Abstract
Context
Weaver syndrome is characterized by tall stature, advanced bone age, characteristic facies, and variable intellectual disability. It is caused by heterozygous mutations in enhancer of zeste homolog 2 (EZH2), a histone methyltransferase responsible for histone H3 at lysine 27 (H3K27) trimethylation. However, no early truncating mutations have been identified, suggesting that null mutations do not cause Weaver syndrome.
Objective
To test alternative hypotheses that EZH2 variants found in Weaver syndrome cause either a gain of function or a partial loss of function.
Design
Exome sequencing was performed in a boy with tall stature, advanced bone age, and mild dysmorphic features. Mutant or wild-type EZH2 protein was expressed in mouse growth plate chondrocytes with or without endogenous EZH2, and enzymatic activity was measured. A mouse model was generated, and histone methylation was assessed in heterozygous and homozygous embryos.
Results
A de novo missense EZH2 mutation [c.1876G>A (p.Val626Met)] was identified in the proband. When expressed in growth plate chondrocytes, the mutant protein showed decreased histone methyltransferase activity. A mouse model carrying this EZH2 mutation was generated using CRISPR/Cas9. Homozygotes showed perinatal lethality, whereas heterozygotes were viable, fertile, and showed mild overgrowth. Both homozygous and heterozygous embryos showed decreased H3K27 methylation.
Conclusion
We generated a mouse model with the same mutation as our patient, found that it recapitulates the Weaver overgrowth phenotype, and demonstrated that EZH2 mutations found in Weaver syndrome cause a partial loss of function.
A de novo missense EZH2 mutation, identified in a boy with Weaver syndrome, was introduced into mice, causing decreased histone H3K27 methylation and overgrowth.
Weaver syndrome (1) is characterized by overgrowth, accelerated skeletal maturation, intellectual disability, and possibly increased susceptibility to malignancies. We studied a boy with features of Weaver syndrome and identified a heterozygous de novo mutation in enhancer of zeste homolog 2 (EZH2). This finding is consistent with recent studies showing de novo germline heterozygous mutations in EZH2 in Weaver syndrome (2, 3). However, the molecular pathophysiology is poorly understood.
EZH2 is a key component of the polycomb repressive complex 2 (PRC2). PRC2 comprises four essential core subunits: EZH1 or 2, embryonic ectoderm development, suppressor of zeste 12 homolog, and retinoblastoma-associated protein 48 (4). PRC2 is an important chromatin modifier, responsible for the trimethylation of histone H3 at lysine 27 (H3K27me3), which then serves as an epigenetic signal for chromatin condensation and transcriptional repression (5).
Interestingly, all EZH2 mutations found to date in Weaver syndrome are either missense mutations or truncating mutations in the last exon (3), which therefore may not initiate nonsense-mediated RNA decay (6). The lack of early truncating mutations suggests that Weaver syndrome is not caused by simple haploinsufficiency of EZH2. We reasoned that EZH2 mutations in Weaver syndrome could be either partial loss-of-function or gain-of-function mutations. We recently showed that complete loss of Ezh1 and Ezh2 in mouse growth plate slows growth (7), suggesting that the increased growth in Weaver syndrome might be due to a gain of function. In contrast, a prior study found that Weaver mutations decreased histone methyltransferase activity in a cell-free system, although it was unable to assess all three methylation steps catalyzed by EZH2 (8). To determine whether Weaver mutations cause a partial loss of function or a gain of function, we transfected wild-type or mutated Ezh2 cDNA into wild-type mouse chondrocytes or chondrocytes lacking Ezh2 and assessed histone methyltransferase activity. We also used CRISPR/Cas9 to generate a mouse model that carried the Weaver EZH2 mutation that we had identified and studied histone methylation and postnatal growth in this mouse model.
Subjects and Methods
Patient report
This study was approved by the Institutional Review Board of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and written informed consent was obtained from the participant and legal guardians. Heights and weights were plotted on the Centers for Disease Control and Prevention growth charts (www.cdc.gov/growthcharts) (9), and Z-scores were calculated using the Third National Health and Nutrition Examination Survey growth data (10).
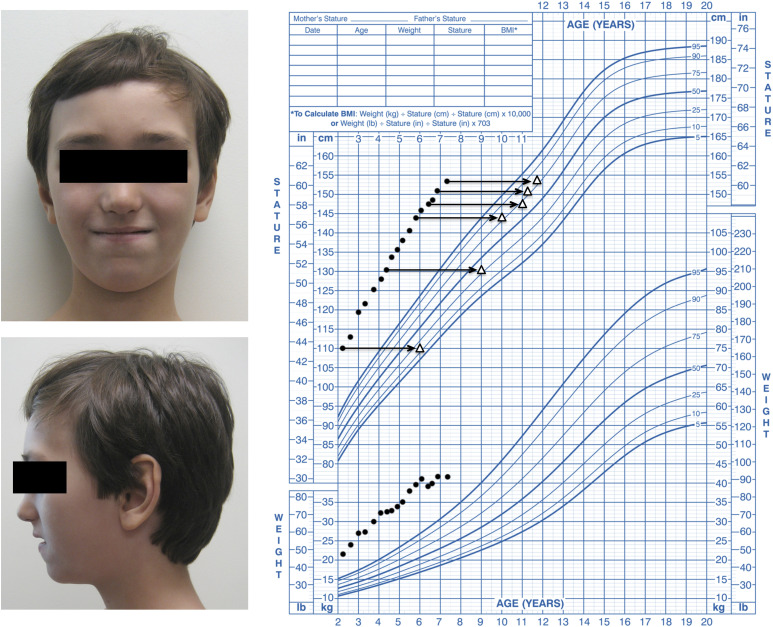
A 7-year 4-month-old boy was evaluated for tall stature at the Division of Pediatric Endocrinology and Diabetes, Ichan School of Medicine at Mount Sinai, and then referred to the National Institutes of Health Clinical Center Pediatric Endocrinology Clinic for further genetic evaluation. He was born at gestational week 36 with a birth length of 54.6 cm [+3.0 standard deviation score (SDS)] and birth weight of 3.15 kg (+0.9 SDS). During childhood, he showed a high growth velocity, causing both height and weight to diverge from the normal range. His bone age was advanced by ∼4 years (Fig. 1). He had moderately delayed motor, speech, and cognitive development. His parents were of normal stature (mother 168.8 cm, +0.85 SDS; father 179.8 cm, +0.4 SDS). On examination at the National Institutes of Health, he was noted to be a tall, proportional, and prepubertal boy with macrocephaly and subtle facial feature including large ears, mild ocular hypertelorism, and relative retrognathia (Fig. 1). He had large hands with thin, deep-set nails. There were no signs of excessive sex steroid exposure. Endocrine evaluations were normal with no indications of growth hormone excess, hyperthyroidism, early puberty, congenital adrenal hyperplasia, or other condition of increased sex steroid production (Supplemental Table S1). Genetic testing for Sotos syndrome was negative, and comparative genomic hybridization microarray did not detect any deletions or duplications.
Figure 1.
A patient with overgrowth, advanced bone age, developmental delay, and facies suggestive of Weaver syndrome. Left: Photograph of proband with Weaver syndrome at age 7 years 4 months. Right: Growth chart (National Center for Health Statistics, Centers for Disease Control and Prevention) of proband showing heights and weights for chronological age (filled circles) and bone age (open triangles).
Animal procedures and tissue processing
All animals were used in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council 2003). Mice were weighed weekly to follow their postnatal growth pattern. Mice were euthanized at various ages, and the heart, liver, lungs, kidneys, and spleen were excised and weighed. Tibiae were excised, separated from adjacent muscle, and their lengths measured using a digital vernier caliper. Both males and females were used in all experiments.
Mouse genome editing to create the Ezh2V626M allele
The Val to Met substitution at codon 626 was created by CRISPR-mediated genome editing directly in fertilized eggs of C57BL/J (The Jackson Laboratory) as described (11). Three target sequences for guide RNAs (gRNAs) near the target codon were initially selected using the ranking tool CRISPR Design (http://crispr.mit.edu). Selected gRNAs were synthesized by T7 in vitro transcription as described (12). Briefly, templates for in vitro transcription were first assembled in a polymerase chain reaction (PCR) with a forward primer containing sequences of the T7 promoter, the gRNA, the first 15 base pairs (bp) of the tracer RNA portion of the guide RNA backbone, and the reverse primer derived from the last 20 bp of the tracer RNA. A plasmid with the backbone sequence of the SpCas9 gRNA was used as the PCR template. gRNAs were then synthesized with the MegaScript T7 kit (Ambion). The gRNAs were compared for their activity to introduce a double-strand break at the target site in mouse embryonic stem cells with Surveyor assay (13) (data not shown). gRNA gE16(57) (Supplemental Table 2) was chosen. A single-stranded DNA oligonucleotide (127-mer) carrying the c.1876G>A transversion (NM_004456.4) was designed, based upon the position and orientation of the guide RNA gE16(57) (14), and synthesized (Integrated DNA Technologies) (Supplemental Table 2) to create the p.V626M substitution. The combination of the guide RNA with the mutagenic oligonucleotide was further tested for its efficiency, introducing the desired point mutation into the mouse genome by digital-droplet PCR (data not shown) with a pair of probes for wild-type and mutated alleles, respectively (Supplemental Table 2). In that system, 4% of the mouse fibroblast cells harboring a tet-inducible Cas9 expression cassette (unpublished data) were correctly edited upon induction of Cas9 expression (data not shown). C57BL6/J donor eggs were microinjected with a mixture of SpCas9 protein (50 ng/µL; PNA BIO), gRNA gE16(57) (50 ng/µL), and the mutagenic oligo (100 ng/µL) as described (11). F0 founders carrying the mutation (c.1876G>A; V626M) were confirmed by Sanger sequencing of a PCR amplicon (418 bp) generated using genotyping forward and reverse primers (Supplemental Table 2). This approach of PCR (using these primers) followed by Sanger sequence was used for routine genotyping of this Weaver mouse line.
Plasmid preparation
Plasmids expressing wild-type or Weaver variants of EZH2 were construct by gene synthesis (GeneArt Gene Synthesis; Life Technologies) using a pcDNA3.1 backbone.
Chondrocyte isolation and transfection
Growth plates from proximal tibias and distal femurs were dissected from 1-week-old EZH1/2 cartilage-knockout mice (7) or EZH1-knockout EZH2 wild-type littermates aseptically and digested in 0.3% collagenase type I (Sigma-Aldrich) in Dulbecco’s modified Eagle medium (DMEM)/F12 medium. The released cells were resuspended, and 1 × 106 chondrocytes was plated each in a 60-mm dish in DMEM/F12 medium (Invitrogen) supplemented with 10% fetal bovine serum, 1% penicillin (100 U/mL)/streptomycin (100 μg/mL), and 50 μg/mL ascorbic acid in a humidified incubator at 37°C, 5% CO2. Transfection was performed as previously described (7). Briefly, monolayer chondrocytes were treated with hyaluronidase (5 U/mL; Sigma-Aldrich) for 6 hours. Prior to transfection, cells were washed once in phosphate-buffered saline (PBS) and changed to 5 mL DMEM/F12 medium without antibiotics. Plasmids expressing wild-type or Weaver variants of EZH2 were transfected (8 μg/dish) into chondrocytes using Lipofectamine 2000 (Life Technologies) following the manufacturer’s standard protocol.
Histone methyltransferase activity assay
Nuclear proteins were extracted from chondrocytes 48 hours after transfection using the EpiQuik Nuclear Extraction Kit (EpiGentek) and tested for H3K27 histone methyltransferase activity using an enzyme-linked immunosorbent assay–based EpiQuik histone methyltransferase activity assay (EpiGentek). EZH2 enzymatic activity is proportional to the amount of biotinylated histone H3 methylated by PRC2 complex in the nuclear extract, detected by a horseradish peroxidase–conjugated trimethyl-H3K27 specific antibody, and quantified by optical density at 450 nm.
Immunostaining
Mouse embryos were fixed in formalin and embedded in paraffin for sectioning (Histoserv, Inc.). Sections of mouse embryos at embryonic day (E)14.5 were baked at 65°C for 45 minutes, deparaffinized in xylene, rehydrated through ethanol series (100%, 100%, 95%, and 95%), and rinsed with PBS. Antigen retrieval was performed using proteinase K (100 μg/mL in PBS, 30 minutes). Endogenous peroxidase activity was blocked by 3% H2O2. Staining was performed using antihistone H3 (1:1000; ab1791; Abcam), anti-H3K27me2 (1:1000; 07-452; Millipore), or anti-H3K27ne3 (1:1000; 07-449; Millipore), with a VECTASTAIN ABC kit (Vector Laboratories) followed by DAB Substrate kit (Vector Laboratories) according to the manufacturer’s instructions. Sections were counterstained with methyl green.
Results
De novo heterozygous EZH2 missense mutations in a child with overgrowth and advanced bone age
To identify the genetic cause of the patient’s overgrowth syndrome, exome sequencing was performed in the patient and parents. The data were filtered for nonsynonymous de novo variants, and a missense mutation in EZH2 [NM_004456.4: c.1876G>A (p.Val626Met)] previously reported to cause Weaver syndrome was identified in the patient but not the parents. The mutation was confirmed by Sanger sequencing. The mutation information was submitted to the ClinVar database (allele ID 168576).
Weaver-associated EZH2 mutations decreased histone methyltransferase activity
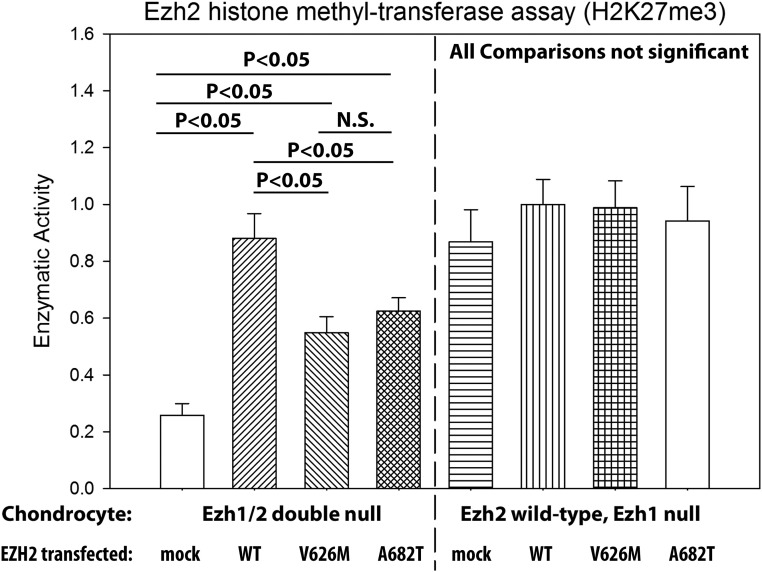
Because EZH1 and EZH2 are known to have partial functional redundancy, we chose to test the enzymatic activity of the mutant EZH2 in cells lacking both endogenous EZH1 and EZH2. We therefore used chondrocytes isolated from mice with ubiquitous deletion of Ezh1 and cartilage-specific deletion of Ezh2 (Ezh1/2 mice) (7). We transfected wild-type EZH2 or two different EZH2 constructs with Weaver-associated mutations, EZH2(p.V626M) and EZH2(p.A682T), into these Ezh1/2-null chondrocytes and measured H3K27 histone methyltransferase activity in their nuclear extracts (Fig. 2, left panel). We found that, compared with mock transfection, cells transfected with wild-type EZH2 had the highest methyltransferase activity, whereas cells transfected with EZH2(p.V626M) and EZH2(p.A682T) demonstrated activity that was greater than mock transfection but less than wild-type EZH2 (Fig. 2, left panel), suggesting partial loss of function. When the same experiment was repeated using chondrocytes lacking Ezh1 but with intact Ezh2, there were no statistically noteworthy differences in methyltransferase activity among cells transfected with wild-type EZH2, EZH2(p.V626M), or EZH2(p.A682T) (Fig. 2, right panel). In particular, we did not observe any tendency for increased activity in the Weaver variants transfected in wild-type chondrocytes, suggesting that these variants are not cooperative gain-of-function mutations in the presence of wild-type EZH2 (15).
Figure 2.
Weaver-associated EZH2 mutations decreased histone methyltransferase activity. Ezh1/2-null chondrocytes (left) or Ezh1-null chondrocytes (right) were transfected with constructs expressing EZH2 (wild-type or variant), and then nuclear extracts were tested for H3K27 methyltransferase activity. The two Weaver-associated EZH2 variants that were tested (V626M and A682T) both showed decreased methyltransferase activity compared with wild-type EZH2 in the Ezh1/2-null chondrocytes. No differences were found in the Ezh1-null chondrocytes, which have endogenous Ezh2 expression. N.S., not significant.
Creation of a mouse model for Weaver syndrome
To further confirm that Weaver EZH2 mutants cause a partial loss of function in H3K27 histone methyltransferase activity and to gain insights into the role of EZH2 mutations in body growth, we next used CRISPR/Cas9 genome editing to generate a Weaver mouse model carrying the Ezh2(p.V626M) variant. Sanger sequencing confirmed the c.1876G>A (p.V626M) substitution (Fig. 3A). The resultant heterozygous pups looked grossly normal and were fertile.
Figure 3.
Heterozygous Weaver mice showed postnatal overgrowth. CRISPR/Cas9 genome editing was used to generate a mouse model carrying a Weaver-associated Ezh2 variant (V626M). (A) Crosses of heterozygotes generated mice that were wild-type, heterozygous, or homozygous for the Weaver variant, which was confirmed by PCR amplification of a genomic region containing the variant followed by Sanger sequencing. (B) Body weight was measured in heterozygous Weaver mice and wild-type (WT) littermates weekly up until 8 weeks of age (homozygotes were perinatally lethal). Two-way analysis of variance (ANOVA) showed that body weights differed significantly between genders and genotypes (both P < 0.001). Pairwise comparison showed that, in both males (M) and females (F), body weights were significantly increased in heterozygous Weaver mice. (C–H) At 8 weeks of age, heterozygote Weaver mice showed increased weights of heart, kidney, and spleen but no differences in weights of liver and lung or in tibia length compared with wild-type mice. *P < 0.05.
Perinatal lethality in homozygous Weaver mice
When mice heterozygous for the Weaver mutation were crossed with other heterozygous mice, the ratio of wild-type to heterozygous offspring was roughly 1:2, consistent with Mendelian ratios (Table 1). However, no surviving homozygous pups were found. A small number of homozygous pups were born dead or died shortly after birth, suggesting perinatal lethality. Histopathology showed diffuse atelectasis of lungs in homozygous pups, suggesting respiratory insufficiency to be the cause of perinatal lethality. Earlier in gestation, at E14.5 or E18.5, homozygous embryos were identified in numbers consistent with Mendelian ratios (Table 1). Homozygous Weaver embryos at E18.5 showed a slightly decreased body size compared with wild-type or heterozygotes (P < 0.05; Table 1), but histopathology identified no gross abnormality.
Table 1.
Genotypes of Live Births and Embryos From Ezh2 (V626M) Heterozygous Intercrosses
| Category | Total Number | Homozygous Wild-Type | Heterozygous | Homozygous V626M |
|---|---|---|---|---|
| Embryos (E14.5) | 33 | 4 | 18 | 11 |
| Embryos (E18.5) [weight (g)] | 23 | 6 (1.13 ± 0.02) | 12 (1.08 ± 0.03) | 5 (1 found dead) (0.92 ± 0.07)a |
| Postnatal mice, age 0 to 1 d | 27 | 9 | 13 | 5 (all found dead) |
| Postnatal mice, age 1–7 d | 64 | 22 | 42 | 0 |
Analysis of variance, homozygous vs wild-type, P = 0.009; homozygous vs heterozygous, P = 0.014. Weight between wild-type and heterozygous not significant.
The Weaver mutation had a milder effect in terms of mortality than a previously described null mutation in Ezh2, which caused embryonic lethality at E8.5 in homozygous embryos and decreased numbers of live heterozygous pups compared with Mendelian ratios (16). The milder effect on mortality in the Weaver model, in both the homozygous and heterozygous state, is consistent with the hypothesis that the Weaver mutation causes only a partial loss of function.
Postnatal overgrowth in heterozygous Weaver mice
To gain further insights into the role of Weaver Ezh2 mutations in postnatal growth, we followed the heterozygous Weaver mice to 8 weeks of age. The growth of heterozygotes was similar to that of wild-type littermates up to 5 weeks of age, when the heterozygotes started showing signs of overgrowth (Fig. 3B). At 8 weeks, overall body weight was increased in heterozygous Weaver mice, more prominently in females (Fig. 3B). Compared with human patients with Weaver syndrome, the overgrowth found in mice appeared relatively mild. To assess the tissue distribution of the overgrowth, we measured the weights of major organs and tibia length in Weaver mice at 8 weeks of age (Fig. 3C–3H). Tibia lengths were not different between wild-type and heterozygotes (Fig. 3H), whereas the weights of heart, kidney, and spleen were found to be increased in heterozygotes. The increased weight of spleen (Fig. 3G) raises the possibility of increased lymphocyte proliferation, which might be related to a possible increased risk of leukemia and lymphoma in Weaver syndrome suggested by some reports (17, 18).
Heterozygous and homozygous Weaver mice showed decreased histone methyltransferase activity
Immunostaining of E14.5 embryos (Fig. 4) demonstrated that dimethyl-H3K27 levels were greatest in wild-type embryos, decreased in heterozygous embryos, and further decreased in homozygous embryos (Fig. 4C). A similar pattern was seen for trimethyl H3K27 (Fig. 4D). These findings provide in vivo confirmation that the Weaver EZH2 variant causes a partial loss-of-function mutation and specifically excludes a cooperative gain of function. The marked decrease in H3K27 methylation also suggests that Ezh1 does not compensate for the decreased Ezh2 activity in mouse embryonic tissues.
Figure 4.
Homozygous and heterozygous Weaver mice showed decreased H3K27 methylation in vivo. Immunostaining of E14.5 embryos was used to examine difference in histone methyltransferase activity in vivo. Formalin-fixed wild-type (WT), heterozygotes (Het), and homozygotes (Homo) were placed on the same slide and stained with (A) hematoxylin and eosin (H&E) for histology, (B) antibody against total H3, (C) antibody against H3K27me2, or (D) antibody against H3K27me3. Brown color indicates immunostaining, and green represents counterstain. We found that both H3K27me2 and me3 levels were greatest in wild-type embryos, decreased in heterozygous embryos, and further decreased in homozygous embryos. No difference was observed for total H3 among different genotypes. Scale bar, 3 mm.
Discussion
We studied a patient with overgrowth, advanced bone age, developmental delay, and facies suggestive of Weaver syndrome. Sequencing revealed a de novo heterozygous mutation in EZH2. Heterozygous de novo mutations in EZH2 have previously been demonstrated in Weaver syndrome. However, the molecular pathogenesis remains unclear. Two alternative possibilities are that these mutations cause a loss of function, as occurs, for example, in myeloid malignancies, or cause a gain of function, similar to somatic mutations found in B-cell lymphomas. To date, the causative mutations have all been missense mutations or else truncating mutations in the last exon; no earlier truncating mutations have been described, suggesting that Weaver syndrome is not caused by complete loss-of-function mutations but still might be caused by partial loss of function. To distinguish these possibilities, we expressed Ezh2 with the mutation that we identified in this patient, and also a second Weaver-associated mutation, in chondrocytes lacking Ezh1 and Ezh2 and found that the mutations partially impaired synthesis of trimethyl H3K27. No effect was seen in cells that had intact normal Ezh2 alleles. We next used CRISPR/Cas to introduce into the mouse genome the mutation that we had identified in Weaver syndrome. Mice that were heterozygous for this mutation showed mild overgrowth, including increased body mass and mass of the heart, kidney, and spleen. This mouse represents an animal model for Weaver syndrome, although, for reasons that are not clear, this mouse model does not show the skeletal overgrowth that is present in human Weaver syndrome. Homozygous mice died in the perinatal period. Immunostaining revealed a decrease in di- and trimethyl H3K27 in both homozygous embryos and, to a lesser degree, heterozygous embryos.
Our findings represent the most definitive proof to date that Weaver-associated mutations cause a partial loss of function rather than a gain of function. One recent study by Cohen et al. (8) reported that EZH2 variants from Weaver syndrome reduced histone methyltransferase activity. However, assessment of function is complicated because EZH2 actually catalyzes three sequential methylation reactions, yielding mono-, di-, and trimethylated H3K27. Somatic mutations in EZH2 identified in B-cell lymphomas (19) modify substrate specificity, leading to decreased activity at the monomethylation step but increased activity at the trimethylation step. Initially, these mutations, studied in isolation, were thought to cause an overall loss of function, but, when studied in the heterozygous state, the mutant proteins act cooperatively with wild-type protein (which efficiently catalyzes the monomethylation reaction) to increase trimethyl H3K27 (15, 20). The prior analysis of Weaver EZH2 variants (8) used a cell-free enzyme assay with a mix of core histones and H3 peptides at different methylation states as substrate and therefore did not distinguish the three sequential methylation reactions. In an alternative assay, biotinylated peptides that were unmethylated, monomethylated, or dimethylated were used separately as substrates, but the assay was only able to detect the first methylation step above background level. As a result, these prior findings did not exclude the possibility that Weaver mutations alter substrate specificity and thereby produce overall gain of function in the heterozygous state through cooperativity with the wild-type protein. In the current study, we addressed this problem in two ways. First, we expressed the mutant protein not only in cells lacking Ezh2 but also in cells already expressing wild-type Ezh2 and did not see increased production of trimethyl H3K27. Second, we created a mouse model of Weaver syndrome, performed immunohistochemistry for di- and trimethyl-H3K27, and found decreased methylation not only in the homozygous embryos but also in the heterozygous embryos, again demonstrating a loss of function, rather than a gain, through cooperativity with the wild-type allele product. This conclusion, that the mutations in Weaver syndrome cause a partial loss of PRC2 histone methyltransferase activity, is also consistent with recent reports that Weaver syndrome can be caused by loss-of-function mutations in embryonic ectoderm development and suppressor of zeste 12 homolog, two other components of the PRC2 complex (21, 22). It is also possible that the missense mutations found in patients with Weaver syndrome result in a protein that is included in the PRC2 complex but abrogates its function and prevents wild-type protein from participating in the complex, whereas, for null mutations, the gene product of the wild-type allele can participate in the complex without interference. Our in vitro findings did not tend to support this possibility; when EZH2 with a Weaver-associated missense mutation was transfected into chondrocytes expressing endogenous Ezh2, the enzyme activity was not diminished.
Our findings pose an interesting question: if EZH2 mutations that cause Weaver are indeed loss of function, why have no early truncating mutations been found in Weaver syndrome? One possible explanation is that heterozygous-null mutations in EZH2 are clinically silent even though heterozygous hypomorphic alleles cause Weaver syndrome. However, truncating mutation in EZH2 have not been observed in the Exome Aggregation Consortium (ExAC; constraint metrics: probability of truncating mutation intolerance pLI = 1.00, Z = 5.45 for missense mutations) (23), suggesting strong selective pressure to avoid null mutations in EZH2 even in the heterozygous state, implying that these mutations confer a survival (or reproductive) disadvantage. A second possible explanation is that heterozygous-null mutations in EZH2 are lethal. In mice, homozygous ablation of Ezh2 is lethal at E8. Some heterozygous pups survive but are present at lower frequency than would be predicted by Mendelian ratios, suggesting partial lethality. In humans, 14 patients heterozygous with deletions that include EZH2 have been reported in the Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER). For two of the heterozygous deletions (DECIPHER patients 1671 and 252404), the submitting clinicians provided us with phenotypic descriptions, which both included multiple anomalies and short stature. In addition, a single patient with a homozygous deletion encompassing EZH2 has been reported, suggesting that homozygous-null mutations in EZH2 in humans, unlike the mouse, are not completely incompatible with life (24). A third possibility is that heterozygous-null mutations in EZH2 lead to a phenotype that is distinct from Weaver syndrome. It is likely that this phenotype results in decreased fitness because null mutations in EZH2 are not found in ExAC. The phenotype of heterozygous-null mutations is likely buried in the phenotype of individuals reported in DECIPHER with deletions encompassing EZH2, but it is difficult to dissect the defects due to deletion of EZH2 from the defects due to deletion of neighboring genes.
Based on our current findings, previous human cases, and mouse models (Table 2), we speculate that mutations in EZH2 may have a complex genotype-phenotype relationship, which reflects a graded relationship between PRC2 histone methyltransferase activity and gene expression. A mild decease in EZH2 activity, due to heterozygous hypomorphic mutations in Weaver syndrome, may lead to derepression of a set of growth-promoting genes in the genome, causing overgrowth. A greater decrease in EZH2 activity, such as occurs in heterozygous-null mutations in EZH2, leads to derepression of additional genes, causing developmental or organ dysfunction and a phenotype, as-yet unknown, distinct from Weaver syndrome. This model is consistent with the slight overgrowth observed in our Weaver mice contrasted with the impaired bone growth in mice lacking Ezh1 and 2 in growth plate cartilage (7). The precise genes that, when derepressed, lead to overgrowth in Weaver syndrome would be difficult to identify. Although we were previously able to identify derepressed genes in mice with homozygous loss of Ezh1 and 2 in growth plate, analogous studies in the Weaver model, with its milder genetic defect and subtle phenotypic, would likely be technically challenging.
Table 2.
Summary of Human and Mouse In Vivo Findings at Different Levels of EZH2 Activity
| Genotype | Mouse Phenotype | Human Phenotype | Predicted Relative Enzyme Activity (%) |
|---|---|---|---|
| Homozygous null | Embryonic lethal (16) | DECIPHER: one 12.26-Mb homozygous deletion found with intellectual disability, abnormal facies, seizures, etc. | 0 |
| Homozygous Weaver mutation | Perinatal lethal (current study) | Not described | ∼30 (speculated based on current study) |
| Heterozygous null | Decreased live births (16) | DECIPHER: various heterozygous deletions found, with a variety of anomalies. No loss-of-function variants found in ExAC, suggesting decreased fitness phenotype | 50 |
| Heterozygous Weaver mutation | Born at normal ratio, mild regrowth | Weaver syndrome: overgrowth, advanced bone maturation, intellectual disability, etc. | ∼65 (assumes mutant has 30% activity) |
DECIPHER refers to findings in the database found at https://decipher.sanger.ac.uk/. ExAC refers to findings in the database found at http://exac.broadinstitute.org/.
In summary, our current findings demonstrate that EZH2 variants found in Weaver syndrome cause a partial loss of function in H3K27 histone methyltransferase activity. Our Weaver mouse model, combined with previous in vivo findings in mice and humans, reveal a complex genotype-phenotype relationship for EZH2 mutations.
Supplementary Material
Acknowledgments
This study makes use of data generated by the DECIPHER community. We thank Prof. David FitzPatrick from the MRC Human Genetics Unit, University of Edinburgh, Scotland, United Kingdom, and Dr. Sara Loddo from the Laboratory of Medical Genetics, Bambino Gesù Children’s Hospital, Rome, Italy, for providing patient information deposited to DECIPHER. A full list of centers that contributed to the generation of the data involving EZH2 is available from http://decipher.sanger.ac.uk and via e-mail from decipher@sanger.ac.uk. Funding for DECIPHER was provided by the Wellcome Trust.
Financial Support: This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health (NIH).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- bp
base pairs
- DECIPHER
Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources
- DMEM
Dulbecco’s modified Eagle medium
- E
embryonic day
- EZH2
enhancer of zeste homolog 2
- ExAC
Exome Aggregation Consortium
- gRNA
guide RNA
- H3K27me3
trimethylation of histone H3 at lysine 27
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PRC2
polycomb repressive complex 2
- SDS
standard deviation score
References
- 1. Tatton-Brown K, Murray A, Hanks S, Douglas J, Armstrong R, Banka S, Bird LM, Clericuzio CL, Cormier-Daire V, Cushing T, Flinter F, Jacquemont ML, Joss S, Kinning E, Lynch SA, Magee A, McConnell V, Medeira A, Ozono K, Patton M, Rankin J, Shears D, Simon M, Splitt M, Strenger V, Stuurman K, Taylor C, Titheradge H, Van Maldergem L, Temple IK, Cole T, Seal S, Rahman N; Childhood Overgrowth Consortium . Weaver syndrome and EZH2 mutations: Clarifying the clinical phenotype. Am J Med Genet A. 2013;161A(12):2972–2980. [DOI] [PubMed] [Google Scholar]
- 2. Gibson WT, Hood RL, Zhan SH, Bulman DE, Fejes AP, Moore R, Mungall AJ, Eydoux P, Babul-Hirji R, An J, Marra MA, Chitayat D, Boycott KM, Weaver DD, Jones SJ; FORGE Canada Consortium . Mutations in EZH2 cause Weaver syndrome. Am J Hum Genet. 2012;90(1):110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tatton-Brown K, Hanks S, Ruark E, Zachariou A, Duarte SV, Ramsay E, Snape K, Murray A, Perdeaux ER, Seal S, Loveday C, Banka S, Clericuzio C, Flinter F, Magee A, McConnell V, Patton M, Raith W, Rankin J, Splitt M, Strenger V, Taylor C, Wheeler P, Temple KI, Cole T, Douglas J, Rahman N; Childhood Overgrowth Collaboration . Germline mutations in the oncogene EZH2 cause Weaver syndrome and increased human height. Oncotarget. 2011;2(12):1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. Molecular architecture of human polycomb repressive complex 2. eLife. 2012;1:e00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298(5595):1039–1043. [DOI] [PubMed] [Google Scholar]
- 6. Maquat LE. When cells stop making sense: effects of nonsense codons on RNA metabolism in vertebrate cells. RNA. 1995;1(5):453–465. [PMC free article] [PubMed] [Google Scholar]
- 7. Lui JC, Garrison P, Nguyen Q, Ad M, Keembiyehetty C, Chen W, Jee YH, Landman E, Nilsson O, Barnes KM, Baron J. EZH1 and EZH2 promote skeletal growth by repressing inhibitors of chondrocyte proliferation and hypertrophy. Nat Commun. 2016;7:13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cohen AS, Yap DB, Lewis ME, Chijiwa C, Ramos-Arroyo MA, Tkachenko N, Milano V, Fradin M, McKinnon ML, Townsend KN, Xu J, Van Allen MI, Ross CJ, Dobyns WB, Weaver DD, Gibson WT. Weaver syndrome-associated EZH2 protein variants show impaired histone mMethyltransferase function in vitro. Hum Mutat. 2016;37(3):301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Centers for Disease Control and Prevention growth charts. Available at: www.cdc.gov/growthcharts. Accessed.
- 10. US Department of Health and Human Services, National Center for Health Statistics. Third National Health and Nutrition Examination Survey, 1988–1994. Hyattsville, MD: National Center for Health Statistics; 1996. [Google Scholar]
- 11. Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Varshney GK, Pei W, LaFave MC, Idol J, Xu L, Gallardo V, Carrington B, Bishop K, Jones M, Li M, Harper U, Huang SC, Prakash A, Chen W, Sood R, Ledin J, Burgess SM. High-throughput gene targeting and phenotyping in zebrafish using CRISPR/Cas9. Genome Res. 2015;25(7):1030–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pilato B, De Summa S, Danza K, Papadimitriou S, Zaccagna P, Paradiso A, Tommasi S. DHPLC/SURVEYOR nuclease: a sensitive, rapid and affordable method to analyze BRCA1 and BRCA2 mutations in breast cancer families. Mol Biotechnol. 2012;52(1):8–15. [DOI] [PubMed] [Google Scholar]
- 14. Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE. Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol. 2016;34(3):339–344. [DOI] [PubMed] [Google Scholar]
- 15. Sneeringer CJ, Scott MP, Kuntz KW, Knutson SK, Pollock RM, Richon VM, Copeland RA. Coordinated activities of wild-type plus mutant EZH2 drive tumor-associated hypertrimethylation of lysine 27 on histone H3 (H3K27) in human B-cell lymphomas. Proc Natl Acad Sci USA. 2010;107(49):20980–20985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O’Carroll D, Erhardt S, Pagani M, Barton SC, Surani MA, Jenuwein T. The polycomb-group gene Ezh2 is required for early mouse development. Mol Cell Biol. 2001;21(13):4330–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Basel-Vanagaite L. Acute lymphoblastic leukemia in Weaver syndrome. Am J Med Genet A. 2010;152A(2):383–386. [DOI] [PubMed] [Google Scholar]
- 18. Usemann J, Ernst T, Schäfer V, Lehmberg K, Seeger K. EZH2 mutation in an adolescent with Weaver syndrome developing acute myeloid leukemia and secondary hemophagocytic lymphohistiocytosis. Am J Med Genet A. 2016;170A(5):1274–1277. [DOI] [PubMed] [Google Scholar]
- 19. Morin RD, Johnson NA, Severson TM, Mungall AJ, An J, Goya R, Paul JE, Boyle M, Woolcock BW, Kuchenbauer F, Yap D, Humphries RK, Griffith OL, Shah S, Zhu H, Kimbara M, Shashkin P, Charlot JF, Tcherpakov M, Corbett R, Tam A, Varhol R, Smailus D, Moksa M, Zhao Y, Delaney A, Qian H, Birol I, Schein J, Moore R, Holt R, Horsman DE, Connors JM, Jones S, Aparicio S, Hirst M, Gascoyne RD, Marra MA. Somatic mutations altering EZH2 (Tyr641) in follicular and diffuse large B-cell lymphomas of germinal-center origin. Nat Genet. 2010;42(2):181–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yap DB, Chu J, Berg T, Schapira M, Cheng SW, Moradian A, Morin RD, Mungall AJ, Meissner B, Boyle M, Marquez VE, Marra MA, Gascoyne RD, Humphries RK, Arrowsmith CH, Morin GB, Aparicio SA. Somatic mutations at EZH2 Y641 act dominantly through a mechanism of selectively altered PRC2 catalytic activity, to increase H3K27 trimethylation. Blood. 2011;117(8):2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cohen AS, Tuysuz B, Shen Y, Bhalla SK, Jones SJ, Gibson WT. A novel mutation in EED associated with overgrowth. J Hum Genet. 2015;60(6):339–342. [DOI] [PubMed] [Google Scholar]
- 22. Imagawa E, Higashimoto K, Sakai Y, Numakura C, Okamoto N, Matsunaga S, Ryo A, Sato Y, Sanefuji M, Ihara K, Takada Y, Nishimura G, Saitsu H, Mizuguchi T, Miyatake S, Nakashima M, Miyake N, Soejima H, Matsumoto N. Mutations in genes encoding polycomb repressive complex 2 subunits cause Weaver syndrome. Hum Mutat. 2017;38(6):637–648. [DOI] [PubMed] [Google Scholar]
- 23. Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG; Exome Aggregation Consortium . Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 2009;84(4):524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.