Abstract
The composition, abundance, and diversity of midgut bacteria in mosquitoes can influence pathogen transmission. We used 16S rRNA microbiome profiling to survey midgut microbial diversity in pooled samples of laboratory colonized dengue-refractory, Cali-MIB, and dengue-susceptible, Cali-S Aedes aegypti (Linnaeus). The 16S rRNA sequences from the sugar-fed midguts of adult females clustered to 63 amplicon sequence variants (ASVs), primarily from Proteobacteria, Firmicutes, Flavobacteria, and Actinobacteria. An average of five ASVs dominated the midguts, and most ASVs were present in both Cali-MIB and Cali-S midguts. No differences in abundance were noted at any phylogenetic level (Phylum, Class, Order, Family, Genus) by analysis of composition of microbiome (w = 0). No community diversity metrics were significantly different between refractory and susceptible mosquitoes. These data suggest that phenotypic differences in the susceptibility to dengue virus between Cali-MIB and Cali-S are not likely due to major differences in midgut bacterial communities.
Keywords: vector competence, microbiome, midgut infection barrier
Tripartite interactions between host, pathogens, and vectors are extremely important in determining the ultimate outcome of pathogen transmission and host disease. A variety of studies has demonstrated the effects of genetic variation, gene expression, and the environment on vector susceptibility to pathogens at both individual and population levels (Palmer et al. 2018). Many of these intriguing relationships have been studied in mosquito systems due to their importance in the field of human health, as vectors for pathogens that cause some of the world’s most prevalent and widespread diseases, such as malaria, dengue, chikungunya, Zika, filariasis, and West Nile fever.
Symbiotic relationships between individuals and their microbiomes have long been known. These relationships have spurred interest into microbiome research as a possible primary and secondary driver of disease and pathogen susceptibility. These interactions manifest in individuals in one of two possible manners: obligate systems, typically older, more evolutionary stable systems where bacterial presence is fundamental for systemic function, or nonobligate systems, typically newer, more diverse and flexible, where bacterial presence is determined by extrinsic factors, such as the environment, or intrinsic variable factors, such as familial lineage and mating partners. As with most symbiotic relationships, these close interactions can increase (nutritionally supplement), impede (increase pathogen susceptibility), or remain neutral with regards to their hosts through resource competition, production of secondary metabolites, and immune priming (Dennison et al. 2014).
Mosquito–microbiome interactions may change a mosquito’s susceptibility to pathogen invasion; increasing bacterial diversity and abundance tends to decrease susceptibility to pathogens (Dennison et al. 2014). Bacterial symbionts that are new in mosquitoes seem to increase resistance in Ae. aegypti to dengue virus (DENv). While the exact mechanisms are not known, this could be accomplished by promoting the defensive abilities of the mosquito via increased antimicrobial peptides (Proteus sp.) and reactive oxygen species production (Wolbachia sp.) (Pan et al. 2012, Ramirez et al. 2012, Rancès et al. 2012). These Ae. aegypti – DENv studies have, thus, far been conducted solely on long established laboratory-reared colonies of susceptible mosquitoes. In contrast, we aimed to investigate these symbiotic relationships in naturally susceptible (Cali-S) and refractory (via a midgut infection barrier [MIB], Cali-MIB) mosquitoes to determine the relationship between mosquito microbiota and vector competence. These two strains originated from field-collected mosquitoes in the city of Cali, Colombia, where ~30% of field-collected mosquitoes are refractory to DENv-2 (Ocampo and Wesson 2004, Caicedo et al. 2013). Subsequent studies determined that the DENv-susceptible or -refractory phenotype was determined in the midgut of these mosquitoes within 48 h of blood being ingested by the mosquitoes (Ocampo et al. 2013, Serrato et al. 2017, Caicedo et al. 2018). Although microbiota may exist in a variety of mosquito tissues and organs (salivary glands, ovaries, Malpighian tubules, and midgut), we examined only the gut microbiome, which serves as the first barrier to virus infection, to determine if changes in abundance or diversity of the microbiota contributed to the differential susceptibility to DENv between the Cali-MIB and Cali-S phenotypes.
Metagenomics projects typically investigate the diversity and presence of a variety of symbionts, including (but not limited to) archaea, bacteria, and various eukaryotic species. Due to their previously identified effects in mosquito–pathogen interactions (Gonzalez-Ceron et al. 2003, Wang et al. 2009, Gusmão et al. 2010, Cirimotich et al. 2011, Chauhan et al. 2012, Pan et al. 2012, Ramirez et al. 2012), bacteria were chosen as the focus of this study. Eukaryotes such as yeast and fungi have been isolated from mosquitoes, but their effects remain unknown, and they were excluded from analysis. Nonculture-dependent techniques were used to gain bacterial abundance and diversity values without technique- or medium-related biases. Sequencing was completed on the hyper-diverse 16S V4-V5 region using the Illumina platform for its superior read depth (at the cost of read length), to tease apart rare operational taxonomic units (OTUs). Midgut bacterial diversity and composition were compared within and between Cali-S and Cali-MIB mosquitoes to determine if differences in microbiota composition could explain differences in susceptibility to DENv.
Materials and Methods
Ethics Statement
All protocols were approved by the Centro Internacional de Entrenamiento e Investigaciones Médicas (CIDEIM) institutional review committee for research in animals (1021), governed by law 84 (1989) and resolution 8430 (1993) of the National Ministry of Agriculture in Colombia.
Mosquito Rearing and Maintenance
Cali-S and Cali-MIB strains of Ae. aegypti were selected from field-collected larvae and pupae from artificial larval habitats in five locations in and around the city of Cali, Colombia (Caicedo et al. 2013). Strains were selected after exposure to DENv-2 using an isofamily selection technique, where only eggs from verified susceptible or refractory females were hatched (Ocampo and Wesson 2004, Caicedo et al. 2013, Ocampo et al. 2013). F<30 mosquitoes were maintained under standard laboratory conditions: 28 ± 2ºC, 70% relative humidity, and a 12:12 (L:D) h cycle. Larvae were maintained at a density of 300 larvae/2 liter of distilled water in plastic pans and were fed daily with 2 ml of a stock solution (8 g/400 ml) of beef liver (DIFCO, Becton, Dickinson and Company, Franklin Lakes, NJ). Adults were fed with 10% sugar solution ad libitum.
Sample Preparation
Four days after emergence, sugar-fed Cali-S and Cali-MIB females were surface sterilized individually through a series of four washes, each performed in a new 1.5-ml microcentrifuge tube. First, each female was placed in a 70% ethanol bath for 5 min, followed by 3 min of vortexing, then a 1-min nuclease-free water bath, followed by 1 min of vortexing, a second 1-min 70% ethanol bath, followed by 1 min of vortexing, and a last 1-min nuclease-free water bath, followed by 1 min of vortexing. Midguts were dissected in 30 µl of sterilized phosphate buffer solution (PBS). Midguts (n = 80) were pooled into groups of five each (eight susceptible pools, S1–8, and eight refractory pools, R1–8) (n = 16). A negative control of 10 µl of PBS used for dissections, and a positive control of 10 µl of a pure Escherichia coli culture were also created. DNA was extracted from each sample using Qiagen’s DNeasy Blood and Tissue Kit (Hilden, Germany) following the manufacturer’s instructions for the extraction of bacteria. DNA concentration was evaluated using a Qubit® 2.0 Fluorometer (ThermoFisher Scientific, Waltham, MA). A portion of the DNA obtained from each sample was used in a polymerase chain reaction (PCR) with two conserved 16S primers (S-D-Bact-0564-a-S-15: 5′-AYT GGG YDT AAA GNG-3′, and S-D-Bact-0785-b-A-18: 5′-TAC NVG GGT ATC TAA TCC-3′) (Klindworth et al. 2013) to verify that bacterial DNA had been extracted from each sample pool. Each PCR contained: 6 µl of nuclease-free water, 10 µl of 5 Prime Hot Master Mix (5 Prime, Gaithersburg, MD), 2 µl of 25 µM forward primer, 2 µl of 25 µM reverse primer, and 5 µl of the template DNA. PCRs were performed in duplicate on a PTC-200 Peltier Thermocycler (Bio-Rad, Hercules, CA) with the following program: hot start of 94°C for 3 min, 35 cycles of 94°C for 45 s, 45°C for 60 s, and 72°C for 90 s, with a final extension of 72°C for 10 min. Any sample with a concentration higher than 20 ng/µl was considered acceptable for sequencing.
Amplicon Library Preparation and Sequencing
All DNA was sequenced at MR DNA (Shallowater, TX) using an Illumina MiSeq platform (San Diego, CA) to sequence the bacterial 16S rRNA gene. Briefly, the V4-V5 hyper variable region was targeted via PCR (see primers in Table 1), adding treatment-specific barcodes to each sample. The PCR was performed using the HotStartTaq Plus Master Mix Kit (Qiagen, Hilden, Germany) under the following conditions: 94°C for 3 min, 28 cycles of 94°C for 30 s, 53°C for 40 s, and 72°C for 1 min, after which a final elongation step at 72°C for 5 min was performed. After amplification, PCR products were checked for relative intensity on a 2% agarose gel. Samples were then purified using calibrated Agencourt AMPure XP beads (Brea, CA). A Nextera DNA Sample Preparation Kit was then used (Illumina, CA) to prepare the samples. Paired end (2 × 300 bp) sequencing was performed on a MiSeq platform by loading pooled samples (at 12 PM) onto a 600 cycle v3 reagent cartridge (Illumina, CA).
Table 1.
Primer sequences used for 16S amplification
| Primer Name | Sequence |
|---|---|
| 515F | GTGCCAGCMGCCGCGGTAA |
| 926R | CCGYCAATTYMTTTRAGTTT |
| NC | GAGATCAG |
| PC | GAGAGTGT |
| R1 | GAGATCTC |
| R2 | GAGATGAC |
| R3 | GAGATGTG |
| R4 | GAGTACAG |
| R5 | GAGTACTC |
| R6 | GAGTAGAC |
| R7 | GAGTAGTG |
| R8 | GAGTCACT |
| S1 | GAGTCAGA |
| S2 | GAGTCTCA |
| S3 | GAGTCTGT |
| S4 | GAGTGACA |
| S5 | GAGTGAGT |
| S6 | GAGTGTCT |
| S7 | GAGTGTGA |
| S8 | GAGTTCAC |
Amplicon Data Analysis
(DA DA2) (Callahan et al. 2016) and Quantitative Insights Into Microbial Ecology 2 (QIIME2) (release 2018.8) (Caporaso et al. 2010) were used to analyze all resultant FASTQ files obtained from MR DNA . First, using QIIME2, FASTQ files were demultiplexed (cutadapt demux-paired), and the adapters were trimmed off (cutadapt trim-paired) (Martin 2011). Next, DADA2 (v1.8.0) was used to create read quality profiles, and reads were truncated to avoid low-quality scores (>240 bp for forward, >200 bp for reverse reads) (maxN = 0, truncQ = 2, maxEE = 2). Error rates were then learned (err) and dereplication was completed (derepFastq). The dada core sample inference algorithm was then applied to the filtered and dereplicated forward and reverse data. Reads were merged (mergePairs) with a minimum overlap of 12 bp. An amplicon sequence variant (ASV) table was made (makeSequenceTable) for all 16 (8 refractory and 8 susceptible) samples, and chimeric samples were removed (removeBimeraDenovo). Silva (v132) (Callahan 2018) was used as the taxonomic training set (via assignTaxonomy and addSpecies). The feature table, representative fasta sequences, taxonomy data, and sample metadata were then imported into QIIME2 (tools import).
In order to complete downstream diversity and composition analyses, rooted and unrooted phylogenetic trees were created (phylogeny align-to-tree-mafft-fasttree) in QIIME2. Samples with a sampling depth of <1,000 sequences per sample were removed to retain an acceptable sequence coverage. ASVs with an abundance of <0.01% were also removed. Alpha rarefraction (McDonald et al. 2012) and diversity (Faith’s phylogenetic diversity, Pielou’s evenness, Shannon and species richness) (Kruskal and Wallis 1952, McKinney 2010), as well as beta diversity (Jaccard, unweighted unifrac, weighted unifrrac, and Bray-Curtis) (Anderson 2001) were calculated. Taxa were grouped by abundance (McKinney 2010, McDonald et al. 2012), and analysis of composition of microbiome (ANCOM) was used to test for differential abundance across multiple taxonomic levels (Phylum, Class, Order, Family, and Genus) (Mandal et al. 2015). Linear discriminant analysis effect size (LEfSe) was used to detect potential phenotype-specific bacterial markers (Segata et al. 2011).
Results
Analyzing Negative and Positive Controls
As expected, our negative PBS control did result in some bacterial reads (Salter et al. 2014); however, the abundance of these reads was substantially lower than those obtained from any of our samples or our positive control. To control for contamination, the read counts for the negative control were subtracted from all sample counts (which resulted in the elimination of negative control-specific ASVs). Furthermore, the composition of bacteria in the negative control was different than the ASVs noted in any of our experimental samples or in our positive control. Our positive control primarily showed one highly abundant Shigella ASV. As 16S sequencing is often unable to differentiate between Shigella and E. coli species (Khot and Fisher 2013), we completed an Escherichia- and Shigella-specific PCR on the positive control DNA, which verified that our positive control was indeed E. coli bacterial DNA.
Bacterial Abundance and Community Composition
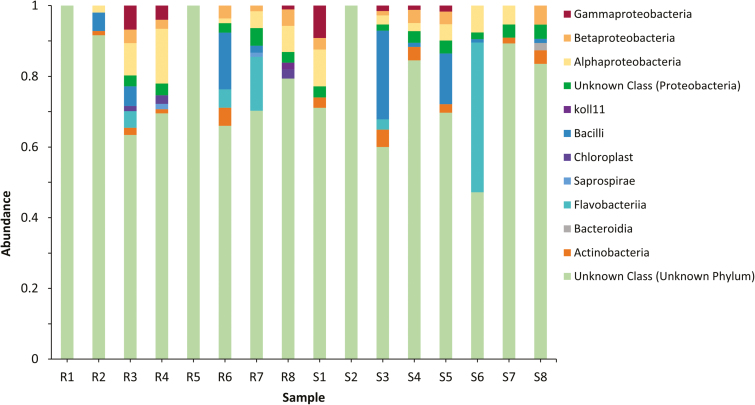
After quality filtering, samples R1, R2, R5, and S2 were removed due to low depth of sampling (≤1,000 sequences per sample), whereas the remaining samples were rarefied to a sampling depth of 1,333 sequences. Chimeric filtering removed 13% of sequences. The remaining sequences clustered into 63 ASVs from 6 phyla, 9 classes, 13 orders, 19 families, and 27 genera (some classified further into sub-species), although only 21 ASVs had an overall abundance >1%. Most of the sequences were from Proteobacteria (40.5%), comprised of Alphaproteobacteria (22%), Betaproteobacteria (10%), and Gammaproteobacteria (8.5%), then Flavobacteria (23%), Bacilli (21%), and Actinobacteria (8.5%) (Fig. 1). As 16S sequencing is unable to differentiate between Swaminathania and Asaia, and Asaia sp. are commonly associated with insect guts, it is likely that the Swaminathania ASVs observed here are actually Asaia sp. (Deutscher et al. 2018).
Fig. 1.
Bacterial taxa organized by order, expressed per sample. Bars on the left are from Cali-MIB samples (R1–R8), whereas bars on the right are from Cali-S samples (S1–S8).
An average of five ASVs dominated both the Cali-S and Cali-MIB midguts (≥5% total abundance). Actinobacteria and Bacilli (Bacillales) dominated the susceptible midguts, whereas Alphaproteobacteria and Flavobacteriia made up most bacteria in refractory midguts. The majority of ASVs were present in both Cali-S and Cali-MIB midguts. Only a few low-abundance OTUs were exclusively found in either phenotype.
The ANCOM tests at all levels of analysis (Phylum, Class, Order, Family, and Genus) did not yield any significantly different results (w = 0). The linear discriminant analysis effect size (LEfSe) did not identify any taxonomic biomarkers specific to the Cali-MIB or Cali-S phenotype (P ≥ 0.00005) at different hierarchical levels.
Alpha and Beta Diversity Metrics
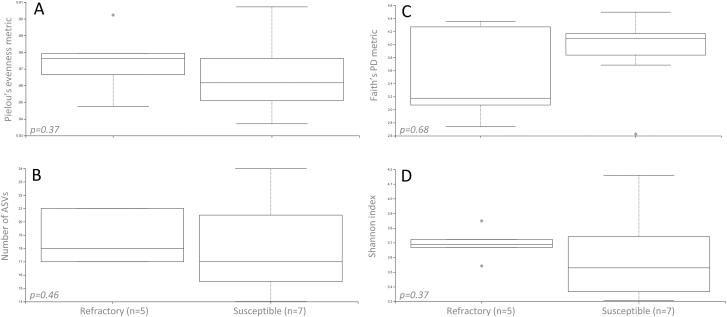
In all measured alpha diversity metrics (Faith’s phylogenetic diversity, species richness, Pielou’s evenness, and Shannon index), no significant differences were found between Cali-MIB and Cali-S mosquitoes (P-value ≥ 0.05, Fig. 2). All alpha rarefraction plots reached saturation. Similarly, no significant differences were observed between Cali-S and Cali-MIB mosquitoes in terms of beta diversity metrics (P-value ≥ 0.05, Table 2).
Fig. 2.
Alpha diversity boxplots. (A) Pielou’s evenness, (B) observed number of ASVs, (C) Faith’s phylogenetic diversity, and (D) Shannon.
Table 2.
Beta diversity statistical outputs, where the composition of bacteria between Cali-S and Cali-MIB midguts was compared using four beta diversity metrics
| Sample Size | Permutations | Pseudo-F | P-value | q-value | |
|---|---|---|---|---|---|
| Bray-Curtis | 12 | 999 | 0.627477 | 0.881 | 0.881 |
| Jaccard | 12 | 999 | 0.714433 | 0.931 | 0.931 |
| Unweighted | 12 | 999 | 3.01566 | 0.098 | 0.098 |
| Weighted | 12 | 999 | 1.08424 | 0.41 | 0.41 |
Discussion
Overall, we found a low number of dominant bacterial species present in each midgut (average of five), a trend that has been observed in other midgut composition studies (Yun et al. 2014, Ngo et al. 2016, Muturi et al. 2017). No diversity differences were found to exist between Cali-S and Cali-MIB mosquitoes using any of the metrics examined here, nor were there any significant differences between the abundance of bacteria in Cali-S and Cali-MIB midguts, identified both by LEfSe, and via ANCOM tests completed in QIIME2. It is possible that these differences are biologically relevant yet masked by the lack of difference in overall interphenotype composition and diversity.
In Cali-S, bacteria from the phylum Firmicutes and Actinobacteria were high in abundance. In other studies, their presence was noted in mosquitoes treated with low doses of Bacillus thuringiensis (Demisse 2013), and species from Firmicutes have been shown to inhibit gut algal growth (Engel and Moran 2017). Firmicutes bacteria are quite common Gram-positive anaerobic insect gut symbionts, although their role in the midgut is largely unknown. Bacteria from the order Burkholderiales were also abundant in Cali-S midguts. These are usually observed in hemipterans, such as kissing bugs (Triatominae species) and bean bugs (Riptortus spp.), and contribute to the detoxification of insecticides, enabling these insects to resist or tolerate insecticides (Kikuchi et al. 2012). Two transcriptome studies identified multiple genes tied to insecticide resistance that were highly expressed in Cali-S but not in Cali-MIB mosquitoes, an observation that may be indirectly linked to the presence of Burkholderiales species in Cali-S midguts. Last, Actinobacteria were highly abundant in the Cali-S midgut. Actinobacteria have been characterized in beetles and hemipterans as important nutrient processors, aiding in the digestion of sugar and blood meals (Zucchi et al. 2012). Actinobacteria are part of the core microbiota of Ae. aegypti (David et al. 2016).
In Cali-MIB mosquitoes, abundance differences were observed in Flavobacteriaceae. Flavobacteria are extremely ubiquitous Gram-negative mosquito gut symbionts, most likely due to their wide range of metabolic activities (Terenius et al. 2012, Yadav et al. 2018). They have no reported links to vector competence in mosquitoes but are a common larval food source (Chen et al. 2014). They are commonly found in laboratory-reared mosquito guts and can be transferred by transstadial means (Boissière et al. 2012, Chen et al. 2015). Due to their high abundance across a variety of mosquito genera, a variety of Flavobacteria have been proposed as a biological control technique, manipulating them to express and deliver B. thuringiensis proteins (Chen et al. 2014, 2015).
To complete functional tests, studies have traditionally used retraction experiments to render the gut aseptic and reintroduce a single species of bacteria (Ramirez et al. 2012, Jupatanakul et al. 2014, Koskella et al. 2017). The introduction of individual species identified through culture-dependant methods did not significantly change the vector competence of Cali-MIB or Cali-S mosquitoes (Molina-Henao and Ocampo, unpublished). Although this gives us useful information about the putative role of a single bacterial species, it negates the large interplay between and among all members of the midgut microbiota (Koskella et al. 2017). We could use a similar technique to functionally assess abundance trends we noted but understanding the indirect effects of individual bacterial species and the rest of the microbiome would be theoretically challenging. Future studies should aim for a more inclusive metagenomic technique, giving a more holistic view of all microbiota (including fungi, protozoans, and viruses), rather than solely bacteria.
Other studies have shown that mosquitoes from different geographical localities harbor different microbiomes (Ngo et al. 2016, Muturi et al. 2017). When mosquitoes from different localities were raised in a laboratory environment, there were no differences in their gut microbiomes, suggesting environmental factors as the primary driver in mosquito gut microbiome differences (Dickson et al. 2018). Multiple studies in mosquitoes and other insects have, however, shown that microbiota contribute to vector competence, even within laboratory settings (Joyce et al. 2011, Ramirez et al. 2012, Jupatanakul et al. 2014, Muturi et al. 2017). This dichotomy could persist due to the intrinsic differences in physiology between different susceptible and refractory strains.
Although bacterial species associated with changes in vector competence for viruses (Proteus sp., Chromobacterium sp., Pseudomonas rhodesiae, Enterobacter ludwigii, and Vagococcus salmoninarium) (Dong et al. 2011, Joyce et al. 2011, Apte-Deshpande et al. 2012, Boissière et al. 2012, Ramirez et al. 2012, Jupatanakul et al. 2014) were not detected in any of the midgut samples in this study, some bacterial ASVs from the same family (Enterobacteriaceae) were found. This suggests that the Cali-MIB refractory phenotype does not stem from previously characterized microbe-dependant factors.
As observed in Fig. 1, intraphenotype variability in midgut microbiota was larger than the variability observed between phenotypes. Despite this intraphenotype variability, the diversity captured within this study seemed to be sufficient, as all diversity index rarefaction curves were at or near saturation. The large disparity between samples observed in this study may have been due, in part, to the make-up of the refractory samples. While the selection process allows for the creation of 100% susceptible lines, only 50% of the Cali-MIB line is 100% refractory to dengue. This means that half of the refractory samples in this study are most likely susceptible in nature.
All the compositional differences examined in this study are based on sugar-fed mosquitoes. There seems to be evidence suggesting that the microbiota composition changes upon blood feeding, as there is an increase in bacterial abundance, which could result in cases of competition among bacterial species (Gaio et al. 2011, Jupatanakul et al. 2014, Coon et al. 2016). As a blood meal provides a different, more proteinaceous nutritional profile, midgut bacterial abundances post-blood feeding would likely be much different than those observed here. DENv moves from the bolus to enter midgut epithelial cells as early as 8 h after ingestion (Junjhon et al. 2014), which may be too short a period for significant proliferation of bacteria to affect DENv viability directly. As a result, the holo-immune concept has been proposed (Dheilly 2014), whereby bacteria exert indirect effects on viral replication and dissemination mediated through host innate immunity, secondary metabolite production, resource competition, or via the secretion of miRNAs. Further research investigating the gut microbiome through blood and virus feeding, as well as basal metabolomics studies to assess the characteristics of the gut environments, would augment our current understanding of differences in Cali-S and Cali-MIB midguts.
Microbiome studies are important tools that help us identify communities and highlight differences between microbiomes and the environments that support microbial proliferation. These studies have helped us identify potential biological control agents, novel RNAi delivery systems, and prospective antiviral compounds. The results from this study help augment our current understanding of mosquito midgut microbiomes, and aid in curating microbiome data from susceptible and refractory Ae. aegypti phenotypes with the aim of identifying factors that might shift the balance toward mosquitoes that do not transmit arboviruses to humans.
Data Availability
The raw sequence data have been uploaded onto NCBI’s SRA database (SRA Accession: SRP149364, Bio Project ID: PRJNA473810).
Acknowledgments
The authors would like to thank the entire Vector Biology and Control team at CIDEIM for training and technical support, as well as Iman Baharmand (SFU) for technical assistance. This research was funded in part by the Departamento Administrativo de Ciencia, Tecnología e Innovación (http://www.colciencias.gov.co/), Contract 2229-519-28645 to CB, and a Discovery Grant to CL (RGPIN261940) from the Natural Sciences and Engineering Research Council of Canada (http://www.nserc-crsng.gc.ca/).
References Cited
- Anderson M. J. 2001. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26: 32–46. [Google Scholar]
- Apte-Deshpande A., Paingankar M., Gokhale M. D., and Deobagkar D. N.. 2012. Serratia odorifera a midgut inhabitant of Aedes aegypti mosquito enhances its susceptibility to dengue-2 virus. PLoS One. 7: e40401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissière A., Tchioffo M. T., Bachar D., Abate L., Marie A., Nsango S. E., Shahbazkia H. R., Awono-Ambene P. H., Levashina E. A., Christen R.,. et al. 2012. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 8: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caicedo P., Barón O., Pérez M., Alexander N., Lowenberger C., and Ocampo C.. 2013. Selection of Aedes aegypti (Diptera: Culicidae) strains that are susceptible or refractory to Dengue-2 virus. Can. Entomol. 145: 273–282. [Google Scholar]
- Caicedo P. A., Serrato I. M., Sim S., Dimopoulos G., Coatsworth H., Lowenberger C., and Ocampo C. B.. 2018. Immune response-related genes associated to blocking midgut dengue virus infection in Aedes aegypti strains that differ in susceptibility. Insect Sci. . [DOI] [PubMed] [Google Scholar]
- Callahan B. 2018. Silva taxonomic training data formatted for DADA2. (Silva version 132) [Data set]. Zenodo . [Google Scholar]
- Callahan B. J., Mcmurdie P. J., Rosen M. J., Han A. W., Johnson A. J. A., and Holmes S. P.. 2016. DADA2 : high-resolution sample inference from Illumina amplicon data. Nat. Methods 13: 581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso J. G., J., Kuczynski J., Stombaugh K., Bittinger F. D., Bushman E. K., Costello N., Fierer A. G., Peña J. K., Goodrich J. I., Gordon, et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan C., S. K. Behura B. Debruyn D. D. Lovin B. W. Harker C. Gomez-Machorro A. Mori J. Romero-Severson, and Severson D. W.. 2012. Comparative expression profiles of midgut genes in dengue virus refractory and susceptible Aedes aegypti across critical period for virus infection. Plos One. 7: e47350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., M. G. Kaufman M. L. Korir, and Walker E. D.. 2014. Ingestibility, digestibility, and engineered biological control potential of Flavobacterium hibernum, isolated from larval mosquito habitats. Appl. Environ. Microbiol. 80: 1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., M. Bagdasarian, and Walker E. D.. 2015. Elizabethkingia anophelis: molecular manipulation and interactions with mosquito hosts. Appl. Environ. Microbiol. 81: 2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirimotich C. M., J. L. Ramirez, and Dimopoulos G.. 2011. Native microbiota shape insect vector competence for human pathogens. Cell Host Microbe. 10: 307–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon K. L., Brown M. R., and Strand M. R.. 2016. Gut bacteria differentially affect egg production in the anautogenous mosquito Aedes aegypti and facultatively autogenous mosquito Aedes atropalpus (Diptera: Culicidae). Parasit. Vectors. 9: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M. R., L. M. Santos A. C. Vicente, and Maciel-de-Freitas R.. 2016. Effects of environment, dietary regime and ageing on the dengue vector microbiota: evidence of a core microbiota throughout Aedes aegypti lifespan. Mem. Inst. Oswaldo Cruz. 111: 577–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demisse D. D. 2013. Influence of nutrients and integrated mosquito management tactics on mosquitoes and their habitat microbiomes. Ph.D. dissertation. UC Riverside, Riverside. [Google Scholar]
- Dennison N. J., N. Jupatanakul, and Dimopoulos G.. 2014. The mosquito microbiota influences vector competence for human pathogens. Curr. Opin. Insect Sci. 3: 6–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutscher A. T., Burke C. M., Darling A. E., Riegler M., Reynolds O. L., and Chapman T. A.. 2018. Near full-length 16S rRNA gene next-generation sequencing revealed Asaia as a common midgut bacterium of wild and domesticated Queensland fruit fly larvae. Microbiome. 6: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheilly N. M. 2014. Holobiont – holobiont interactions : redefining host – parasite interactions. PLoS Pathog. 10: e1004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson L. B., A., Ghozlane S., Volant C., Bouchier L., Ma A., Vega-Rúa I., Dusfour D., Jiolle C., Paupy M. N., Mayanja, et al. 2018. Diverse laboratory colonies of Aedes aegypti harbor the same adult midgut bacterial microbiome. Parasit. Vectors. 11: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Das S., Cirimotich C., Souza-Neto J. A., McLean K. J., and Dimopoulos G.. 2011. Engineered Anopheles immunity to Plasmodium infection. PLoS Pathog. 10: e1004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel P., and Moran N. A.. 2017. The gut microbiota of insects – diversity in structure and function. FEMS Microbiol Rev. 37: 699–735. [DOI] [PubMed] [Google Scholar]
- Gaio A., Gusmão D., Santos A., Berbert-Molina M., Pimenta P., and Lemos F.. 2011. Contribution of midgut bacteria to blood digestion and egg production in Aedes aegypti (diptera: culicidae) (L.). Parasit. Vectors. 4: 105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Ceron L., F. Santillan M. H. Rodriguez D. Mendez, and Hernandez-Avila J. E.. 2003. Bacteria in midguts of field-collected Anopheles albimanus block Plasmodium vivax sporogonic development. J. Med. Entomol. 40: 371–374. [DOI] [PubMed] [Google Scholar]
- Gusmão D. S., A. V. Santos D. C. Marini M. Bacci M. A. Jr Berbert-Molina, and Lemos F. J.. 2010. Culture-dependent and culture-independent characterization of microorganisms associated with Aedes aegypti (Diptera: Culicidae) (L.) and dynamics of bacterial colonization in the midgut. Acta Trop. 115: 275–281. [DOI] [PubMed] [Google Scholar]
- Joyce J. D., J. R. Nogueira A. A. Bales K. E. Pittman, and Anderson J. R.. 2011. Interactions between La Crosse virus and bacteria isolated from the digestive tract of Aedes albopictus ( Diptera : Culicidae ). Interactions between La Crosse virus and bacteria isolated from the digestive tract of Aedes albopictus (Diptera : Culicidae ). J. Med. Entomol. 48: 389–394. [DOI] [PubMed] [Google Scholar]
- Junjhon J., J. G. Pennington T. J. Edwards R. Perera J. Lanman, and Kuhn R. J.. 2014. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 88: 4687–4697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupatanakul N., S. Sim, and Dimopoulos G.. 2014. The insect microbiome modulates vector competence for arboviruses. Viruses. 6: 4294–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khot P. D. and Fisher M. A.. 2013. Novel approach for differentiating Shigella species and Escherichia coli by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51: 3711–3716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi Y., M. Hayatsu T. Hosokawa A. Nagayama K. Tago, and Fukatsu T.. 2012. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. U. S. A. 109: 8618–8622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M., and Glöckner F. O.. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskella B., L. J. Hall, and Metcalf C. J. E.. 2017. The microbiome beyond the horizon of ecological and evolutionary theory. Nat. Ecol. Evol. 1: 1606–1615. [DOI] [PubMed] [Google Scholar]
- Kruskal W. H., and Wallis W. A.. 1952. Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47: 583–621. [Google Scholar]
- Mandal S., W. Van Treuren R. A. White M. Eggesbø R. Knight, and Peddada S. D.. 2015. Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26: 27663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequecning reads. EMBnet J. 17: 10–12. [Google Scholar]
- McDonald D., J. C., Clemente J., Kuczynski J. R., Rideout J., Stombaugh D., Wendel A., Wilke S., Huse J., Hufnagle F., Meyer, et al. 2012. The biological observation matrix (BIOM) format or: how I learned to stop worrying and love the ome-ome. Gigascience. 1: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. 2010. Data Structures for Statistical Computing in Python, pp. 51–56. Invan der Walt S., Millman, J (eds.), Proceedings of the 9th Python Science Conference. 28 June 2010. Austin, Texas. [Google Scholar]
- Muturi E. J., J. L. Ramirez A. P. Rooney, and Kim C. H.. 2017. Comparative analysis of gut microbiota of mosquito communities in central Illinois. Plos Negl. Trop. Dis. 11: e0005377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo C. T., Romano-Bertrand S., Manguin S., and Jumas-Bilak E.. 2016. Diversity of the bacterial microbiota of Anopheles mosquitoes from binh Phuoc Province, Vietnam. Front. Microbiol. 7: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ocampo C. B. and Wesson D. M.. 2004. Population dynamics of Aedes aegypti from a dengue hyperendemic urban setting in Colombia. Am. J. Trop. Med. Hyg. 71: 506–513. [PubMed] [Google Scholar]
- Ocampo C. B., Caicedo P. A., Jaramillo G., Ursic Bedoya R., Baron O., Serrato I. M., Cooper D. M., and Lowenberger C.. 2013. Differential expression of apoptosis related genes in selected strains of Aedes aegypti with different susceptibilities to dengue virus. PLoS One. 8: e61187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer W. H., Varghese F. S., and Van Rij R. P.. 2018. Natural variation in resistance to virus infection in dipteran insects. Viruses. 10: 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., G. Zhou J. Wu G. Bian P. Lu A. S. Raikhel, and Xi Z.. 2012. Wolbachia induces reactive oxygen species (ROS)-dependent activation of the Toll pathway to control dengue virus in the mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 109: E23–E31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J. L., Souza-Neto J., Cosme R. T., Rovira J., Ortiz A., Pascale J. M., and Dimopoulos G.. 2012. Reciprocal tripartite interactions between the Aedes aegypti midgut microbiota, innate immune system and dengue virus influences vector competence. PLoS Negl. Trop. Dis. 6: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancès E., Ye Y. H., Woolfit M., McGraw E. A., and O’Neill S. L.. 2012. The relative importance of innate immune priming in Wolbachia-mediated dengue interference. PLoS Pathog. 8: e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salter S. J., Cox M. J., Turek E. M., Calus S. T., Cookson W. O., Moffatt M. F., Turner P., Parkhill J., Loman N. J., and Walker A. W.. 2014. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biology. 12: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N., Izard J., Waldron L., Gevers D., Miropolsky L., Garrett W. S., and Huttenhower C.. 2011. Metagenomic biomarker discovery and explanation. Genome Biol. 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrato I. M., P. A. Caicedo Y. Orobio C. Lowenberger, and Ocampo C. B.. 2017. Vector competence and innate immune responses to dengue virus infection in selected laboratory and field-collected Stegomyia aegypti (= Aedes aegypti). Med. Vet. Entomol. 31: 312–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terenius O., J. M. Lindh K. Eriksson-Gonzales L. Bussière A. T. Laugen H. Bergquist K. Titanji, and Faye I.. 2012. Midgut bacterial dynamics in Aedes aegypti. FEMS Microbiol. Ecol. 80: 556–565. [DOI] [PubMed] [Google Scholar]
- Wang J., Y. Wu G. Yang, and Aksoy S.. 2009. Interactions between mutualist Wigglesworthia and tsetse peptidoglycan recognition protein (PGRP-LB) influence trypanosome transmission. Proc. Natl. Acad. Sci. U. S. A. 106: 12133–12138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav K. K., Chandel K., Organisation D., and Bhagyawant S. S.. 2018. Midgut bacterial microbiota of important mosquito disease vectors and its role in control of parasite transmission, pp. 245–299. InKishore Tyagi B. and Dhanasekaran, D (eds.), Microbial. Control of vector-borne diseases. CRC Press, Boca Raton, FL. [Google Scholar]
- Yun J. H., S. W., Roh T. W., Whon M. J., Jung M. S., Kim D. S., Park C., Yoon Y. D., Nam Y. J., Kim J. H., Choi, et al. 2014. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 80: 5254–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchi T. D., Prado S. S., and Cônsoli F. L.. 2012. The gastric caeca of pentatomids as a house for actinomycetes. BMC Microbiol. 12: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The raw sequence data have been uploaded onto NCBI’s SRA database (SRA Accession: SRP149364, Bio Project ID: PRJNA473810).