of Wisconsin-Madison. Cells used for all experiments were at <12 passages and were cultured in medium containing Dulbecco’s modified Eagle’s medium (DMEM; Caisson Laboratories, Inc., Smithfield, UT), fetal bovine serum (FBS 10%; Atlanta Biologicals, Inc., Flowery Branch, GA), insulin (10 µg/mL; Sigma-Aldrich Corporation, St. Louis, MO), and antibiotics (penicillin, 100 IU/mL; Sigma-Aldrich Corporation and streptomycin, 100 µg/mL; Caisson Laboratories, Inc.) at 37 °C, with 5% CO2, in a humidified environment. Upon confluence (80%), MAC-Ts were treated as indicated and lysed in pH 7.4 PBS solution containing Triton X-100 (0.5%), NaCl (300 mM) and protease and phosphatase inhibitor cocktail (HALT, Thermo Fisher Scientific Inc., Waltham, MA. #1861280). Cell protein lysates were sonicated and clarification by centrifugation (at 16,000 × g for 5 min) was performed. Concentration of protein was determined by BCA Assay Kit (Pierce, Thermo Fisher Scientific, Inc., #23225). Concentrations of SCFAs were chosen based on a previous study that observed effects of SB on histone acetylation in bovine primary mammary epithelial cells (Ochoa-Zarzosa et al., 2009) and also on a study that reported that butyrate acts as HDACi in bone marrow-derived macrophages (Chang et al., 2014). The assays described below were performed at least twice.
HDAC Activity Assays
Assays for HDAC activity evaluation were performed according to Lemon et al. (2011). To access class I, IIa, and IIb HDAC activity. MAC-T protein lysates (15 µg protein per well) were diluted into PBS (100 μL per well) in a white-walled, 96-well plate. Vehicle control (Veh; distilled H2O) or increasing concentrations (0, 0.0001, 0.0003, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, and 5 mM) of sodium propionate (SP; Sigma-Aldrich Corporation. #P1880) or sodium butyrate (SB; Sigma-Aldrich Corporation. #303410) were added to cell protein lysates and incubated at 37 °C for 2 h. Apicidin (300 μM, class I HDAC inhibitor; Enzo Life Sciences, Inc., Farmingdale, NY. #BML-GR340) was added to the class IIb HDAC substrate wells and plater were incubated for 30 min at 37 °C prior to addition of class-specific HDACs substrates. Class-specific HDAC substrates (Class I, IIa, and IIb) were then added (50 µM per well) and plates were incubated for 2 h at 37 °C. Next, developer/stop solution (50 μL per well) containing Triton X-100 (1.5%), trichostatin A (TSA, 3 μM; Sigma-Aldrich Corporation. #T-8552), and trypsin (0.75 mg/mL; Sigma-Aldrich Corporation. #T-0303) in PBS was added and plates incubated for 20 min at 37 °C. Trypsin is added after deacetylation because it is active only against deacetylated substrate, it cleaves the fluorophore 7-amino-4-methycoumarin (AMC) resulting in increased fluorescence emission. AMC fluorescence was measured via Synergy 2 plate reader (BioTek Instruments, Inc., Winooski, VT), with excitation–emission at 360/460 nm.
To determine which class I HDAC(s) are regulated by SB and SP, recombinant proteins for the class I HDACs (Reaction Biology Corp.), HDAC1 (#KDA-21-365), HDAC2 (#KDA-21-277), HDAC3 (#KDA-22-278), and HDAC8 (#KDA-21-285) were incubated in a white-walled, 96-well plate against vehicle control (Veh; distilled H2O), SP (1 mM) or SB (1 mM) for 24 h at 37 °C. Recombinant proteins were then incubated against the class I specific HDAC substrate (ZLPA, 50 µM) for 2 h at 37 °C. Next, developer/stop solution and trypsin were added and HDAC activity was then read as described previously. Biological quadruplicate were used for each treatment.
Immunoblotting
Upon confluence (80%), MAC-Ts cultured in medium previously described were treated with increasing concentrations (0, 0.3, 1.0, and 3.0 mM) of SP or SB for 2 h at 37 °C prior to stimulation with LPS (1 µg/mL Escherichia coli O111:B4, Sigma-Aldrich Corporation. #L4130,). Biological triplicates were used for each treatment.
SDS-PAGE electrophoresis was used to resolve MAC-T protein lysates, which were then transferred to nitrocellulose membranes (Bio-Rad Laboratories, Inc., Hercules, CA) and probed with specific primary antibodies for total histone 3 (H3; Cell Signaling Technology, Inc., Danvers, MA. #4499), acetyl-histone H3 lysine9/lysine14 (acetyl-H3K9/14; Cell Signaling Technology, Inc., #9677), acetyl-histone H3 lysine18 (acetyl-H3K18; Cell Signaling Technology, Inc., #13998), and acetyl-histone H3 lysine27 (acetyl-H3K27; Cell Signaling Technology, Inc., #8173). Horseradish peroxidase-conjugated secondary antibodies (SouthernBiotech, Birmingham, AL) were used at a concentration of 1:2,000. Chemiluminescence reagent (SuperSignal West Pico; Thermo Fisher Scientific Inc., #34075) and imaging equipment (ChemiDoc XRS+ imager; Bio-Rad Laboratories, Inc.) were used to detect and image protein. Densitometry of Immunoblots was conducted via software (Image J; Wayne Rasband, Research Services Branch, National Institute of Mental Health, Bethesda, MA).
Real-Time Quantitative PCR
To assess inflammatory gene expression, upon confluence (80%) MAC-Ts cultured in medium previously described were pretreated with vehicle control (Veh; distilled H2O), SP or SB (1 mM) for 24 h at 37 °C prior to LPS stimulation (1 µg/mL). Following LPS treatment for 2 h at 37 °C, total RNA was harvested using TRI Reagent (Life Technologies, Thermo Fisher Scientific Inc., #15596026) according to the manufacturer’s instructions. RNA samples were diluted to 100 ng/μL (500 ng) and converted to cDNA using the commercial kit (RT2 First-Strand cDNA Synthesis Kit; Qiagen, Hilden, Germany. #330401). PCR array analysis was conducted via SABioscence RT2 Profiler Cow Inflammatory Cytokines & Receptors (Qiagen, Hilden, Germany. #PABT-011ZA) and PCR array data was verified with quantitative PCR (qPCR). Biological triplicates were used for each treatment and data normalization was performed using three endogenous controls geometric mean on the array platform.
Validation of PCR array data was performed by qPCR of five inflammatory genes. For that, RNA (500 ng) was converted to cDNA via commercial kit (Verso cDNA Synthesis Kit; Thermo Fisher Scientific Inc., #AB-1453) and qPCR analysis conducted using master mix (Apex qPCR GREEN Master Mix; Genesee Scientific Corp., San Diego, CA. #42-120) on a qPCR equipment (Bio-Rad CF96X; Bio-Rad Laboratories, Inc.).
PCR primers for the genes tumor necrosis factor α (TNFα) (Fwd: 5′-GCCAACT CCCTCTGTTTATGT-3′) and (Rev: 5′-GACACC TTGACCTCCTGAATAA-3′), bone morphogenetic protein 2 (BMP2) (Fwd: 5′-GCTGGACT GGCCAAAGTCTTA-3′) and (Rev: 5′-TTCACGA GGGCCACTAGATA-3′), chemokine (C-X-C motif) ligand 9 (CXCL9) (Fwd: 5′-ATTTG CTCCAAGCCCTTCT-3′) and (Rev: 5′-GACCTG TTTCTCCCACTCTTT-3′), CXC-chemokine receptor 1 (CXCR1) (Fwd: 5′-GTATGC TGTGGTCGTCATCTAT-3′) and (Rev: 5′-CGAC CAATCCGGCTGTATAA-3′), lymphotoxin-α (LTA) (Fwd: 5′-GAGCTTGGGT GGATGACTAAA-3′) and (Rev: 5′-CCTTCTCT CCATGCTCCATAAA-3′), and 18S ribosomal RNA (Fwd: 5′-GCCGCTAGAG GTGAAATTCTTG-3′) and (Rev: 5′-CTTTC GCTCTGGTCCGTCTT-3′) were designed via Integrated DNA Technologies Primer Quest Tool (https://www.idtdna.com/Primerquest/Home/Index, accessed 18 June 2017) with bovine sequences available from the GenBank database. Under the experimental conditions of the present study, 18S was validated as a suitable reference gene, thus was used on the data normalization. Gorzelniak et al. (2001) and Ferguson et al. (2010) previously described the suitability for reference gene validation. The 2−ΔΔCT method was used to determine relative differences of level of gene expression among treatments (Livak and Schmittgen, 2001; Ferguson et al., 2010), and one-cycle difference was associated to a two-fold increase or decrease in mRNA expression.
Statistical Methods
GraphPad Prism software (GraphPad InStat Software, San Diego, CA) was used to analyze data and produce graphs. One-way ANOVA followed by Tukey post hoc analysis was conducted to assess statistical significance (declared at P ≤ 0.05).
RESULTS AND DISCUSSION
Short-Chain Fatty Acids Inhibit HDAC Activity in MAC-Ts
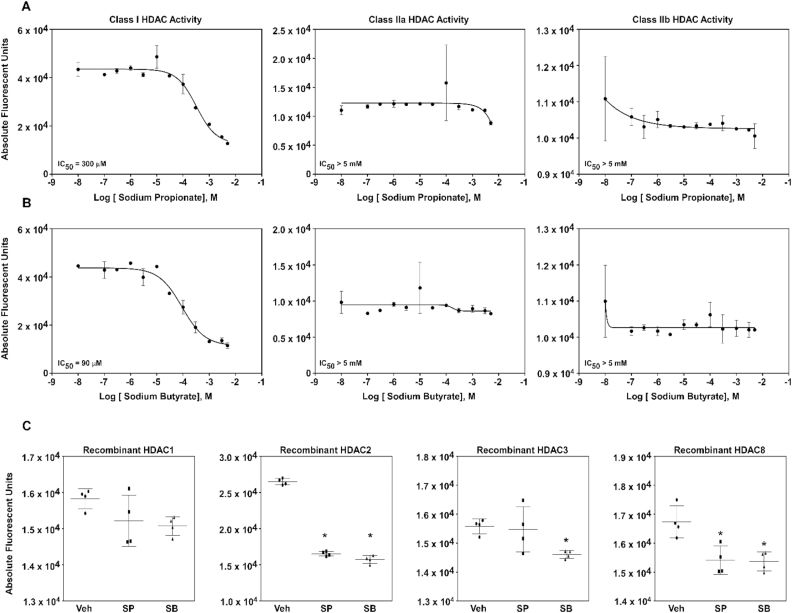
To determine whether SP and SB would inhibit HDAC activity in bovine mammary epithelial cells, we first investigated whether these SCFAs could inhibit zinc-dependent HDAC enzymes by incubating MAC-Ts protein lysates with increasing concentrations of SP or SB (Figure 1A and B). Incubation with SP and SB selectively and concentration-dependently decreased class I HDAC activity, however, had no effect on class IIa and IIb HDAC activity.
Figure 1.
Sodium propionate (SP) and sodium butyrate (SB) effects on histone deacetylase (HDAC) activity in bovine mammary epithelial cells (MAC-Ts). MAC-Ts were cultured in medium containing Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS, 10%), insulin (10 µg/mL), and antibiotics (100 IU/mL of penicillin and 100 µg/mL of streptomycin). At 80% confluence, MAC-Ts were lysed and collected. Biological quadruplicates were used for each treatment. (A) Cell lysates were incubated with increasing concentrations (0, 0.0001, 0.0003, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, and 5 mM) of SP for 2 h at 37 °C prior to incubation with class-specific HDAC substrates for 2 h at 37 °C. Next, cells were incubated with developer/stop solution for 20 min at 37 °C and HDAC activity was determined by fluorescence detection. (B) Cell lysates were incubated with increasing concentrations (0, 0.0001, 0.0003, 0.001, 0.003, 0.01, 0.03, 0.1, 0.3, 1, 3, and 5 mM) of SB for 2 h at 37 °C prior to incubation with class-specific HDAC substrates for 2 h at 37 °C. Next, cells were incubated with developer/stop solution for 20 min at 37 °C and HDAC activity was determined by fluorescence detection. (C) Recombinant proteins for the class I HDACs (HDAC1, 2, 3, and 8) were incubated with vehicle control (Veh; distilled H2O), SP (1 mM) or SB (1 mM) for 24 h at 37 °C. Recombinant proteins were then incubated against the class I-specific HDAC substrate for 2 h at 37 °C. Next, cells were incubated with developer/stop solution for 20 min at 37 °C and HDAC activity was determined by fluorescence detection. One-way ANOVA followed by Tukey post hoc analysis was conducted to assess statistical significance set at P < 0.05. *Significantly different from Veh.
To confirm the effects of SP and SB on zinc-dependent HDAC activity, we next investigated which class I HDACs SP and SB were able to inhibit by incubating recombinant proteins for class I HDAC1, HDAC2, HDAC3, and HDAC8 against SP or SB (Figure 1C). The results showed that inhibition of class I HDAC activity differed between the SCFAs tested, where SB inhibited HDACs 2, 3, and 8, while SP inhibited HDACs 2 and 8.
Propionate and butyrate are SCFAs derive from ruminal microbial fermentation of dietary carbohydrates, they are readily absorbed through the rumen wall and serve as the main energy source to ruminant animals (Bergman, 1990). Nutrition widely influences the immune system of an individual and in humans different functions of SCFAs have been identified, independent of energy synthesis, such as immune regulation (Meijer et al., 2010). Diet-derived compounds have received increased consideration as potential cell functions modulators due to their potential effects on gene expression modulation via epigenetic changes (Kussmann and Van Bladeren, 2011). We report that the SCFAs SP and SB inhibited HDAC activity in MAC-Ts, with selectively directed against class I HDACs. In addition, we showed that SP and SB differentially inhibited class I HDAC isoforms that was concomitant with differential histone H3 hyperacetylation profiles. Thus, SCFAs potentially regulate gene expression via hyperacetylation of nucleosomal chromatin that contributes to DNA relaxation. Taken together, our results suggest that both SP and SB differentially inhibit class I HDACs in MAC-Ts.
Short-Chain Fatty Acids Increase Histone H3 Acetylation in LPS-Stimulated MAC-Ts
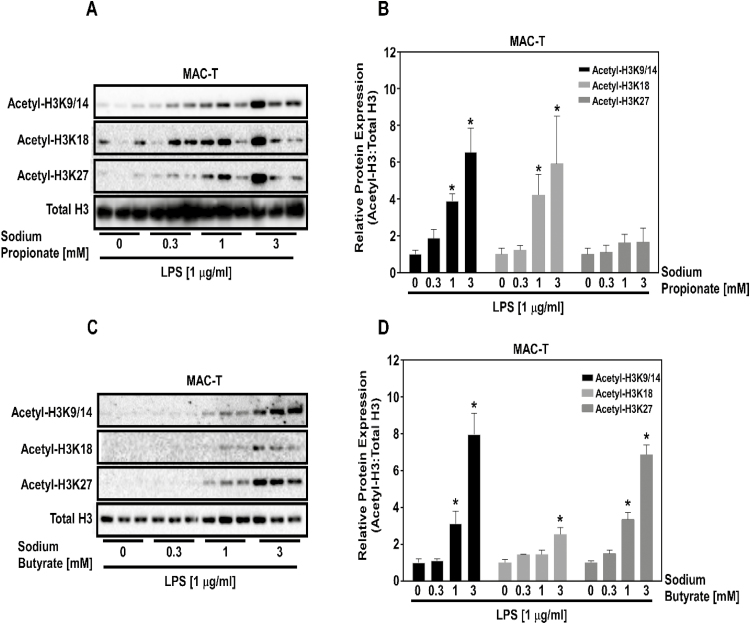
Inhibition of HDACs would predict an increase histone acetylation. To test this postulate, MAC-Ts were treated with increasing concentrations of SP (Figure 2A and B) and SB (Figure 2C and D) prior to LPS stimulation. As predicted, SP (Figure 2A) and SB (Figure 2C) concentration-dependently increased histone H3 acetylation in MAC-Ts. Quantitation of histone H3 acetylation in response to SP (Figure 2B) and SB (Figure 2D) mirrored differences observed in HDAC isoform inhibition (Figure 1), where SB increased H3K9/14, H3K18, and H3K27 acetylation, while SP increased H3K9/14 and H3K18 acetylation.
Figure 2.
SP and SB effects on histone H3 acetylation in bovine mammary epithelial cells (MAC-Ts). MAC-Ts were cultured in medium containing DMEM, FBS (10%), insulin (10 µg/mL), and antibiotics (100 IU/mL of penicillin and 100 µg/mL of streptomycin). Biological triplicates were used for each treatment. (A) MAC-Ts were treated with increasing concentrations (0, 0.3, 1.0, and 3.0 mM) of SP for 2 h at 37 °C prior to LPS (1 µg/mL) stimulation. MAC-Ts were lysed and collected 2 h post-LPS stimulation and immunoblotted for H3K9/14, H3K18, H3K27, and total H3. (B) Data were quantified. (C) MAC-Ts were treated with increasing concentrations (0, 0.3, 1.0, and 3.0 mM) of SB for 2 h at 37 °C prior to LPS (1 µg/mL) stimulation. MAC-Ts were lysed and collected 2 h post-LPS stimulation and immunoblotted for H3K9/14, H3K18, H3K27, and total H3. (D) Data were quantified. One-way ANOVA followed by Tukey post hoc analysis was conducted to assess statistical significance set at P < 0.05. *Significantly different from SP and SB 0 mM concentration.
Acetylation and deacetylation of histone proteins play a major role in chromatin dynamics and in gene expression (Ruijter et al., 2003). Histone deacetylation results in condensation of chromatin, which is associated to transcriptional repression. HDACs are enzymes that control this epigenetic process (Buchwald et al., 2009) and the dysregulation of their activity is associated with many inflammatory diseases. On the other hand, HDAC inhibition has been shown to have anti-inflammatory effects in various inflammatory models including rheumatoid arthritis and pulmonary hypertension (Zhao et al., 2012; Angiolilli et al., 2017).
In this study, we demonstrated that SP and SB increased histone H3 acetylation. In agreement with our findings, Candido et al. (1978) evaluated different cell line responses to 24 h incubation with 5 mM of SB and reported that all the cultured cell lines tested responded to SB treatment by increasing histone H3 acetylation. In addition, Wang et al. (2017) showed that SP concentration-dependently (0.125 to 1 mM) increased histone H3 acetylation in mouse mammary epithelial cells that were incubated with 1 μg/mL of LPS for 3 h. Our results showed that SP and SB differentially act as class I HDAC inhibitors, resulting in differential histone H3 hyperacetylation in MAC-Ts.
Short-Chain Fatty Acids and Inflammatory Gene Expression in LPS-Stimulated MAC-Ts
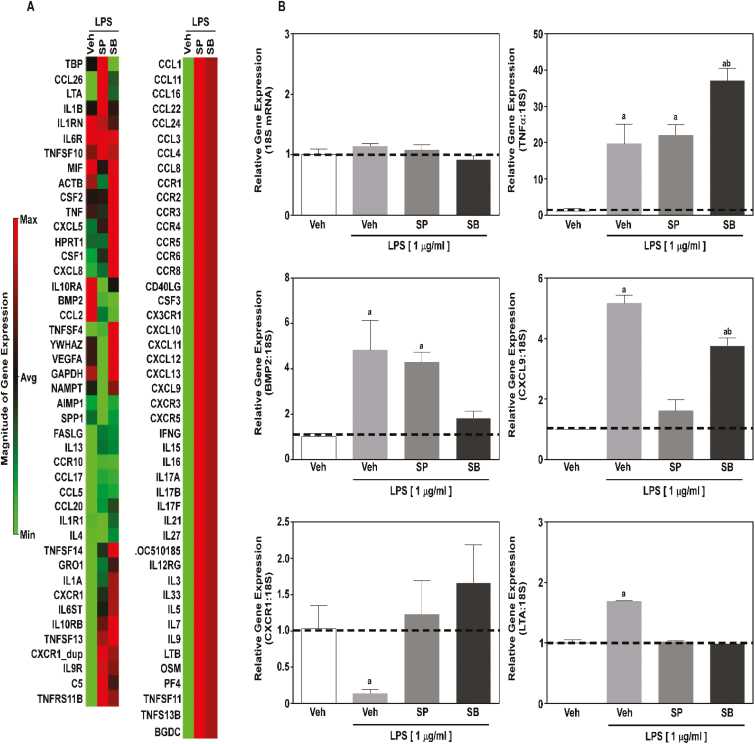
Based on our findings, we postulated that SCFAs HDAC inhibitors would elicit anti-inflammatory actions in MAC-Ts in response to LPS. To determine the anti-inflammatory impact for SCFAs, MAC-Ts were treated with SP or SB prior to LPS stimulation. RNA was isolated 2 h post-LPS, representing maximal induction for TNFα and IL-1α previously tested, and the expression of 83 inflammatory genes was evaluated by PCR array (Figure 3A).
Figure 3.
SP and SB effects on inflammatory gene expression in bovine MAC-Ts. MAC-Ts were cultured in medium containing DMEM, FBS (10%), insulin (10 µg/mL), and antibiotics (100 IU/mL of penicillin and 100 µg/mL of streptomycin), were treated with vehicle control (Veh; distilled H2O), SP (1 mM) or SB (1 mM) for 24 h at 37 °C prior to LPS (1 µg/mL) stimulation, and RNA was isolated 2 h post-LPS stimulation. Biological triplicates were used for each treatment. (A) Heatmap generated from a quantitative polymerase chain reaction (qPCR) array was conducted to examine inflammatory gene expression (83 genes). (B) Real-time qPCR was used to validate BMP2, CXCR1, TNFα, CXCL9, and LTA gene expression from mRNA identified in the array and 18S was used as internal control. One-way ANOVA followed by Tukey post hoc analysis was conducted to assess statistical significance set at P < 0.05. a,b Significantly different from Veh not treated with LPS.
In contrast to our postulate, overall inflammatory gene expression was not decreased by SCFAs. While treatment with SP and SB did normalize LPS-mediated inflammatory gene expression of select genes, SCFAs increased the expression of other inflammatory genes in MAC-Ts (Figure 3A). Validation of select inflammatory gene expression via qPCR confirmed our PCR array data (Figure 3B) and demonstrated that SP and SB (1) did not decrease TNFα gene expression in MAC-Ts; (2) differentially regulated BMP2 and CXCL9 gene expression, and (3) normalized LPS-mediated CXCR1 and LTA gene expression.
Butyrate is one of the main SCFA that can elicits anti-inflammatory effects (Meijer et al., 2010) and has already been used in clinical practice on the treatment of different inflammatory disease (Luhrs et al., 2002). It has been demonstrated that butyrate and propionate act as HDAC inhibitors and exert effects on inflammatory process by suppression of inflammatory response (Segain et al., 2000; Wang et al., 2017). Previous findings showed that butyrate (0.25 to 0.5 mM) and propionate (0.25 to 1 mM) up-regulated the antimicrobial peptide gene expression in bovine mammary epithelial cells (Alva-Murillo et al., 2012). In addition, it was demonstrated that SP (0.125 to 1 mM) suppressed cytokines production (TNFα) in mouse mammary epithelial cells affected by LPS-induced inflammation (Wang et al., 2017).
In contrast to what was postulated, our results showed that inhibition of HDAC by SP and SB was not followed by inhibition of inflammatory gene expression in MAC-Ts. Consistent with our findings; Tarrerias et al. (2002) reported that butyrate and propionate fail to decrease inflammation in rat colonic inflammation. The inconsistent results may be due to differences in the SCFA concentration used and the inflammation models. While our data demonstrate that SCFAs inhibited HDAC activity resulting in histone H3 hyperacetylation, these macromolecules did not inhibit overall LPS-mediated inflammatory gene expression in our system. Under these experimental conditions, these data collectively suggests that SP and SB likely have off-target actions independent of HDAC activity that can elicit an anti- or pro-inflammatory gene response in MAC-Ts.
CONCLUSION
Our results showed that SP and SB inhibited HDAC activity and increased histone H3 acetylation in bovine mammary epithelial cells. However, contrary to our postulate, these short-chain fatty acids did not attenuate LPS-mediated inflammation, suggesting that SP and SB can elicit anti- and pro-inflammatory actions independent of HDAC activity (e.g. receptor-mediated signaling). It should be noted that our experimental conditions were acute (<24 h) and cell and LPS based. This study is a groundbreaking work useful for newcomers in the field to seek the holes of HDAC inhibitors on bovine mammary gland inflammation process. Thus, examination of SCFAs and/or other HDACs inhibitors in different bovine mammary inflammation systems is worth being further investigated to delineate the potential impact of these compounds on HDAC activity, histone hyperacetylation, and inflammatory process of bovine mammary gland tissue.
ACKNOWLEDGMENTS
We would like to show our gratitude to Laura Hernandez (University of Wisconsin-Madison) for kindly donating the MAC-T cells used in this study.
Footnotes
This research was partially supported by the USDA National Institute of Food and Agriculture—NIFA (project number: 1003089 and HATCH-NEV00727). The core facilities used in this study were supported by the National Institute of General Medical Sciences of the National Institute of Health (grant number: P20 GM103554). L.G. Silva, the first author of this study, was funded by the National Council for Scientific and Technological Development—CNPq (process number: 233478/2014-0) and A. S. Avila was funded by the Coordination for the Improvement of Higher Education Personnel—CAPES (process number: 88881.135508/2016-01).
LITERATURE CITED
- Adcock I. M. 2007. HDAC inhibitors as anti-inflammatory agents. Br. J. Pharmacol. 150:829–831. doi: 10.1038/sj.bjp.0707166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alva-Murillo N., Ochoa-Zarzosa A., and López-Meza J. E.. 2012. Short chain fatty acids (propionic and hexanoic) decrease Staphylococcus aureus internalization into bovine mammary epithelial cells and modulate antimicrobial peptide expression. Vet. Microbiol. 155:324–331. doi: 10.1016/j.vetmic.2011.08.025 [DOI] [PubMed] [Google Scholar]
- Angiolilli C., Kabala P. A., Grabiec A. M., Van Baarsen I. M., Ferguson B. S., García S., Malvar Fernandez B., McKinsey T. A., Tak P. P., Fossati G.,. et al. 2017. Histone deacetylase 3 regulates the inflammatory gene expression programme of rheumatoid arthritis fibroblast-like synoviocytes. Ann. Rheum. Dis. 76:277–285. doi: 10.1136/annrheumdis-2015-209064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann H., and Gauldie J.. 1994. The acute phase response. Immunol. Today 15:74. [DOI] [PubMed] [Google Scholar]
- Bergman N. E. 1990. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 70:567–590. doi: 10.1152/physrev.1990.70.2.567 [DOI] [PubMed] [Google Scholar]
- Buchwald M., Krämer O. H., and Heinzel T.. 2009. HDACi—targets beyond chromatin. Cancer Lett. 280:160–167. doi: 10.1016/j.canlet.2009.02.028 [DOI] [PubMed] [Google Scholar]
- Bush E. W., and McKinsey T. A.. 2009. Targeting histone deacetylases for heart failure. Expert Opin. Ther. Targets 13:767–784. doi: 10.1517/14728220902939161 [DOI] [PubMed] [Google Scholar]
- Candido E. P., Reeves R., and Davie J. R.. 1978. Sodium butyrate inhibits histone deacetylation in cultured cells. Cell 14:105–113. doi: 10.1016/0092-8674(78)90305-7 [DOI] [PubMed] [Google Scholar]
- Chang P. V., Hao L., Offermanns S., and Medzhitov R.. 2014. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA. 111:2247–2252. doi: 10.1073/pnas.1322269111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier R. J., Stiening C. M., Pollard B. C., VanBaale M. J., Baumgard L. H., Gentry P. C., and Coussens P. M.. 2006. Use of gene expression microarrays for evaluating environmental stress tolerance at the cellular level in cattle. J. Anim. Sci. 84(Suppl):E1–13. [DOI] [PubMed] [Google Scholar]
- Ferguson B. S., Nam H., Hopkins R. G., and Morrison R. F.. 2010. Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in adipocytes. PLoS One 5:e15208. doi: 10.1371/journal.pone.0015208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzelniak K., Janke J., Engeli S., and Sharma A. M.. 2001. Validation of endogenous controls for gene expression studies in human adipocytes and preadipocytes. Horm. Metab. Res. 33:625–627. doi: 10.1055/s-2001-17911 [DOI] [PubMed] [Google Scholar]
- Grozinger C. M., and Schreiber S. L.. 2002. Deacetylase enzymes: biological functions and the use of small-molecule inhibitors. Chem. Biol. 9:3–16. doi: 10.1016/S1074-5521(02)00092-3 [DOI] [PubMed] [Google Scholar]
- Halili M. A., Andrews M. R., Sweet M. J., and Fairlie D. P.. 2009. Histone deacetylase inhibitors in inflammatory disease. Curr. Top. Med. Chem. 9:309–319. doi: 10.2174/156802609788085250 [DOI] [PubMed] [Google Scholar]
- Huynh H. T., Robitaille G., and Turner J. D.. 1991. Establishment of bovine mammary epithelial cells (MAC-T): an in vitro model for bovine lactation. Exp. Cell Res. 197:191–199. doi: 10.1016/0014-4827(91)90422-Q [DOI] [PubMed] [Google Scholar]
- Kussmann M., and Van Bladeren P. J.. 2011. The extended nutrigenomics – understanding the interplay between the genomes of food, gut microbes, and human host. Front. Genet. 2:21. doi: 10.3389/fgene.2011.00021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemon D. D., Horn T. R., Cavasin M. A., Jeong M. Y., Haubold K. W., Long C. S., Irwin D. C., McCune S. A., Chung E., Leinwand L. A.,. et al. 2011. Cardiac HDAC6 catalytic activity is induced in response to chronic hypertension. J. Mol. Cell. Cardiol. 51:41–50. doi: 10.1016/j.yjmcc.2011.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Luhrs H., Gerke T., Muller J. G., Melcher T., Schauber J., Boxberger F., Scheppach W. Menzel T.. 2002. Butyrate inhibits NF-kB activation in lamina propria macrophages of patients with ulcerative colitis. Scand. J. Gastroenterol. 37:458–466. [DOI] [PubMed] [Google Scholar]
- Mattila T., and Frost A. J.. 1989. Induction by endotoxin of the inflammatory response in the lactating and dry bovine mammary gland. Res. Vet. Sci. 46:238–240. PMID: 2784862. [PubMed] [Google Scholar]
- Meijer K., de Vos P., and Priebe M. G.. 2010. Butyrate and other short-chain fatty acids as modulators of immunity: what relevance for health?Curr. Opin. Clin. Nutr. Metab. Care 13:715–721. doi: 10.1097/MCO.0b013e32833eebe5 [DOI] [PubMed] [Google Scholar]
- Ochoa-Zarzosa A., Villarreal-Fernández E., Cano-Camacho H., and López-Meza J. E.. 2009. Sodium butyrate inhibits staphylococcus aureus internalization in bovine mammary epithelial cells and induces the expression of antimicrobial peptide genes. Microb. Pathog. 47:1–7. doi: 10.1016/j.micpath.2009.04.006 [DOI] [PubMed] [Google Scholar]
- Patnala R., Arumugam T. V., Gupta N., and Dheen S. T.. 2017. HDAC inhibitor sodium butyrate-mediated epigenetic regulation enhances neuroprotective function of microglia during ischemic stroke. Mol. Neurobiol. 54:6391–6411. doi: 10.1007/s12035-016-0149-z [DOI] [PubMed] [Google Scholar]
- Ruijter A. J. M. D., Gennip A. H. V., Caron H. N., Kemp S., and Kuilenburg A. B. P. V.. 2003. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem. J. 370:737–749. doi: 10.1042/bj20021321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segain J. P., Raingeard de la Blétière D., Bourreille A., Leray V., Gervois N., Rosales C., Ferrier L., Bonnet C., Blottière H. M., and Galmiche J. P.. 2000. Butyrate inhibits inflammatory responses through NF-kappaβ inhibition: implications for Crohn’s disease. Gut 47:397–403. doi: 10.1136/gut.47.3.397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva L. G., Ferguson B. S., and Faciola A. P.. 2017. Rapid communication: prolactin and hydrocortisone impact TNFα-mediated mitogen-activated protein kinase signaling and inflammation of bovine mammary epithelial (MAC-T) cells. J. Anim. Sci. 95:5524–5531. doi: 10.2527/jas2017.2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarrerias A. L., Millecamps M., Alloui A., Beaughard C., Kemeny J. L., Bourdu S., Bommelaer G., Eschalier A., Dapoigny M., and Ardid D.. 2002. Short-chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS-induced colonic inflammation in rats. Pain 100:91–97. doi: 10.1016/S0304-3959(02)00234-8 [DOI] [PubMed] [Google Scholar]
- Wang J., Wei Z., Zhang X., Wang Y., Yang Z., and Fu Y.. 2017. Propionate protects against lipopolysaccharide-induced mastitis in mice by restoring blood–milk barrier disruption and suppressing inflammatory response. Front. Immunol. 8:1–9. doi: 10.3389/fimmu.2017.01108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Chen C. N., Hajji N., Oliver E., Cotroneo E., Wharton J., Wang D., Li M., McKinsey T. A., Stenmark K. R.,. et al. 2012. Histone deacetylation inhibition in pulmonary hypertension: therapeutic potential of valproic acid and suberoylanilide hydroxamic acid. Circulation 126:455–467. doi: 10.1161/CIRCULATIONAHA.112.103176 [DOI] [PMC free article] [PubMed] [Google Scholar]