Abstract
Polyglutamine (polyQ) repeat diseases are a class of neurodegenerative disorders caused by CAG-repeat expansion. There are diverse cellular mechanisms behind the pathogenesis of polyQ disorders, including transcriptional dysregulation. Interestingly, we find that levels of the long isoform of nuclear paraspeckle assembly transcript 1 (Neat1L) are elevated in the brains of mouse models of spinocerebellar ataxia types 1, 2, 7 and Huntington's disease (HD). Neat1L was also elevated in differentiated striatal neurons derived from HD knock-in mice and in HD patient brains. The elevation was mutant Huntingtin (mHTT) dependent, as knockdown of mHTT in vitro and in vivo restored Neat1L to normal levels. In additional studies, we found that Neat1L is repressed by methyl CpG binding protein 2 (MeCP2) by RNA-protein interaction but not by occupancy of MeCP2 at its promoter. We also found that NEAT1L overexpression protects from mHTT-induced cytotoxicity, while reducing it enhanced mHTT-dependent toxicity. Gene set enrichment analysis of previously published RNA sequencing data from mouse embryonic fibroblasts and cells derived from HD patients shows that loss of NEAT1L impairs multiple cellular functions, including pathways involved in cell proliferation and development. Intriguingly, the genes dysregulated in HD human brain samples overlap with pathways affected by a reduction in NEAT1, confirming the correlation of NEAT1L and HD-induced perturbations. Cumulatively, the role of NEAT1L in polyQ disease model systems and human tissues suggests that it may play a protective role in CAG-repeat expansion diseases.
Introduction
Polyglutamine (polyQ) repeat expansion diseases encompass at least nine inherited, fatal neurodegenerative disorders associated with CAG expansion, including Huntington's disease (HD), six spinocerebellar ataxias (SCA) types 1/2/6/7/17, dentatorubral-pallidoluysian atrophy and Machado–Joseph disease (1). The CAG-repeat expansion diseases target varying populations of neurons, resulting in different clinical onset and manifestations. In HD, for example, mutant Huntingtin (mHTT) initially affects neurons in the caudate, putamen and cortex and compromises many cellular pathways including the mechanistic target of rapamycin, mitochondrial biogenesis, axonal growth and cholesterol homeostasis (2). Previous studies using HD cells, HD mouse model tissues and postmortem HD brain tissues have revealed that these varied pathologies may result from widespread transcriptome dysregulation (3–6).
Long non-coding RNAs (lncRNAs) are transcripts of over 200 nucleotides (nt) long that have poor protein-coding potential (7). LncRNAs are functional in diverse cellular processes, including regulation of gene expression, genomic imprinting, nuclear organization and nuclear–cytoplasmic trafficking (8). Emerging evidence has shown that lncRNAs are associated with brain aging and the pathophysiology of neuropsychiatric as well as neurodegenerative disorders (9). For example, the nuclear paraspeckle assembly transcript 1 (NEAT1) lncRNA is elevated in brain tissues from patients with frontotemporal lobar degeneration with TAR DNA-binding protein 43 proteinopathy (10). Additionally, studies show that NEAT1 drives oncogenic growth by altering the epigenetic landscape of target gene promoters (11).
NEAT1 is transcribed by RNA polymerase II to produce two distinct isoforms, NEAT1S (NEAT1_1, at 3735 nt) and NEAT1L (NEAT1_2, at 22 741 nt), referring to the short and long isoforms, respectively. NEAT1 is an essential architectural component of nuclear paraspeckle structure, and it is ubiquitously expressed in human tissues (12) and plays a functional role in hypoxia, adipogenesis, immune response and organogenesis (11). Although NEAT1S has been reported to be dysregulated in HD (13,14), the impact of the disease allele on NEAT1L remains unknown. Here, we find that NEAT1L is elevated in human HD brain tissues, brain tissues from HD mouse models and HD cell lines as well as in the brain tissues from mouse models of SCA1, SCA2 and SCA7. Together, the in vitro and in vivo works suggest that NEAT1L may play a protective role in the setting of CAG-repeat expansion disease.
Results
Previous studies using microarrays and RNA sequencing (RNA-seq) reported that NEAT1S is dysregulated in HD (14), but these works did not ascertain NEAT1L levels. In addition, the impact of polyQ disease on NEAT1L levels in general is unknown. To address this, total RNA from mouse brain was prepared and transcribed with random hexamer primers. cDNAs were further amplified with primer sets specifically targeted to Neat1L to avoid amplification of the 3.7 kb of Neat1S (Fig. 1A; Supplementary Material, Table S1). We found that Neat1L was elevated ∼1.5-fold in cerebellar tissues isolated from mouse models of SCA1, SCA2 and SCA7 and in striatal tissues harvested from HD mice, relative to wild-type (WT) littermates from each of the respective mouse colonies (Fig. 1B–E). Two other neuronal lncRNAs, Tcl1 Upstream Neuron-Associated long intergenic noncoding RNAs and PINKY were not altered (Supplementary Material, Fig. S1A–D).
Figure 1.
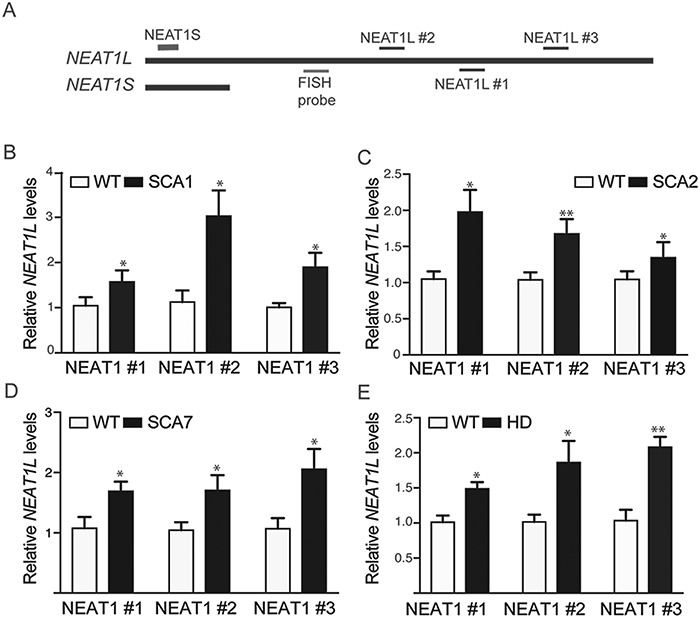
NEAT1L is upregulated in mouse models of polyQ repeat disease. (A) Cartoon of NEAT1L and NEAT1L transcripts with primers depicted (not drawn to scale). Shown are the relative positions for Quantitative PCR (qPCR) primers for NEAT1S/NEAT1L and the probe for FISH detection of NEAT1L. (B–E) Assessment of Neat1L expression in brain lysates from SCA1, SCA2, SCA7 and HD mice with each primer pair. (B) WT (n = 5) versus transgenic SCA1 mice (n = 5); (C) WT (n = 6) versus transgenic SCA2 mice (n = 6); (D) WT (n = 5) versus transgenic SCA7 mice (n = 5); (E) WT (n = 5) versus transgenic BACHD mice (n = 5). For all panels, results are expressed as mean ± SEM and data analyzed by the Mann–Whitney t-test. *P < 0.05; **P < 0.01.
Next, we isolated cytoplasmic and nuclear RNA from striatal of age-matched WT and zQ175 HD knock-in mice to assess the relative expression levels of Neat1S and Neat1L. For this, oligo (dT)18 (ThermoFisher Scientific, Waltham, MA) was used to reverse transcribe the polyadenylated Neat1S mRNA (Neat1L is not polyadenylated). In WT or HD knock-in zQ175 mice, Neat1S levels were more abundant than Neat1L but did not change significantly with disease state (Supplementary Material, Fig. S1E and F). However, nuclear Neat1L was upregulated in HD mice compared with that found in WT mice (Supplementary Material, Fig. S1G). Given these findings, we focused further work on Neat1L.
Neat1L levels were also assessed in differentiated immortalized striatal cells derived from Q111 knock-in mice (Q111 cells), a model of HD. Immortalized striatal cells derived from normal mice (Q7 cells) were used as control. Similar to the results in mice, Neat1L was elevated 2-fold in Q111 cells (Fig. 2A). Consistent with the observations in mouse model systems and cell lines, we found a significant elevation of NEAT1L in postmortem brain samples from HD patients (Fig. 2B), while other lncRNAs, including NEAT1S, were similar between disease and normal states (Supplementary Material, Fig. S2).
Figure 2.
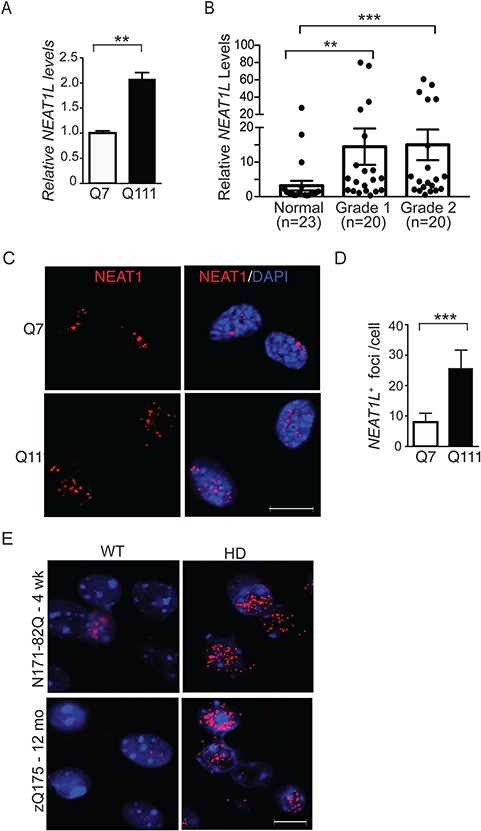
NEATL levels are upregulated in HD models and patient brain tissue. (A) RT-qPCR analysis of NEAT1L transcript levels in differentiated Q7 and Q111 immortalized striatal neurons. Data represent mean ± SEM. Significance was determined by unpaired, two-tailed t-test. **P < 0.01, ***P < 0.001. A minimum of three biological replicates with triple technical replicates per experiment were done. (B) RNA was isolated from the BA9 region from control donors or HD patients that had been pathologically scored as Von Sattal grade 1 or 2. RNA levels were quantified by RT-qPCR. Data were normalized to control human samples. Significance was determined using the Kruskal–Wallis test with Dunn’s post hoc analysis. ns = not significant. (C and D) Representative images (C) and quantification (D) of Neat1L positive foci in Q7 and Q111 cells assessed by FISH. Scale bar = 10 μm. For statistical analyses of the Neat1L RNA foci, ∼120 differentiated striatal neurons were counted randomly under the microscope. All images of the same experiment were processed with the same excitation power and exposure time and processed similarly using ImageJ software (National Institutes of Health, Bethesda, MD). Data represent mean ± SEM. Significance was determined by unpaired, two-tailed t-test. ***P < 0.001. (E)Neat1L RNA-FISH in tissues collected from presymptomatic 4-week-old N171-82Q mice (upper panels) and symptomatic 12-month-old zQ175 mice (lower panels). Scale bar = 10 μm.
Neat1L transcripts were next assessed by RNA fluorescence in situ hybridization (FISH) in Q7 and Q111 cells as well as in sections from HD mice. Neat1L-positive RNA foci were more abundant in Q111 cells than Q7 cells (Fig. 2C and D). RNA-FISH on striatal sections from pre-symptomatic 4-week-old N171-82Q HD mice and symptomatic 12-month-old zQ175 mice showed more Neat1L+ RNA foci in sections harvested from animals at both time points compared with their respective age-matched WT littermates (Fig. 2E).
To determine whether mHTT directly impacts NEAT1L levels, mHTT knockdown studies were performed. BACHD mice, which express full-length human mHTT, were transduced with adeno-associated virus (AAV) vectors expressing artificial miRNA sequences targeting human mHTT (AAV.miHTT) or a control miRNA sequence (AAV.miCtl) (15). This treatment resulted in an ∼50% reduction in mHTT (Fig. 3A). mHTT knockdown restored Neat1L to WT levels (Fig. 3B). A control lncRNA, Hotair, was not altered in BACHD mice and did not change with AAV.miHTT or AAV.miCtrl treatment (Fig. 3C). Taken together, these data imply that Neat1L levels are responsive to mHTT expression levels.
Figure 3.
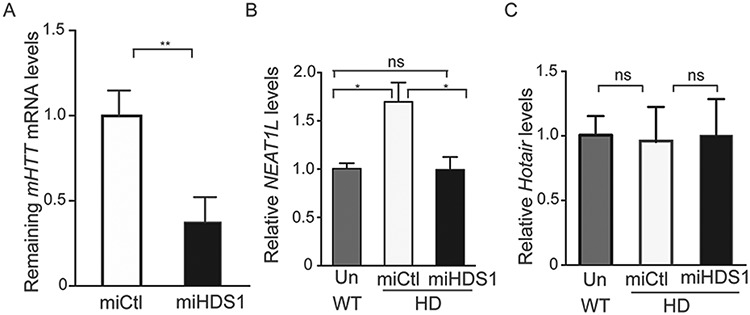
mHTT knockdown restores Neat1L expression. Eight-week-old BACHD mice were injected into the striatal with AAV vectors encoding artificial miRNAs targeting mHTT or with AAVs expressing a non-targeting control microRNA (miCtl) sequence. Striatal tissues were harvested 16 weeks later for RNA extraction. (A)mHTT mRNA levels in striatal tissue lysates as assessed by RT-qPCR (n = 3). Data are mean ± SEM and significance was analyzed by unpaired, two-tailed t-test, **P < 0.01. (B)Neat1L RNA levels in striatal tissue lysates from untreated WT mice or AAV.miHDS1 or AAV.miCtl-treated HD mice as assessed by RT-qPCR (n = 3). Data are mean ± SEM and significance analyzed by one-way ANOVA with Bonferroni post hoc analysis, *P < 0.05; ns = not significant. (C)Hotair RNA levels in striatal tissue lysates as assessed by RT-qPCR (n = 3). Data are mean ± SEM and significance analyzed by one-way ANOVA with Bonferroni post hoc analysis for panel B and C, ns = not significant.
To investigate the correlation between elevated NEAT1L levels and HD pathogenesis further, publicly available RNA-seq data sets from Neat1L knockout mice (16) were evaluated and compared with RNA-seq data from HD patient-derived induced pluripotent stem cell (iPSC) lines (17). Key biological processes regulated by NEAT1L include cell development, proliferation, mitosis, differentiation and the cell cycle (Fig. 4A; Supplementary Material, Table S2). Overlapping gene sets include those involved in synaptic transmission, nervous system development and neurological system processes (Fig. 4B). To assess the relevance of the implicated genes and pathways, we overexpressed NEAT1L and quantified genes that are altered in Neat1L knockout mice and reported to be beneficial to HD (2,17,18). NEAT1L overexpression increased the expression of transcripts encoding peroxisome proliferator activated receptor-γ (PPARG), Nuclear Factor Kappa B Subunit 1(NFκB1), and brain-derived neurotrophic factor as well as other genes (Fig. 4C). To confirm the observation from our overexpression studies, we transfected SH-SY5Y cells with a control Gapmer antisense oligonucleotide (ASO) or a Gapmer ASO specifically targeting NEAT1L (Supplementary Material, Fig. S3). As expected, genes that were upregulated were repressed after NEAT1L knockdown (Fig. 4D). Overall, these results suggest that NEAT1L upregulation may be a protective response of cells harboring polyQ repeat expanded transcripts.
Figure 4.
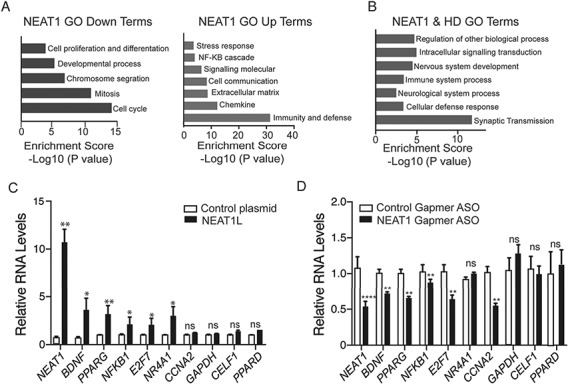
Evaluation of Neat1-KD and mHTT-induced transcriptional pathways. (A) RNA-Seq data from Neat1L knockout mouse embryonic fibroblasts was analyzed by PANTHER (Protein ANalysis THrough Evolutionary Relationships, http://pantherdb.org) using default settings to evaluate pathways affected by the absence of Neat1L in down-(left) or up-(right) regulated gene sets. (B) Overlapping GO analysis terms derived from the RNA-seq data sets from Neat1L knockout mouse embryonic fibroblasts and HD patient-derived iPSC lines. (C) Genes reported beneficial to HD cells increase with NEAT1 overexpression. Plasmids overexpression NEAT1L or GFP were transfected into neural N2A cells and the RNA levels of the indicated genes assessed by RT-qPCR 24 h later (n = 6 biological replicates with two or three technical replicates). ACTB was used as a reference gene, with data normalized to control plasmid-transfected cells. Data are mean ± SEM and significance determined by Mann–Whitney two-tailed t-test for all comparisons. *P < 0.05, **P < 0.01; ns = not significant. (D) Genes compromised in HD are repressed by NEAT1L knockdown. SH-SY5Y cells were transfected with control Gapmer ASOs or NEAT1L specific Gapmer ASOs. Twenty-four hours post transfection, RNA was harvested and transcript levels quantified by RT-qPCR. Data were normalized to the control ASO-treated group (n = 6 biological replicates, performed in triplicate). β-Actin was used as a reference gene. Data are mean ± SEM with significance determined Mann–Whitney two-tailed t-test for all comparisons. *P < 0.05, **P < 0.01; ns = not significant.
We next tested if elevated NEAT1L levels were protective. SH-SY5Y cells were transfected with GFP-tagged HTT expression plasmids (mHTT (104Q) or normal (25Q); Supplementary Material, Fig. S4A and B), along with ASOs targeting NEAT1L or control ASOs. As expected, cells transfected with mHTT (104Q) have lower viability than HTT (25Q)-transfected cells (Fig. 5A), and NEAT1L KD by ASOs exacerbated this difference (Fig. 5A). To verify the correlation between NEAT1 levels and mHTT toxicity, ASO-transfected SH-SY5Y cells were challenged with the mitochondrial toxin 3-nitropropionic acid (3-NP), a compound that induces neuronal cell death (19). Fewer NEAT1L knockdown cells (Fig. 5B) survived the 3-NP challenge relative to those pre-treated with a control ASO. We confirmed the sensitivity of HTT-expressing cells to NEAT1L levels by ASOs using a second knockdown approach with siRNAs. Similar to the ASO treatment, NEAT1L knockdown by siRNAs enhanced the toxicity in mHTT versus HTT transfected cells (Supplementary Material, Fig. S4C and D). The dose effect of ASO treatment (greater than 50% silencing) versus siRNA knockdown (∼25% silencing) suggests that NEAT1L levels moderates toxicity induced by mHTT.
Figure 5.
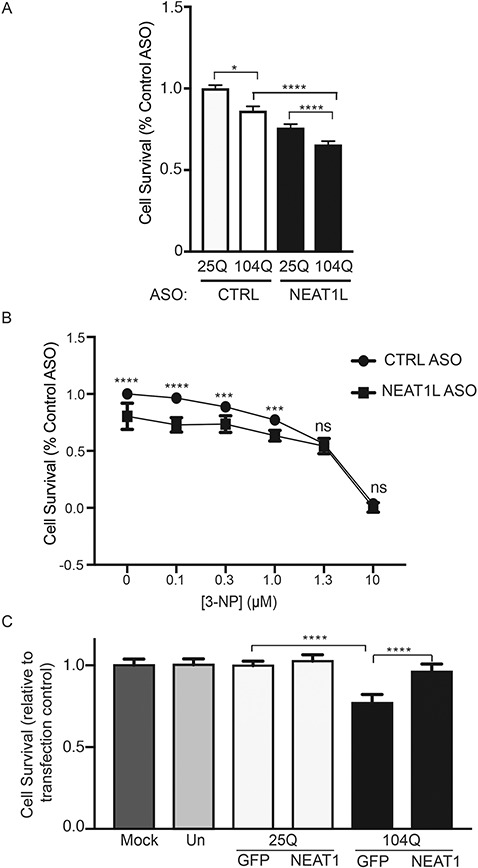
NEAT1L protects cells from mHTT-induced toxicity. (A) SH-SY5Y cells were transfected with NEAT1L-targeting or control ASOs, followed by transfection with HTT (25Q) or mHTT (104Q) expression plasmids and cell viability assessed 48 h later (n = 10 technical replicates). Data were normalized to control ASO/HTT (25Q) treated cells. Data represent mean ± SEM with significance determined by one-way ANOVA with Bonferroni post hoc comparison. **P < 0.01, ***P < 0.001, ****P < 0.000. (B) SH-SY5Y cells were transfected with NEAT1L targeting or control ASOs as described above and then 3-NP administered at the indicated doses 48 h post-transfection. Cell viability was assessed 48 h later (n = 10 technical replicates). Data are normalized to control ASO-treated groups. For each concentration, comparisons were made by Mann–Whitney two-tailed t-test. Data represent mean ± SEM, ***P < 0.001, ****P < 0.000, ns = not significant. (C) SH-SY5Y cells were co-transfected with HTT (25Q) or mHTT (104Q) expression plasmids with NEAT1L or control overexpression plasmids (n = 12 technical replicates) and cell viability assessed 48 h later. Data are mean ± SEM with significance determined by one-way ANOVA, ****P < 0.0001.
To further assess a cytoprotective role for NEAT1L, gain-of-function studies were done. SH-SY5Y cells were co-transfected with plasmids expressing HTT (25Q) or mHTT (104Q) along with NEAT1L over expression plasmids. Controls cells were either mock transfected (reagent only) or left untreated. NEAT1L levels, as assessed by qPCR, were approximately eight times greater than control-treated cells (data not shown). As expected, mHTT (104Q) transfection reduced cell survival, while HTT (25Q) did not. The effect of mHTT on cell viability was significantly rescued with NEAT1L overexpression (Fig. 5C).
LncRNAs are regulated in multiple ways, including transcription factor (TF) promoter binding, epigenetic modification and chromatin remodeling (20). The UCSC browser (University of California Santa Cruz, Santa Cruz, CA) (https://genome.ucsc.edu/) and the TRANSFAC 7.0 (BIOBASE GmbH, Wolfenbuettel, Germany) online module (http://genexplain.com/transfac/) were used to assess possible mechanisms underlying NEAT1L upregulation in HD. TF binding sites 0–10 kb upstream of the promoter include Transcription Factor 12 (TCF12), CCAAT/Enhancer Binding Protein Beta (CEBPB), CCCTC-Binding Factor (CTCF) and methyl CpG binding protein 2 (MeCP2) (Supplementary Material, Table S3; Fig. S5A). To determine their role in NEAT1L expression, transcripts encoding these factors were knocked down in human SH-SY5Y cells using DsiRNAs and NEAT1L transcript levels measured by quantitative reverse transcription PCR (RT-qPCR). NEAT1L was unaltered with SP1, CTCF, TCF12 or CEBPB knockdown (Supplementary Material, Fig. S5A). In contrast, MeCP2 knockdown (Fig. 6A) induced NEAT1L upregulation 2–6-fold in several cell types (Fig. 6B). Interestingly, human HD brain samples showed an ∼20% decrease in MECP2 mRNA and an ∼40% decrease in protein levels relative to control samples (Fig. 6C and D), indicating that reduced MeCP2 levels may underlie NEAT1L upregulation in HD.
Figure 6.
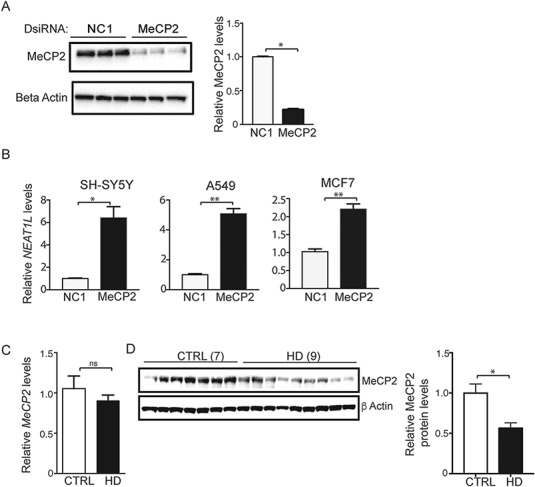
NEAT1L is repressed by MeCP2. (A) Immunoblot analysis of MeCP2 knockdown in SH-SY5Y cells. β-Actin was used as loading control. Right panel, quantification after densitometry of blots. N = 3 biological replicates with three technical replicates. Data are mean ± SEM with significance determined by Mann–Whitney two-tailed t-test, *P < 0.05. (B)NEAT1L RNA levels after MeCP2 knockdown in SH-SY5Y, A549 or MCF7 cells. Data are normalized to HPRT1 and control dsiRNA-treated cells (n = 3 biological replicates with three technical replicates). Data are mean ± SEM with significance determined by unpaired, two-tailed t-test with Welch’s correction, *P < 0.05; **P < 0.01. (C) RT-qPCR analysis of MeCP2 mRNA levels in human brain samples (BA9) from normal (7) and HD (9) individuals. Data are mean ± SEM and are normalized to HPRT1. Significance was determined by Mann–Whitney two-tailed t-test, ns = not significant. (D) Left panel, Representative western blot for MeCP2 protein levels in human brain samples from normal (n = 7) and HD patients (n = 9). β-Actin was used as loading control. Right panel, Quantification after densitometry of MeCP2 protein levels in human brain tissues using ImageJ software. Data are mean ± SEM and are normalized to MeCP2 protein level in control samples. Unpaired, two-tailed t-test with Welch’s correction. *P < 0.05.
MeCP2 is essential for central nervous system function and canonically activates or represses gene expression through direct interactions at promoter regions (21). To evaluate MeCP2 binding at the NEAT1L promoter, Chromatin Immunoprecipitation (ChIP) coupled with quantitative PCR (ChIP-qPCR) was done. Eighteen qPCR primer pairs were designed to cover the entire NEAT1L gene body plus 0–10 kb upstream of the NEAT1 transcription start site. RNF4, a validated target gene of MeCP2, was used as a positive control. We observed significant enrichment at the RNF4 promoter but not in the NEAT1L promoter (Supplementary Material, Fig. S5B). These data suggest that NEAT1L is not directly regulated by MeCP2 through promoter binding and transcriptional activation.
MeCP2 also possesses RNA-binding features (22,23) and recent studies using RNA immunoprecipitation (RIP)-seq have shown that MeCP2 binds lncRNAs, including NEAT1, metastasis associated lung adenocarcinoma transcript 1 and X inactive specific transcript (22). To test if MeCP2 binds NEAT1L transcripts in HD model systems, several different RIP protocols were performed including immunoprecipitation with native lysate, reversible crosslinking with formaldehyde and ultraviolet crosslinking at 254 nm (Supplementary Material, Fig. S6A). Additionally, for the native lysate method, we used two different approaches to prepare the samples [low and high stringency conditions (24,25)]. We observed consistent MeCP2-NEAT1L enrichment across the three protocols and both native lysate methods (Supplementary Material, Fig. S6B), supporting that MeCP2 interacts either directly or indirectly with NEAT1L. Using the native lysate method, we next tested if this interaction was dependent on mHTT. Human embryonic kidney cells 293 (HEK293) transfected with plasmids expressing either normal HTT (Q25) or mHTT (104Q) (18) were lysed and RNA precipitated with MeCP2 antibody. NEAT1L in the immunocomplex was reduced in mHTT (104Q) cells relative to HTT (25Q) cells, reflecting the reduced MeCP2 levels in the setting of mHTT (Fig. 7A and B). FISH-immunohistochemistry on striatal neurons also showed that NEAT1L partially co-localizes with MeCP2 in striatal neurons (Fig. 7C, white triangle). Overall, our data indicate that MeCP2 interacts with NEAT1L and represses its expression in normal conditions and that reduced MeCP2 expression may allow for elevated NEAT1L levels (Fig. 7D).
Figure 7.
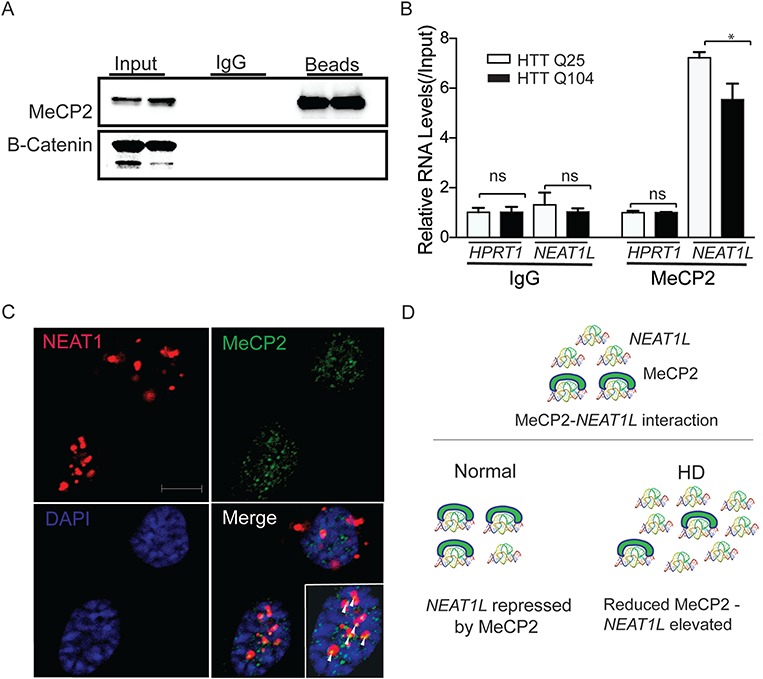
NEAT1L interacts with MeCP2 via RNA-protein interactions. (A) Endogenous MeCP2 was immunoprecipitated from whole-cell extracts of HEK293 cells transfected with HTT (25Q) or mHTT (104Q) expression plasmids. Input and bead-bound RNA-protein complexes were analyzed by western blot (representative of two independent experiments) using the indicated antibodies. Secondary antibodies were MeCP2 (upper) or β-catenin (lower). (B) Assessment of RNAs associated with immunoprecipitated MeCP2 in HTT (25Q) or mHTT (104Q) expressing HEK293 cells. Data are from two independent immunoprecipitation (IP) experiments. For each qPCR (NEAT1L or HPRT1 in Immunoglobulin G (IgG) isotype control or MeCP2 IP), GAPDH was used as a control housekeeping gene, and data were normalized to HTT (25Q) transfected groups and significance determined by two-tailed Mann–Whitney test. *P < 0.05, ns = not significant. (C) Co-localization of NEAT1L RNA (red) and MeCP2 protein (green) in striatal neurons. NEAT1L RNA-FISH was performed on Q111 striatal neurons. Nuclei were stained by DAPI (blue). Arrows indicate co-localization of NEAT1L and MeCP2. Scale bar = 10 μm. (D) Diagram of NEAT1L and MeCP2 interactions in HD. In normal conditions, NEAT1L is bound by MeCP2. In the setting of HD, MeCP2 is reduced, releasing NEAT1L.
Discussion
Emerging reports have shown that lncRNAs play a vital role in cell physiology or serve as modifiers in diverse diseases such as cancer and neurodegenerative disorders (26). NEAT1L has also been found to be beneficial to cell survival and its expression increases with viral infection (27) and cancer progression (28). In this work, we demonstrate that the lncRNA, NEAT1L, is elevated in model systems of polyQ-repeat expansion diseases including HD and several SCA diseases and human HD brain tissue. In cells and mouse models of HD, NEAT1L transcripts interact with and are regulated by MeCP2 levels. Moreover, we show that NEAT1L confers a protective role in cells, which may help alleviate mHTT-induced toxicity. Cumulatively, the data support a role for NEAT1L as a layer of defense against polyQ disease-associated toxicity.
NEAT1 lncRNAs are comprised of two isoforms, NEAT1S and NEAT1L. The human NEAT1 gene encodes the 3.7 kb NEAT1S and the 23 kb NEAT1L, while mouse Neat1 transcripts are 3.2 kb and 20 kb, respectively (29). For sequence structure, NEAT1S and NEAT1L isoforms share the same promoter and 5’ region, and the 3’ end of the NEAT1S isoform is polyadenylated. However, the 3’ end of NEAT1L lacks a canonical poly(A) tail. Both isoforms are predominantly retained in the nucleus and it remains to be determined whether the mechanism of NEAT1S nuclear export is via the canonical mRNA biogenesis pathway. Besides the difference in size and sequence structure, NEAT1S and NEAT1L have distinct expression profiles in cells. In situ hybridization analysis of mouse tissues shows that NEAT1S is ubiquitously expressed, while NEAT1L is restricted to specific subpopulations of cells within individual tissues (16). Additionally, NEAT1S expression is 8-fold higher than NEAT1L (30), and in our experiments using brain tissues and cell lines, we show that NEAT1S is more abundant than NEAT1L in nuclear and cytoplasmic compartments. However, nuclear NEAT1L levels, but not NEAT1S, increase significantly in polyQ repeat expansion disease model systems.
Regulation of lncRNAs is controlled by multiple factors, including RNA decay and epigenetic modification (31). The noted transcriptional dysregulation in HD (3) prompted us to assess TF binding at the NEAT1 promoter. We focused on MeCP2 because knockdown of the other putative TFs predicted to bind the NEAT1L promoter did not impact expression. Our data do not support canonical repression through MeCP2 binding but instead indicate that MeCP2 may regulate NEAT1L through direct or indirect RNA-binding. In the setting of reduced MeCP2, NEAT1L secondary structure may be altered, providing the opportunity to bind to other RNAs or other proteins that may stabilize the transcript in subcellular structures such as paraspeckles or RNA-rich liquid compartments (32).
NEAT1L, but not NEAT1S, is indispensable for nuclear paraspeckle assembly by interacting with ∼60 RNA-binding proteins, including SFPQ and NONO (33). Interestingly, we observed enlarged paraspeckles in HD cells. Also, NEAT1L alone or in combination with NONO-SFPQ can enhance pri-miRNA processing by the Drosha-DGCR8 Microprocessor (34). In HD models and patient tissues, MeCP2 levels are reduced, which could contribute to the notable dysregulation of miRNAs (35–37).
Our analysis of previously published RNA-seq data shows that lack of NEAT1 alters expression of genes associated with development and differentiation pathways as well as genes serving a neuroprotective role. Both NEAT1S and NEAT1L are known components of nuclear paraspeckles (38). NEAT1S and the 5' and 3' regions of NEAT1L are on the paraspeckle surface, while the central portion of NEAT1L is embedded in the paraspeckle (29). NEAT1L expression is rapidly regulated in response to stress, such that the nuclear compartmentalization provides an operational and dynamic manner for regulating gene expression and cellular responses. Our results as well as those from others show that reducing components of the paraspeckle complex, including NEAT1L, NONO or SFPQ, reduces cell viability (34). Interestingly, a recent study shows that RNAs such as NEAT1 can specifically bind paraspeckle proteins and drives the formation, identity and composition of phase-separated granules (38). Another group showed that RNAs, which phase-separate in repeat expansion disorders, including HD, do so independently of RNA-binding proteins (39). Whether elevated NEAT1L in HD is linked to the altered phase separation profiles of proteins and RNAs or protects from polyQ disease pathogenesis requires further study.
Materials and Methods
Human patient samples
Postmortem brain samples were obtained from the following sources: the New York Brain Bank (Columbia University, New York, NY), the Harvard Brain Tissue Resource Center (Belmont, MA), the New Zealand Neurological Foundation Brain Bank (University of Auckland, Auckland, New Zealand) and Johns Hopkins University (Baltimore, MD). Patient data have been described previously (3).
Animal studies
Animal protocols were approved by the Children’s Hospital of Philadelphia Institutional Animal Care and Use Committee. BACHD, zQ175 and N171-82Q mice were purchased from Jackson Laboratory (Bar Harbor, ME). Mice were housed in a temperature-controlled environment on a 12 h light–dark cycle. Food and water were provided ad libitum. Mouse brain slice preparation was described previously (40). mHTT knockdown in BACHD mice was as described previously (15), and striatum was harvested for RNA extraction as described below.
Cell culture and differentiation
The human dopaminergic neuroblastoma cell line, SH-SY5Y, was purchased from ATCC (Rockford, MD). Cells were routinely maintained in Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich, Saint Louis, MO), supplemented with 10% heat-inactivated fetal calf serum, penicillin (100 units/ml), streptomycin (100 μg/ml), L-glutamine (2 mm), sodium pyruvate (1 mm), in 5% CO2 humidified incubator at 37°C. Conditionally immortalized WT STHdhQ7/Q7 striatal neuronal progenitor cells expressing endogenous normal HTT and homozygous mutant STHdhQ111/Q111 striatal neuronal progenitor cell lines expressing endogenous mHTT with 111-glutamines were kindly provided by Dr Marcy MacDonald (41). The Q7 and Q111 cells were grown at 37°C in DMEM supplemented with 10% fetal bovine serum (FBS), 1% nonessential amino acids, 2 mm L-glutamine and 400 μg/ml Geneticin (Invitrogen, Carlsbad, CA). A549 and MCF7 cells were maintained in DMEM medium supplemented with 10% heat-inactivated FBS, 4 mm L-glutamine, 1 mm sodium pyruvate, 1 mm HEPES, penicillin (100 U/ml) and streptomycin (100 μg/ml).
Q7 and Q111 striatal neuron differentiation was described previously (42). Briefly, 400 μl of the dopamine cocktail (Forskolin 50 μmol/l, 3-isobutyl-1-methylxanthine 250 μmol/l, TPA 0.2 μmol/l, dopamine 10 μmol/l and α-FGF 10 ng/ml in DMEM) was added to striatal neurons seeded on a 6-well plate following brief rinsing with phosphate-buffered saline (PBS, pH 7.4). After 6 h, cells were harvested in ice-cold PBS at 4°C. The cell suspension was centrifuged for 5 min at 800 × g. The supernatant was discarded and the cell pellets were stored at −20°C until RNA extraction.
Knockdown and overexpression
All siRNAs were synthesized by Integrated DNA Technologies, Inc. (Coralville, IA), and oligo sequences are provided in Supplementary Material, Table S1. Unless otherwise specified, all siRNAs were double-transfected with Lipofectamine® RNAiMAX Reagent or DharmaFECT 1 Transfection Reagent (GE Dharmacon, Lafayette, CO). In brief, cells were split 1 day before seeding on 12-well plates to 50% confluency and reverse-transfected with 50 nM siRNAs per manufacturer’s instructions. Twenty hours post-transfection, cells were rinsed with PBS and treated with another 50 nM siRNA with the forward transfection protocol per manufacturer's instructions. Fourty-eight hours later, cells were harvested for RNA extraction or downstream assays. Gapmer ASOs were ordered from Qiagen (Exiqon, Vedbaek, Denmark). For NEAT1L ASO transfection, SH-SY5Y cells were transfected with 100 nM control Gapmer or NEAT1L Gapmer ASO (Exiqon, Germantown, MD) with Lipofectamine 2000 Transfection Reagent (42). RNA extraction and RT-qPCR were performed 24 h post-transfection. mHTT knockdown in BACHD mice was as described previously (15). Briefly, AAVs were generated that expressed artificial miRNAs that silence human mHTT or express a control miRNA using standard methods (40). Vectors (5 × 109 vector genomes total) were injected stereotactically into mice (three per genotype per treatment) at 8-week of age, and brains harvested 16 weeks later and RNA isolated for RT-qPCR analysis.
pCMV-NEAT1L was described previously (16). For cell viability assays, SH-SY5Y cells in 96-wells were treated with transfection reagent alone, 50 nM control siRNA or NEAT1L siRNA (10 samples per treatment). Fourty-eight hours later, 100 ng NEAT1L-expressing plasmid was transfected into siRNA-treated cells with X-tremeGENE™ 9 DNA Transfection reagent (Roche Diagnostics, Indianapolis, IN) by following the manufacturer's protocol. Empty vector pEGFP-N1 was used as a control. Twenty-four hours post-transfection, cells were lysed for viability assays. To quantify NEAT1L-targeting genes, N2A cells were seeded on 6-well plates and transfected with control plasmid or the NEAT1L overexpression plasmid with X-tremeGENE™ 9 DNA Transfection reagent as per the manufacturer's protocol. For immunoprecipitation studies, 45 μg of HTT (25Q) or mHTT (104Q) plasmids (18) were transfected with Lipofectamine® 2000 Reagent as per the manufacturer’s instructions (Invitrogen) into HEK293 cells seeded on two 150 mm plate at ∼90% confluency the day prior. Transfection medium was replaced 4 h post-transfection. Cells were examined under an immunofluorescence microscopy to check GFP expression 24 h post-transfection and collected for western blotting or immunoprecipitation 72 h post-transfection. For cell viability assays, GFP-tagged HTT expression plasmids (18) were used. For this, 100 ng HTT (25Q) or mHTT(104Q) expression plasmids were co-transfected with 100 ng NEAT1L overexpression plasmid or control plasmid by X-tremeGENE™ 9 DNA Transfection reagent. Fourty-eight hours after transfection, cells were harvested for viability analyses. A minimum of three biological replicates with triple technical replicates per experiment were done.
RNA extraction, reverse transcription and quantitative real-time PCR
RNA was extracted from human brain samples, mouse brain samples and cultured or transfected cells with Trizol reagent (Invitrogen) according to the manufacturer's protocol. Unless noted otherwise, 2–4 μg extracted RNA as quantified by NanoDrop was extensively treated with Turbo DNase I (Invitrogen) at 37°C for 30 min to 2 h to deplete genomic DNA. The DNase reaction was terminated by phenol/chloroform extraction and DNA-free RNA was further purified with Zymo RNA cleanup kit. Total RNA was quantified by NanoDrop and tested for 260/280 ratio greater than 1.9.
To extract cytoplasmic and nuclear RNA, ∼20 mg of striatal tissue was taken from 18-month-old WT mice or zQ175 mice (n = 3 each) and snap frozen in liquid nitrogen. The frozen tissue was pulverized and washed twice with cold PBS (minus Calcium and Magnesium). Cytoplasmic and nuclear RNA were prepared as described (43).
For reverse transcription, Superscript III or High capacity cDNA reverse transcription kit (Life Technologies, Carlsbad, CA) was used according to the manufacturer’s recommendations and 2 μg RNA used for each RT reaction. To discriminate expression profiles of NEAT1S and NEAT1L, RNA was reverse-transcribed using random hexamer primers and amplified with primers specific to NEAT1L. Assays were performed on a CFX384 Real Time System (Bio-Rad, Hercules, CA) using TaqMan primer/probe sets or SYBR Green primers (IDT) specific for human/mouse NEAT1L, MECP2 and all other genes. HPRT1, GAPDH or ACTB was used as a reference gene to normalize the data. Mouse U1 snRNA or HMBS was used to normalize nuclear RNA or cytoplasmic RNA, respectively. Relative gene expression was determined using the delta delta cycle threshold (ddCt) method.
Protein extraction and western blot analysis
Cultured cells were washed twice with cold PBS, scraped and homogenized in crosslinking immunoprecipitation (CLIP) lysis buffer (50 mm Tris-HCl (pH 7.4), 100 mm NaCl, 1 mm MgCl2, 0.1 mm CaCl2, 1% NP40, 0.5% sodium deoxycholate and 0.1% SDS) supplemented with Protease inhibitor Cocktail III (Merck Millipore, Darmstadt, Germany) as previously described (44). Cell lysates were briefly sonicated and centrifuged at 16 100 × g for 20 min at 4°C. The supernatants were collected and protein concentration measured with a DC Protein Assay Kit (Bio-Rad). Afterwards, equal amounts of protein were separated on 4–12% Bis-Tris gels and blotted onto polyvinylidene fluoride membrane pre-activated with methanol. After blocking with 5% (w/v) non-fat dry milk or bovine serum albumin, Fraction V in TBS with 0.1% Tween® 20, blots were incubated with rabbit anti-MeCP2 (#3456S, 1:1000; Cell Signaling Technology, Danvers, MA), rabbit anti-GFP (#GTX113617, 1:500; GeneTex, Irvine, CA) or rabbit anti beta-catenin (#Ab2365, 1:5000; Abcam, Cambridge, United Kingdom) antibodies followed by horseradish peroxidase-coupled antibodies (goat anti-rabbit: #111-035-144, 1:20 000; Jackson ImmunoResearch, West Grove, PA). In case of post-IP western blotting, TrueBlot®: Anti-Rabbit IgG HRP (#18-8816-31, 1:4000; Rockland Immunochemicals Inc., Limerick, PA) was used to avoid interference from IgG heavy and light chains. Blots were developed with ECL Plus reagents (Amersham Pharmacia Biotech Inc., Piscataway, NJ) and densitometry of protein levels determined with the ChemiDoc Imaging System (Bio-Rad) and Image Lab analysis software. A minimum of three biological replicates with triple technical replicates per experiment were done.
RNA and protein immunoprecipitation
RIP was as described previously with minor modifications (25,44–46). Briefly, 1.3 × 107 HEK293T cells were seeded on 150 mm plates and transfected with 45 μg HTT (25Q) or mHTT (104Q)—expressing plasmids. At 72 h post-transfection, media was discarded and cells rinsed with ice-cold PBS. Transfected cells or mock-treated cells were directly collected for RIP (25), or ultraviolet-crosslinked at 254 nm, 0.4 J cm-2 (45), or treated with 1% formaldehyde for 10 min and then quenched with 0.25 M Glycine (46). Cells were collected by scraping followed by centrifugation at 200 × g for 5 min at 4°C. To the cell pellet, CLIP lysis buffer (25) or polysome lysis buffer (24) supplemented with protease inhibitor (10 μL Murine RNAse Inhibitor per ml; New England Biolabs, Ipswich, MA) was added. Samples to which CLIP lysis buffer was added were then sonicated on ice at 10–15% amplitude for 30 s (5 s on and 10 s off). Samples to which polysome lysis buffer were added were triturated with a pipette for cell lysis. For each 1 ml lysate, 7 μL Turbo DNAse I was added and incubated at 37°C for 5 min. Cell lysates were rapidly cooled on ice for >1 min, and spun at 16 100 × g for 20 min at 4°C, after which supernatants were collected. Dynabeads® M-280 Sheep Anti-Rabbit IgG (Life Technologies) were washed twice with Phosphate Buffered Saline with Tween 20 (PBST) and incubated in 250 μL lysis buffer. A 50 μL aliquot of the beads was used to preclear the lysate supernatant by rotating at room temperature (RT) for 30 min and the cleared lysate was saved. For immunoprecipitation, 200 μL of the remaining beads were incubated with 10 μg of rabbit anti-MeCP2 polyclonal antibody or rabbit IgG control and rotated for 2 h at RT. Beads were washed thrice with 900 μL cold lysis buffer (with RNAse Inhibitor for RIP), resuspended in the cleared lysate and incubated in a cold room overnight. Unbound impurities were removed and beads were washed 3× with High Salt wash buffer (50 mm Tris-HCl (pH 7.4), 1 M NaCl, 1 mm EDTA, 1% NP40, 0.5% deoxycholate and 0.1% SDS; RNAse inhibitor added for RIP) followed by 5× with PNK buffer (20 mm Tris-HCl (pH 7.4), 10 mm MgCl2 and 0.2% Tween-20; RNAse inhibitor added for RIP). Beads were resuspended in 100 μL TEDS buffer (50 mm Tris-HCl (pH7.4), 5 mm EDTA, 1% SDS and fresh 10 mm DTT) and incubated at 70°C for 30 min on a heat block and then spun at 1200 rpm. For RIP, magnets were used to collect the supernatant, 900 μL Trizol was then added and RNA isolated as described above. RNA was dissolved in 44 μL water and 6 μL DNAse I mix (5 μL 10× DNAse buffer and 1 μL DNAse I) and incubated at 37°C for 15 min. DNAse I was inactivated by phenol-chloroform extraction and RNA cleaned with Phase Lock Gel Heavy. RNA was precipitated with 2.5 volume of 100% ethanol in the presence of 0.85 μL Glycoblue at −20°C for at least 2 h, centrifuged and dissolved in 24 μL water. Total RNA recovery was measured by Qubit assay (Invitrogen) and reverse transcription was performed with 4 μL of RNA with Superscript III Reverse Transcriptase in 20 μL volume for qPCR with Taqman primer sets and probe. Fold enrichment was calculated by -ddCt normalization with input and HPRT as the reference gene. For protein immunoprecipitation, 1/10 volume of reducing reagent was added and ¼ volume of NuPAGE 4X Loading dye after which the sample was boiled and loaded onto polyacrylamide gels. A minimum of three biological replicates with triple technical replicates per experiment were done.
RNA FISH
Striatal Q7 and Q111 cells were seeded on poly-L-ornithine-coated 4-well chamber slides (Lab-Tek II, #155382) and differentiated for 4 h as described above. Cell differentiation buffer was aspirated and chamber slides were washed 3× with cold PBS and fixed in 4% formaldehyde. The Affymetrix QuantiGene ViewRNA ISH cell assay kit was used to detect NEAT1L expression according to the manufacturer's protocols (Affymetrix Inc., Santa Clara, CA). Briefly, Q7 and Q111 cells were permeabilized with Detergent Solution QC for 5 min at RT, followed by digestion for 10 min at RT by Protease QS (1:4000 in PBS). NEAT1L-specific probe sets (Affymetrix Inc.) were diluted in buffer QF (1:100). RNA hybridization was conducted for 3 h at 40 ± 1°C. Neurons were then washed with the kit Wash Buffer and sequential hybridization steps conducted for signal amplification-PreAmplifier Mix (1:25 in Diluent QF), Amplifier Mix (1:25 in Diluent QF) and Label Probe Mix (1:25 in Diluent QF), each incubated for 30 min at 40 ± 1°C. RNA-protein co-staining was performed as described (33). After two 10 min washes in Wash Buffer, 200 μL blocking buffer (10% goat serum, 3% BSA and 0.3% Triton X-100) was added to the wells and incubated at RT for 1 h. MeCP2 antibody was diluted 1:200 in blocking buffer and applied to cells for another 1 h at RT or overnight at 4°C in the dark. Cells were washed 5× for 5 min and 200 μL 1:1000 diluted Alexa Fluor® 488-conjugated antibodies added to visualize MeCP2. Neurons were stained with 4',6-diamidino-2-phenylindole (DAPI) and cover-slipped with Prolong Gold Antifade Reagent (Life Technologies) prior to visualization using a Zeiss LSM 710 confocal microscope (Zeiss, Germany) and a 60× (1.4) objective. For statistical analyses of the NEAT1L RNA foci, ∼120 differentiated striatal neurons were counted randomly under the microscope. All images of the same experiment were processed with same excitation power and exposure time and processed similarly using the ImageJ software. For NEAT1L in situ hybridization in mouse brain, the RNAscope® Probe- Mm-Neat1 probe (Cat. No. 440351) was used as per manufacturer’s instructions (Advanced Cell Diagnostics, Newark, CA).
CHIP-qPCR assay
SH5-SY5Y cells were harvested at 90% confluency and counted. For each immunoprecipitation, 2 × 106 cells were crosslinked with 4% formaldehyde solution supplied in TruChIP Chromatin Shearing Reagent kit (Covaris, Woburn, MA) at RT for 5 min. The reaction was quenched and nuclei were isolated according to the manufacturer's manual. Cell nuclei were resuspended in 300 μL 1× Covaris shearing buffer and sonicated with a Covaris S2 at the following setting: duty factor, 5%; intensity, 4; cycles per burst, 200; time, 240 s. Whole lysates were spun at 14 000 × g at 4°C for 10 min and 10% supernatant was saved as input control to normalize the data. The cleared lysate was used for immunoprecipitation with rabbit anti-MeCP2 polyclonal antibody with EZ-Magna ChIP™ A-Chromatin Immunoprecipitation Kit as per manufacturer's instructions. Genomic DNA was extracted for SYBR Green-based qPCR. PCR primers span nine sites at upstream of the NEAT1 locus and nine sites within the NEAT1 gene body. Fold enrichment was calculated with -ddCt by normalization with input and GAPDH as a reference control. The known MeCP2 target RNF4 was used as a positive control (primers are listed in Supplementary Material, Table S1).
Cell viability assay
For all studies, a minimum of three biological replicates with triple technical replicates per experiment were done. Cells were seeded at 7500 cells on 96-well plates and the following day NEAT1L-expressing or pEGFP-N1-expressing plasmids were co-transfected along with HTT (25Q) or mHTT (104Q) plasmids expression plasmids using the X-tremeGENE 9 DNA transfection reagent (Roche Applied Science, Penzberg, Germany) per the manufacturer's instructions. Fourty-eight hours post-transfection, cell viability was assessed using the ATPLite luminescence-based assay (Perkin Elmer Biosciences, Meriden, CT) in accordance with the manufacturer's instructions and data were normalized to untreated control cells.
For NEAT1L depletion, two siRNAs targeting NEAT1L were transfected into 7500 SH-SY5Y cells seeded on 96-well plates using Oligofectamine using reverse transfection methods per the manufacturer's protocol. Gapmer ASO targeting NEAT1L or control ASO was transfected as described above. After 48 h, siRNA or ASO-transfected cells were transfected with HTT (25Q) or mHTT (104Q) expression plasmids using Fugene HD or challenged with 3-NP (Sigma-Aldrich) as described (47). Fourty-eight hours post-transfection or 3-NP treatment, cell survival was determined using the ATPlite assay as described above. Data were normalized to the control ASO or control transfection group (siRNA-plasmid sequential transfection) or PBS control (3-NP study).
Bioinformatic analyses
Genes differentially expressed between WT mice and NEAT1 knockout mice (16), and genes altered in HD (5), were retrieved from public databases and imported into PANTHER (http://www.pantherdb.org) for gene ontology (GO) analysis based on molecular functions (Supplementary Material, Table S2). The number of genes in each functional classification category was compared against the number of genes from NCBI’s mouse genome in that category. The binomial test was used to statistically determine overrepresentation of PANTHER classification categories. Bonferroni-corrected P-values < 0.05 were considered significant.
To search TF binding sites within the NEAT1 promotor, the 10 kb region upstream of the NEAT1 transcriptional start site and entire gene body of NEAT1 were scanned using the UCSC genome browser (48) or the TRANSFAC® database (www.biobase-international.com/transcription-factor-binding-sites from BIOBASE Corporation (Wolfenbuettel, Germany)) (Supplementary Material, Table S3). The background sequences were randomly generated and default settings were used for all other parameters. The significance was set at a P-value < 0.05.
Statistical analysis
Data were analyzed with GraphPad Prism 6 (GraphPad Software, La Jolla, CA) using unpaired two-tailed t-tests when comparing two groups or one-way analysis of variance (ANOVA) for multiple comparisons. Comparison between different data sets was done based on data distribution tested by the D’Agostiono normality test. When the normality assumption was violated, nonparametric tests were done using Mann–Whitney for two groups and Kruskal–Wallis for three or more groups. A significance level of P < 0.05 was considered significant. Values shown represent means ± standard error of the mean (SEM) of at least three biological replicates (with three or more technical triplicates per experiment), unless otherwise stated.
Supplementary Material
Acknowledgements
The authors acknowledge Xiang Yu in the Department of Biology of the University of Pennsylvania for bioinformatics assistance and Dr Albert R La Spada at Duke School of Medicine for providing GFP-tagged HTT (25Q) or mHTT (104Q) plasmids.
Conflict of Interest statement. None declared.
Funding
National Institutes of Health (NS076631 and NS084475 to B.L.D.) and the Children’s Hospital of Philadelphia Research Institute.
References
- 1. Adegbuyiro A., Sedighi F., Pilkington A.W.t., Groover S. and Legleiter J. (2017) Proteins containing expanded polyglutamine tracts and neurodegenerative disease. Biochemistry, 56, 1199–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lee J.H., Tecedor L., Chen Y.H., Monteys A.M., Sowada M.J., Thompson L.M. and Davidson B.L. (2015) Reinstating aberrant mTORC1 activity in Huntington’s disease mice improves disease phenotypes. Neuron, 85, 303–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lin L., Park J.W., Ramachandran S., Zhang Y., Tseng Y.-T., Shen S., Waldvogel H.J., Curtis M.A., Faull R.L.M., Troncoso J.C. et al. (2016) Transcriptome sequencing reveals aberrant alternative splicing in Huntington's disease. Hum. Mol. Genet., 25, 3454–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Labadorf A., Hoss A.G., Lagomarsino V., Latourelle J.C., Hadzi T.C., Bregu J., MacDonald M.E., Gusella J.F., Chen J.-F., Akbarian S. et al. (2015) RNA sequence analysis of human Huntington disease brain reveals an extensive increase in inflammatory and developmental gene expression. PLoS One, 10, e0143563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. iPSC Consortium H.D. (2017) Developmental alterations in Huntington's disease neural cells and pharmacological rescue in cells and mice. Nat. Neurosci., 20, 648–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wake C., Labadorf A., Dumitriu A., Hoss A.G., Bregu J., Albrecht K.H., DeStefano A.L. and Myers R.H. (2016) Novel microRNA discovery using small RNA sequencing in post-mortem human brain. BMC Genomics, 17, 776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geisler S. and Coller J. (2013) RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat. Rev. Mol. Cell Biol., 14, 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang K., Shi Z.-M., Chang Y.-N., Hu Z.-M., Qi H.-X. and Hong W. (2014) The ways of action of long non-coding RNAs in cytoplasm and nucleus. Gene, 547, 1–9. [DOI] [PubMed] [Google Scholar]
- 9. Salta E. and DeStrooper B. (2017) Noncoding RNAs in neurodegeneration. Nat. Rev. Neurosci., 18, 627–640. [DOI] [PubMed] [Google Scholar]
- 10. Lourenco G.F., Janitz M., Huang Y. and Halliday G.M. (2015) Long noncoding RNAs in TDP-43 and FUS/TLS-related frontotemporal lobar degeneration (FTLD). Neurobiol. Dis., 82, 445–454. [DOI] [PubMed] [Google Scholar]
- 11. Lo P.K., Wolfson B. and Zhou Q. (2016) Cellular, physiological and pathological aspects of the long non-coding RNA NEAT1. Front Biol (Beijing), 11, 413–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li S., Li J., Chen C., Zhang R. and Wang K. (2018) Pan-cancer analysis of long non-coding RNA NEAT1 in various cancers. Genes Dis., 5, 27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Johnson R. (2012) Long non-coding RNAs in Huntington's disease neurodegeneration. Neurobiol. Dis., 46, 245–254. [DOI] [PubMed] [Google Scholar]
- 14. Sunwoo J.-S., Lee S.-T., Im W., Lee M., Byun J.-I., Jung K.-H., Park K.-I., Jung K.-Y., Lee S.K., Chu K. et al. (2016) Altered expression of the long noncoding RNA NEAT1 in Huntington’s disease. Mol. Neurobiol., 54, 1577–1586. [DOI] [PubMed] [Google Scholar]
- 15. Monteys A.M., Spengler R.M., Dufour B.D., Wilson M.S., Oakley C.K., Sowada M.J., McBride J.L. and Davidson B.L. (2014) Single nucleotide seed modification restores in vivo tolerability of a toxic artificial miRNA sequence in the mouse brain. Nucleic Acids Res., 42, 13315–13327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nakagawa S., Naganuma T., Shioi G. and Hirose T. (2011) Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J. Cell Biol., 193, 31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Geva M., Kusko R., Soares H., Fowler K.D., Birnberg T., Barash S., Wagner A.M., Fine T., Lysaght A., Weiner B. et al. (2016) Pridopidine activates neuroprotective pathways impaired in Huntington disease. Hum. Mol. Genet., 25, 3975–3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dickey A.S., Pineda V.V., Tsunemi T., Liu P.P., Miranda H.C., Gilmore-Hall S.K., Lomas N., Sampat K.R., Buttgereit A., Torres M.-J.M. et al. (2016) PPAR-δ is repressed in Huntington's disease, is required for normal neuronal function and can be targeted therapeutically. Nat. Med., 22, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brouillet E., Jacquard C., Bizat N. and Blum D. (2005) 3-Nitropropionic acid: a mitochondrial toxin to uncover physiopathological mechanisms underlying striatal degeneration in Huntington's disease. J. Neurochem., 95, 1521–1540. [DOI] [PubMed] [Google Scholar]
- 20. Quinn J.J. and Chang H.Y. (2016) Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet., 17, 47–62. [DOI] [PubMed] [Google Scholar]
- 21. Qiu Z. (2017) Deciphering MECP2-associated disorders: disrupted circuits and the hope for repair. Curr. Opin. Neurobiol., 48, 30–36. [DOI] [PubMed] [Google Scholar]
- 22. Maxwell S.S., Pelka G.J., Tam P.P. and El-Osta A. (2013) Chromatin context and ncRNA highlight targets of MeCP2 in brain. RNA Biol., 10, 1741–1757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Castello A., Fischer B., Frese C.K., Horos R., Alleaume A.-M., Foehr S., Curk T., Krijgsveld J. and Hentze M.W. (2016) Comprehensive identification of RNA-binding domains in human cells. Mol. Cell, 63, 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nicholson C.O., Friedersdorf M. and Keene J.D. (2017) Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. RNA, 23, 32–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M. and Jaffrey S.R. (2016) m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature, 537, 369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matsui M. and Corey D.R. (2017) Non-coding RNAs as drug targets. Nat. Rev. Drug Discov., 16, 167–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang Q., Chen C.-Y., Yedavalli V.S.R.K. and Jeang K.-T. (2013) NEAT1 long noncoding RNA and paraspeckle bodies modulate HIV-1 posttranscriptional expression. MBio, 4, e00596–e00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Idogawa M., Ohashi T., Sasaki Y., Nakase H. and Tokino T. (2017) Long non-coding RNA NEAT1 is a transcriptional target of p53 and modulates p53-induced transactivation and tumor-suppressor function. Int. J. Cancer, 140, 2785–2791. [DOI] [PubMed] [Google Scholar]
- 29. Yamazaki T. and Hirose T. (2015) The building process of the functional paraspeckle with long non-coding RNAs. Front. Biosci., 7, 1–41. [DOI] [PubMed] [Google Scholar]
- 30. Mao Y.S., Sunwoo H., Zhang B. and Spector D.L. (2011) Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat. Cell Biol., 13, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dong X., Muppani N.R. and Wu J. (2016) Long noncoding RNAs: critical regulators for cell lineage commitment in the central nervous system. In Wu, J. (ed.), Transcriptomics and Gene Regulation Springer Netherlands, Dordrecht, pp. 73–97. [Google Scholar]
- 32. Langdon E.M., Qiu Y., Ghanbari Niaki A., McLaughlin G.A., Weidmann C.A., Gerbich T.M., Smith J.A., Crutchley J.M., Termini C.M., Weeks K.M. et al. (2018) mRNA structure determines specificity of a polyQ-driven phase separation. Science, 360, 922–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chujo T., Yamazaki T., Kawaguchi T., Kurosaka S., Takumi T., Nakagawa S. and Hirose T. (2017) Unusual semi-extractability as a hallmark of nuclear body-associated architectural noncoding RNAs. EMBO J., 36, 1447–1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jiang L., Shao C., Wu Q.J., Chen G., Zhou J., Yang B., Li H., Gou L.T., Zhang Y., Wang Y. et al. (2017) NEAT1 scaffolds RNA-binding proteins and the microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol., 24, 816–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Packer A.N., Xing Y., Harper S.Q., Jones L. and Davidson B.L. (2008) The bifunctional microRNA miR-9/miR-9* regulates REST and CoREST and is downregulated in Huntington's disease. J. Neurosci., 28, 14341–14346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gaughwin P.M., Ciesla M., Lahiri N., Tabrizi S.J., Brundin P. and Bjorkqvist M. (2011) Hsa-miR-34b is a plasma-stable microRNA that is elevated in pre-manifest Huntington's disease. Hum. Mol. Genet., 20, 2225–2237. [DOI] [PubMed] [Google Scholar]
- 37. Rodriguez-Lebron E., Liu G., Keiser M., Behlke M.A. and Davidson B.L. (2013) Altered Purkinje cell miRNA expression and SCA1 pathogenesis. Neurobiol. Dis., 54, 456–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamazaki T., Souquere S., Chujo T., Kobelke S., Chong Y.S., Fox A.H., Bond C.S., Nakagawa S., Pierron G. and Hirose T. (2018) Functional domains of NEAT1 architectural lncRNA induce paraspeckle assembly through phase separation. Mol. Cell, 70, 1038–1053. [DOI] [PubMed] [Google Scholar]
- 39. Jain A. and Vale R.D. (2017) RNA phase transitions in repeat expansion disorders. Nature, 546, 243–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keiser M.S., Kordower J.H., Gonzalez-Alegre P. and Davidson B.L. (2015) Broad distribution of ataxin 1 silencing in rhesus cerebella for spinocerebellar ataxia type 1 therapy. Brain, 138, 3555–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Reis S.A., Thompson M.N., Lee J.-M., Fossale E., Kim H.-H., Liao J.K., Moskowitz M.A., Shaw S.Y., Dong L., Haggarty S.J. et al. (2011) Striatal neurons expressing full-length mutant huntingtin exhibit decreased N-cadherin and altered neuritogenesis. Hum. Mol. Genet., 20, 2344–2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Adriaens C., Standaert L., Barra J., Latil M., Verfaillie A., Kalev P., Boeckx B., Wijnhoven P.W.G., Radaelli E., Vermi W. et al. (2016) p53 induces formation of NEAT1 lncRNA-containing paraspeckles that modulate replication stress response and chemosensitivity. Nat. Med., 22, 861–868. [DOI] [PubMed] [Google Scholar]
- 43. Lubelsky Y. and Ulitsky I. (2018) Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature, 555, 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spengler R.M., Zhang X., Cheng C., McLendon J.M., Skeie J.M., Johnson F.L., Davidson B.L. and Boudreau R.L. (2016) Elucidation of transcriptome-wide microRNA binding sites in human cardiac tissues by Ago2 HITS-CLIP. Nucleic Acids Res., 44, 7120–7131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dai W., Li W., Hoque M., Li Z., Tian B. and Makeyev E.V. (2015) A post-transcriptional mechanism pacing expression of neural genes with precursor cell differentiation status. Nat. Commun., 6, 7576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Zhang X.O., Chen T., Xiang J.F., Yin Q.F., Xing Y.H., Zhu S., Yang L. and Chen L.L. (2013) Circular intronic long noncoding RNAs. Mol. Cell, 51, 792–806. [DOI] [PubMed] [Google Scholar]
- 47. Solesio M.E., Saez-Atienzar S., Jordan J. and Galindo M.F. (2013) 3-Nitropropionic acid induces autophagy by forming mitochondrial permeability transition pores rather than activating the mitochondrial fission pathway. Br. J. Pharmacol., 168, 63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S. et al. (2012) GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res., 22, 1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


