High blood pressure, i.e., hypertension, affects 46% of U.S. adults and is recognized as the leading risk factor for coronary artery diseases, stroke, and chronic kidney diseases in the United States and the world.1 Hypertension is directly or indirectly responsible for more cardiovascular disease (CVD) deaths than any other modifiable CVD risk factors.1 Although the pathogenesis of hypertension has been extensively investigated in animal models for hypertension and in patients with hypertension, the underlying mechanisms remain incompletely understood. Among all experimental hypertension models, salt-sensitive hypertension is perhaps the most popular and most extensively studied hypertensive model.2 In humans, salt-sensitive hypertension is generally defined as increased salt sensitivity of the blood pressure response to changes in dietary salt intake; whereas in salt-sensitive hypertensive rats, it is defined by a sustained hypertensive response to salt-loading.2 The mechanism by which salt increases the blood pressure in some, but not other, animals or humans is not well understood. In humans, genome-wide association studies (GWAS) have identified several candidate genes, such as CYP17A1, which encodes the cytochrome P450 enzyme CYP17A1; NPPA and NPPB, which encode the natriuretic peptides;3 and SH2B3 (LNK), which plays a regulatory role in immune responses and cytokine signaling, as important candidate genes for salt-sensitive hypertension.4,5 However, the estimated effects of these “candidate genes” in salt-sensitive or other forms of hypertension are so small that only 1 mmHg of systolic blood pressure variation per allele and 0.5 mmHg of diastolic blood pressure variation per allele are impacted. The efforts to uncover new mechanisms for the salt sensitivity of blood pressure are further complicated by the observation that nearly 100 genes are directly or indirectly associated with the altered salt sensitivity of the blood pressure in experimental animals.2 More importantly, there are no specific drugs or strategies to date for the prevention and treatment of salt-sensitive hypertension in humans, other than the recommendation of a low salt diet.1,2
Against this background, there is perhaps a need for a paradigm shift for the study of new mechanisms and treatments for salt-sensitive hypertension. Recently, there has been emerging evidence suggesting that the epigenetic and epigenomic mechanisms or their dysregulation may play an important role in the development of salt-sensitive hypertension.6,7 Epigenetic modifications via DNA methylation at specific sites, posttranslational histone modifications, miRNAs, or chromatin modifications may respond to a high salt diet by altering the expression of candidate genes, neural and humoral factors, and their downstream signaling pathways, which can lead to salt-sensitive hypertension. However, whether DNA methylation or (de)methylation functionally contributes to the development of salt-sensitive hypertension, and whether DNA methylation or (de)methylation should be targeted to treat salt-sensitive hypertension are virtually unknown. In this issue of Hypertension, Liu et al. reported for the first time that de novo DNA (de)methylation in the renal medulla of the kidney plays an important role in the development of hypertension in salt-sensitive rats in response to a 4% high salt diet, and that direct intrarenal administration of anti-Dnmt3a/Tet3 GapmeRs attenuated the salt-sensitive hypertension by blocking DNA methylation.8 The study by Liu et al. is considered to be novel, because the epigenetic and epigenomic mechanisms of salt-sensitive hypertension have not been well studied. In this study, a novel approach, reduced representation bisulfide sequencing (RRBS), was used to first map genome-wide DNA methylation profiles at a single-base resolution in the renal outer medulla of salt-sensitive rats in response to the normal or 4% high salt diet. DNA methylation profiling in the renal medulla was rightly justified, because the renal medulla plays a key role in the development salt-sensitive hypertension and consists primarily of non-dividing cells, in which DNA methylation changes usually occur. The authors identified broadly different DNA methylation profiles, with significantly higher DNA methylation levels, in two different colonies of salt-sensitive rats: MCWHS (Medical College of Wisconsin colony, fed a 4% NaCl diet) and CRLHS (Charles River Lab. colony, fed a 4% NaCl diet). Furthermore, the higher methylation levels of the salt-sensitive rats were significantly associated with increases in the blood pressure or increased blood pressure salt-sensitivity. In further experiments, the authors determined whether the robust changes in DNA methylation in the renal medulla of the salt-sensitive rats in response to a high salt diet were due to de novo methylation or de-methylation by measuring the expression of the Dnmt3 (Dnmt3a and Dnmt3b) and Tet (Tet1, Tet2, and Tet3) families of enzymes in the renal medulla of the salt-sensitive rats. Dnmt3 and Tet are two key enzymes that catalyze de novo DNA methylation and de-methylation, respectively. 9,10 When fed a 0.4% NaCl diet, no difference in the Dnmt3a expression was detected between the salt-sensitive rats and salt-resistant SS.13BN rats, but when fed a 4% high salt diet, Dnmt3a expression in the renal medulla increased significantly in the salt-sensitive rats, compared to the salt-resistant SS.13BN rats.8 However, the authors did not detect significant changes in Tet expression in the renal medulla of the salt-sensitive rats.8 This suggests that Dnmt3a, but not Tet, is the enzyme that most likely contributes to de novo DNA methylation in the renal medulla of salt-sensitive rats during the development of salt-sensitive hypertension. Another important finding from the report by Liu et al. is translationally relevant. The authors demonstrated that the intramedullary administration of anti-Dnmt3a/Tet3 GammeR’s to block de novo DNA methylation in the medulla significantly attenuated the development of salt-sensitive hypertension in the salt-sensitive rats, by altering the DNA methylation profiles and blocking high salt-induced gene expression in the outer renal medulla. However, there is a significant limitation of the experimental approach of simultaneously knocking-down Dnmt3a and Tet3 using the anti-Dnmt3a/Tet3 GammeR’s approach. The Dnmt3 enzyme increases the DNA methylation levels at some genomic loci, while the Tet enzyme decreases it.9,10 Moreover, the authors found no difference in the Tet expression in the salt-sensitive rats in response to a high salt diet during the development of salt-sensitive hypertension. The authors argued that the anti-Dnmt3a/Tet3 GammeR’s could have attenuated the effects of both Dnmt3a and Tet3 on the DNA methylation of certain gene loci. Indeed, this approach significantly knocked-down Dnmt3a by 38% in the renal outer medulla of the SS rats and attenuated hypertension in the salt-sensitive rats. However, a 38% knockdown of Dnmt3a by the anti-Dnmt3a/Tet3 GammeR’s in the renal outer medulla of the salt-sensitive rats is considered to be relatively low. Nevertheless, the RNA-seq and RRBS analyses of the gene expression in the renal medulla of the salt-sensitive rats found that 76% of the 1712 genes that were differentially expressed in these rats in response to the high salt diet were altered by the anti-Dnmt3a/Tet3 GammeR’s. Three hundred and twenty-eight of the genes inhibited by the anti-Dnmt3a/Tet3 GammeR’s were genes that are associated with cellular metabolism, inflammation, and the extracellular matrix and cardiovascular, metabolic, and inflammatory kidney diseases. 8
Therefore, what are the potential implications for future directions or strategies for studying the mechanisms and treatments for salt-sensitive hypertension? Most of the previous and current studies on salt-sensitive hypertension have primarily focused on a) whether a cluster of genes in a particular chromosome, b) whether a particular neural or hormonal peptide or its receptor, c) whether the deficiency of nitric oxide, or d) whether increased oxidative stress are associated with salt-sensitive hypertension in response to a 4%−8% high salt diet.5–8 An additional challenge is that there is no specific drug or strategy to prevent and treat salt-sensitive hypertension in humans, with the exception of salt restriction. Salt-sensitive hypertension is a multifactorial and very complex disorder. No single candidate gene, humoral factor, cellular mechanism, or signaling pathway can adequately explain, or be targeted in salt-sensitive hypertension. The key challenges in this particular research field remain to be how candidate genes interact with a high salt level to cause salt-sensitive hypertension, and how we can effectively prevent and treat salt-sensitive hypertension. Thus, uncovering novel epigenetic and epigenomic mechanism(s) and molecularly and pharmacologically targeting these mechanisms may represent new directions in the research field of salt-sensitive hypertension. Despite its limitations, the study by Liu et al. should help shift the paradigm of current DNA methylation research, specifically in salt-sensitive hypertension, and broadly in other cardiovascular, hypertensive, and kidney diseases from association analyses to functional, mechanistic, and therapeutic studies in the future.
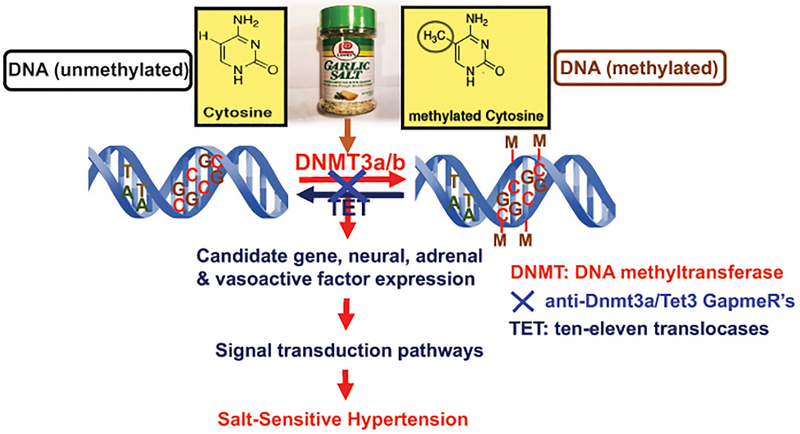
A schematic hypothesis for the pathophysiological role of de novo DNA methylation (or demethylation) in the development of salt-sensitive hypertension. In salt-sensitive hypertension rats, treatment with a 4% or 8% high salt diet induces the DNA methylation (or demethylation) of the candidate genes for salt-sensitive hypertension and genes encoding neural peptides, adrenal hormones, and vasoactive peptides through the actions of DNA methyltransferase 3 (Dnmt3) and ten-eleven translocases (Tet). DNA methylation (or demethylation) is expected to alter the expression and activities of these genes and vasoactive peptides and their downstream signal transduction, which leads to the development of salt-sensitive hypertension. Conversely, molecular or pharmacological inhibition of Dnmt3a/Tet3 with anti-Dnmt3a/Tet3 GapmeRs may theoretically prevent the changes in the DNA methylation profiles of these candidate genes or vasoactive peptides. This in turn blocks the up- or downregulation of their gene expression, biological activities, and downstream signaling pathways, and attenuates salt-sensitive hypertension and target tissue injury.
Sources of Funding
This work was supported in part by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), 2R01DK067299–10A1 and 2R01DK102429–03A1, and the National Heart, Lung, and Blood Institute (NHLBI), 1R56HL130988–01, to Dr. Jia L. Zhuo.
Footnotes
Disclosures
None.
References
- (1).Muntner P, Carey RM, Gidding S, Jones DW, Taler SJ, Wright JT, Jr., Whelton PK. Potential US Population Impact of the 2017 ACC/AHA High Blood Pressure Guideline. Circulation 2018; 137(2):109–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Elijovich F, Weinberger MH, Anderson AM, Appel LJ, Bursztyn M, Cook NR, et al. Salt sensitivity of blood pressure: a scientific statement from the American Heart Association. Hypertension. 2016, 68:e7–e46. [DOI] [PubMed] [Google Scholar]
- (3).John SW, Krege JH, Oliver PM, Hagaman JR, Hodgin JB, Pang SC, Flynn TG, Smithies O. Genetic decreases in atrial natriuretic peptide and salt-sensitive hypertension. Science 1995; 267(5198):679–681. [DOI] [PubMed] [Google Scholar]
- (4).Levy D, Ehret GB, Rice K, Verwoert GC, Launer LJ, Dehghan A, et al. Genome-wide association study of blood pressure and hypertension. Nat Genet 2009; 41(6):677–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rudemiller NP, Lund H, Priestley JR, Endres BT, Prokop JW, Jacob HJ, Geurts AM, Cohen EP, Mattson DL. Mutation of SH2B3 (LNK), a genome-wide association study candidate for hypertension, attenuates Dahl salt-sensitive hypertension via inflammatory modulation. Hypertension 2015; 65(5):1111–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Mu S, Shimosawa T, Ogura S, Wang H, Uetake Y, Kawakami-Mori F, Marumo T, Yatomi Y, Geller DS, Tanaka H, Fujita T. Epigenetic modulation of the renal beta-adrenergic-WNK4 pathway in salt-sensitive hypertension. Nat Med 2011; 17(5):573–580. [DOI] [PubMed] [Google Scholar]
- (7).Liu Y, Liu P, Yang C, Cowley AW, Jr., Liang M. Base-resolution maps of 5-methylcytosine and 5-hydroxymethylcytosine in Dahl S rats: effect of salt and genomic sequence. Hypertension 2014; 63(4):827–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Liu P, Liu YPX, Li Y, Usa K, Nie J, Liang M. DNA de novo (de)methylation in the kidney contributes to salt-induced hypertension. Hypertension. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 1999; 99(3):247–257. [DOI] [PubMed] [Google Scholar]
- (10).Wu X, Zhang Y. TET-mediated active DNA demethylation: mechanism, function and beyond. Nat Rev Genet 2017; 18(9):517–534. [DOI] [PubMed] [Google Scholar]