Abstract
Birdshot Uveitis (Birdshot) is a rare eye condition that affects HLA-A29-positive individuals and could be considered a prototypic member of the recently proposed ‘MHC-I (major histocompatibility complex class I)-opathy’ family. Genetic studies have pinpointed the endoplasmic reticulum aminopeptidase (ERAP1) and (ERAP2) genes as shared associations across MHC-I-opathies, which suggests ERAP dysfunction may be a root cause for MHC-I-opathies. We mapped the ERAP1 and ERAP2 haplotypes in 84 Dutch cases and 890 controls. We identified association at variant rs10044354, which mediated a marked increase in ERAP2 expression. We also identified and cloned an independently associated ERAP1 haplotype (tagged by rs2287987) present in more than half of the cases; this ERAP1 haplotype is also the primary risk and protective haplotype for other MHC-I-opathies. We show that the risk ERAP1 haplotype conferred significantly altered expression of ERAP1 isoforms in transcriptomic data (n = 360), resulting in lowered protein expression and distinct enzymatic activity. Both the association for rs10044354 (meta-analysis: odds ratio (OR) [95% CI]=2.07[1.58–2.71], P = 1.24 × 10(−7)) and rs2287987 (OR[95% CI]: =2.01[1.51–2.67], P = 1.41 × 10(−6)) replicated and showed consistent direction of effect in an independent Spanish cohort of 46 cases and 2103 controls. In both cohorts, the combined rs2287987-rs10044354 haplotype associated with Birdshot more strongly than either variant alone [meta-analysis: P=3.9 × 10(−9)]. Finally, we observed that ERAP2 protein expression is dependent on the ERAP1 background across three European populations (n = 3353). In conclusion, a functionally distinct combination of ERAP1 and ERAP2 are a hallmark of Birdshot and provide rationale for strategies designed to correct ERAP function for treatment of Birdshot and MHC-I-opathies more broadly.
Introduction
‘MHC-I(major histocompatibility complex class I)-opathy’ is a relatively new term proposed to unify a group of severe inflammatory diseases, each characterized by strong association with a unique MHC-I allele and shared immune characteristics (1). Recent genetic association studies revealed a vast enrichment for polymorphisms in the endoplasmic reticulum aminopeptidase (ERAP) genes, ERAP1 and ERAP2, in patients with ankylosing spondylitis (AS), psoriasis or Behçet’s disease (BD) (2–5). Importantly, in these conditions, ERAP is in epistasis with the risk alleles of MHC-I allele. However, not all patients with AS or BD carry a MHC-I risk allele, an observation that has provoked discussions on this unifying concept (6,7) and whether it reflects shared underlying disease etiology.
The mechanism through which ERAP function contributes to MHC-I-opathies is still under intense discussion and the discovery of the underlying mechanism is hampered by several unique characteristics of these specialized enzymes: the peptidases ERAP1 and ERAP2 closely interact with MHC-I by contributing to the generation and destruction of peptides considered for presentation by MHC-I on the cell surface. This key function indicates that aberrant peptide processing by ERAP may be a root cause for MHC-I-opathies (8). Similar to MHC-I, genetic diversity of ERAP genes is conserved by balancing selection (9). Balancing selection maintains ERAP2 deficiency in one-quarter of the population (9) and drives the polymorphic ERAP1 gene to encode various haplotypes (their protein products are also termed ‘allotypes’) (10), few of which paradoxically confer or reduce risk for developing MHC-I-opathies (8). To date, ERAP haplotype studies have been limited to the relatively more common MHC-I-opathies (11–12). In contrast, Birdshot uveitis or Birdshot chorioretinopathy (from here on ‘Birdshot’) is a severe, extremely rare eye condition (∼1–5 cases/500 000) (13,14) that is characterized by enhanced IL-23-IL-17 pathway activation (15–17), and occurs exclusively in HLA-A29-positive individuals (18). A previous genome-wide association study in Birdshot highlighted strong involvement of the ERAP region on chromosome 5q15 (19). The unique (diagnostic) prerequisite of HLA-A29 (20) and ERAP association may classify Birdshot patients within the MHC-I-opathy cluster and, thus, provide an attractive model to define the genetic architecture of this superfamily of conditions (18).
Therefore, we mapped ERAP1 haplotypes and other key variants at 5q15 across a Dutch discovery cohort and validated our findings in a Spanish replication cohort. We interrogated the downstream implications of associated variants using transcriptomic data, molecular cloning and functional assays.
Results
Two independent variants at 5q15 associate with Birdshot
To test for genetic variants that associate with Birdshot, we obtained genotype data for 13 key variants at 5q15 in 84 cases and 890 controls from the Netherlands (Supplementary Table 1). Using a logistic regression model, adjusted for sex and the top two principle components, we identified a top association at the T allele of rs10044354 in leucyl and cystinyl aminopeptidase (LNPEP) (odds ratio (OR) = 2.42[95% CI: 1.69–3.45], P = 1.21 × 10−6, Figure 1 and Supplementary Table 1). We accounted for the effect of rs10044354 using conditional analysis, and identified the C allele of rs2287987 (encoding valine at amino acid position 349) in ERAP1 as a secondary (independent) association to Birdshot (OR = 1.99 [95% CI: 1.36–2.90], P = 3.97 × 10−4, Fig. 1). Due to linkage disequilibrium (LD), several other single nucleotide polymorphisms (SNPs) across the locus also showed strong association, but conditioning on rs2287987 in ERAP1 removed the remainder of the association signal in the region (Supplementary Table 1).
Figure 1.
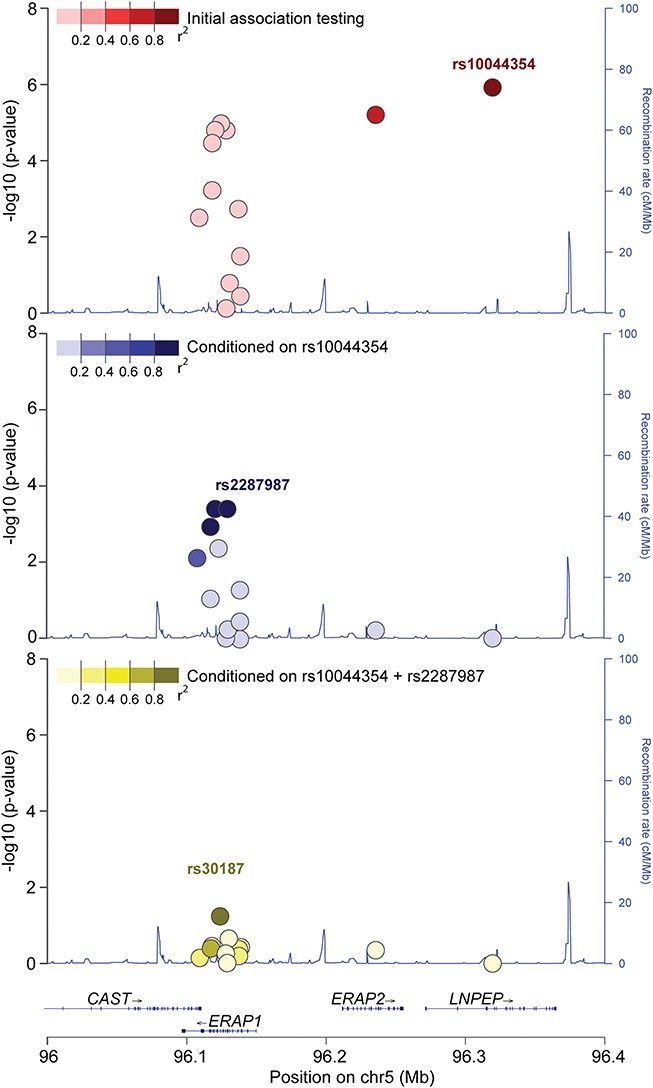
Association and conditional testing for the 13 polymorphisms at 5q15. (A) Initial association testing mapped the strongest association to rs10044354 in LNPEP (Dark red). (B) Conditioning on rs10044354 revealed independent association for the C allele of rs2287987 (amino acid position 349 in ERAP1) and variants in tight LD (rs10050860 and rs17482075 in dark blue) in ERAP1. (C) Conditioning on both rs10044354 and rs2287987 removed the bulk of the signal (rs30187, P = 0.05). Plot framework was generated by LocusZoom (53) and edited for graphical purposes.
We sought to replicate the associations with rs2287987 and rs10044354 using a set of independent samples drawn from an independent Spanish cohort of 46 cases and 2103 controls. Both associations replicated (Table 1 and Supplementary Table 1) and show consistent direction of effect (meta-analysis, rs10044354, OR [95% CI: = 2.07[1.58–2.71], P = 1.24 × 10−7; meta-analysis rs2287987, OR [95% CI]: = 2.01[1.51–2.67], P = 1.41 × 10−6). Interestingly, the combined haplotype rs2287987-rs10044354 showed disease association exceeding the association for each individual SNP in both cohorts (Supplementary Table 2). The strongest association mapped to the protective haplotype rs2287987-T/rs10044354-C (meta-analysis, OR [95% CI]: =0.39 [0.30–0.55],P = 3.9 × 10−9), closely followed by the risk haplotype rs2287987-C/rs10044354-T (OR [95% CI]: = 2.75 [1.92–3.92], P = 2.6 × 10−8), the latter present in 50% of all 130 cases and 26% of all 2993 controls. The frequency of the CT-risk haplotype (0.24 in Dutch cases and 0.19 of Spanish cases) is ∼5–6x less common in African (0.027) and Asian (0.021) populations compared with the European population (0.13, Fig. 2A).
Table 1.
Common ERAP1 haplotypes based on 10 missense variants of 260 Birdshot haplotypes and 5986 control haplotypes from the Netherlands and Spain
| SNP | rs72773968 | rs3734016 | rs26653 | rs26618 | rs27895 | rs2287987 | rs30187 | rs10050860 | rs17482078 | rs27044 | Netherlands | Spain | Meta-analysis | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Frequency (%) | OR | P-valueb | Frequency (%) | OR | P value | Meta OR (R) | Meta P-value (R) |
|||||||||||||
| Residue change | Thr > Ile | Glu > Lys | Pro > Arg | Ile > Met | Gly > Asp | Met > Val | Lys > Arg | Asp > Asn | Arg > Gln | Gln > Glu | ||||||||||
| Haplotypea | Amino acid position ERAP1 protein | Cases (n = 84) | Control (n = 890) | Cases (n=46) | Controls (n=2103) | |||||||||||||||
| 12 | 56 | 127 | 276 | 346 | 349 | 528 | 575 | 725 | 730 | |||||||||||
| Hap1 | I | E | P | I | G | M | K | D | R | Q | 6 | 13 | 0.48 | 0.032 | 14 | 15 | 0.91 | 0.745 | 0.67 | 0.220 |
| Hap2 | T | E | R | I | G | M | K | D | R | Q | 7 | 15 | 0.42 | 6.43 x 10-3 | 10 | 16 | 0.56 | 0.096 | 0.48 | 1.70 × 10-3 |
| hap3 | T | E | R | I | G | M | K | D | R | E | 3 | 6 | 0.46 | 0.097 | 11 | 8 | 1.41 | 0.318 | 0.84 | 0.757 |
| hap5 | T | E | R | I | D | M | R | D | R | E | 7 | 7 | 0.93 | 0.834 | 4 | 6 | 0.69 | 0.47 | 0.85 | 0.562 |
| hap6 | T | E | P | I | G | M | R | D | R | E | 4 | 9 | 0.46 | 0.056 | 7 | 8 | 0.76 | 0.528 | 0.59 | 0.070 |
| hap7 | T | K | P | I | G | M | R | D | R | E | 5 | 4 | 1.41 | 0.355 | 4 | 3 | 1.76 | 0.27 | 1.52 | 0.163 |
| hap8 | T | E | P | M | G | M | R | D | R | E | 29 | 25 | 1.32 | 0.155 | 20 | 22 | 0.85 | 0.528 | 1.10 | 0.678 |
| Hap10 | T | E | P | I | G | V | R | N | Q | E | 36 | 19 | 2.20 | 3.45 × 10 -5 | 29 | 19 | 1.71 | 0.0195 | 1.99 | 2.88 × 10 -6 |
| Total | 97 | 97 | 99 | 98 | ||||||||||||||||
Associations of the ERAP1 coding haplotypes with Birdshot were evaluated by logistic regression and the effect size for association of each haplotype is given as the OR. Meta-analysis (meta-OR and meta-P-value) was performed as described in the method section. (a) Haplotype numbers according to Ombrello et al, (2015) (21). The eight haplotypes listed account for >95% of all the ERAP1 haplotypes in the Dutch and Spanish controls.(b) Significance (P < 1.49 × 10−4, see methods). Only Hap10 is significant. We also note a lower frequency of Hap2 in cases.
Figure 2.
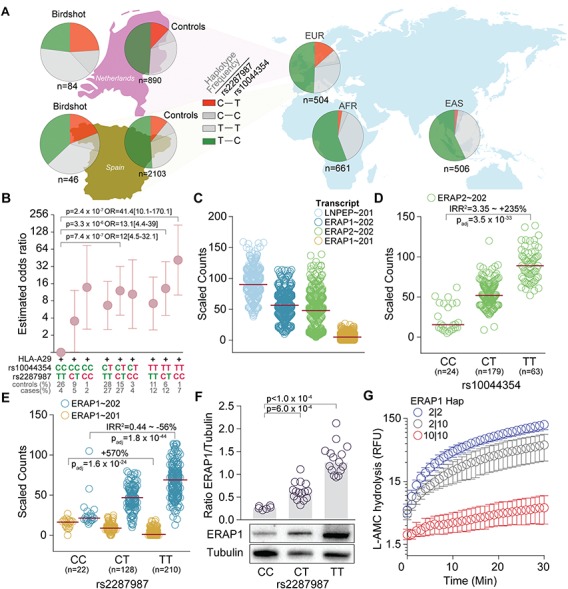
Birdshot associated variants affects ERAP1 and ERAP2 expression and function. (A) The frequency of the rs2287987-rs10044354 haplotypes in Dutch and Spanish cases and controls, and the EUR, EAS and AFR super populations of the 1000 Genomes project (48). The frequency of the haplotypes is indicated as the percentage of all haplotypes for each population. (B) We tested for association of rs2287987-rs10044354 genotypes in 129 HLA-A29-positive cases and 439 HLA-A29-positive controls in the combined Dutch and Spanish samples. ORs (and 95% confidence intervals) for each genotype combination were calculated relative to lowest risk genotype: the rs2287987-TT/rs10044354-CC (set to an OR of 1). The risk allele (red) and protective alleles (green) are highlighted. (C) Expression data for transcripts at 5q15 in 360 lymphoid cell lines from the European populations from the GEUVIDAS consortium (48). Red lines indicate the median expression. (D) The lead SNP rs10044354 is a strong eQTL for ERAP2 independent of rs2248374 (details eQTL analysis in Supplementary Table 4). (E) The Birdshot-associated rs2287987 is associated with changes in the two major ERAP1 transcripts. (F) Quantitative Western blot analysis of the protein expression of ERAP1 in HLA-A29-positive cell lines according to the distribution of rs2287987 (more details see Supplementary Fig. 4). (G) Hydrolysis [expressed as relative fluorescent units of the substrate Leu-AMC by immunoprecipitated ERAP1 protein from lymphoid cell lines homozygous Hap10 (risk haplotype), homozygous Hap2 (protective haplotype) and a heterozygous donor for these two ERAP1 haplotypes. Error bars indicate the mean (range) of three independent experiments.
Since Birdshot is associated with HLA-A29, we investigated the association for the rs2287987-rs10044354 haplotype in the context of HLA-A29 status in the combined 129 HLA-A29-positive cases and 439 HLA-A29-positive controls (15% of controls). ORs for each haplotype’s association to Birdshot were calculated relative to the haplotype carrying the two lowest risk genotypes: rs2287987-TT-rs10044354-CC. In the HLA-A29-postive population, only genotypes that harbor risk alleles from both rs2287987 and rs10044354 associated with Birdshot (P < 1.49 × 10−4), with the strongest association observed for the rs2287987-CC-rs10044354-TT genotype (OR [95% CI]: = 41.4 [10.08–170.08], P = 2.4 × 10-7) (Fig. 2B). Considered collectively, our association testing demonstrates that rs10044354 in LNPEP and rs2287987 in ERAP1 explained the bulk of the association with Birdshot at 5q15 and that combinations of these ERAP variants significantly increase the risk in the HLA-A29-positive population.
ERAP1 Hap10 is associated with Birdshot
Common ERAP1 haplotypes have been previously linked to MHC-I-opathies (8). Considering 10 non-synonymous variants (i.e. 10 amino acid positions) in ERAP1 together (21), we identified eight common (frequency >1%) haplotypes (Table 1). The Birdshot-associated C allele of rs2287987 (349V) resides in the ERAP1 haplotype previously labelled ‘Hap10’ (21). Indeed, testing for association of phased haplotype data revealed that Hap10 was associated with Birdshot and replicated in both cohorts (meta-analysis OR = 1.99 [95% CI: 1.49–2.65], P = 2.88 x 10−6, Table 1). We also noted that Hap2 was less frequent in cases (Table 1). Sixty-three percent of Dutch cases and 46% of Spanish cases had at least one copy of Hap10 compared with 37.2% and 34.7% of controls, respectively (Supplementary Table 3).
The primary association signal at 5q15 associates with increased ERAP2 expression
Next, we aimed to predict the local transcriptional regulation of the Birdshot-associated loci at 5q15 using RNA-sequencing data from 360 individuals of European ancestry and 82 individuals from African ancestry. We used lymphoid cell lines due to their high gene and protein expression of ERAPs and representation of expression quantitative trait loci (eQTLs) cis effects of blood cell populations (Supplementary Fig. 6). We applied a linear mixed model controlling for sex, ancestry, and batch to model if the SNPs imposed eQTLs effects on transcripts encoded by 5q15 (Fig. 2C); The leading SNP (rs10044354) in LNPEP was not associated with the expression of its major transcript (Supplementary Table 4). We extracted common missense and splice region variants of LNPEP in high LD with rs10044354 from the European populations of the 1000 Genomes (22) and identified the splice region variant rs3836862 (r2 > 0.96 in five populations) located near the exons encoding the zinc-binding motif of the enzyme and is predicted to influence splicing. However, we detected no evidence for potential alternative splicing of LNPEP by this variant in lymphoid cell lines (detailed experiments provided under Supplementary Fig. 5). Correcting for the expression of ERAP2 mediated by rs2248374, we found the risk allele of rs10044354 to be strongly associated with ERAP2 in samples from European ancestry. Further, this allele associated to a 235% increase in expression of ERAP2 independent from rs2248374 (Incidence rate ratio (IRR2) [95% CI] = 3.35[2.76–4.06], P = 3.48 × 10−33, Supplementary Table 4 and Fig. 2D). In line with this, conditioning on rs2248374 in ERAP2 in the 130 cases and 2993 controls combined, we found the association for rs10044354 with Birdshot to be independent from rs2248374 (P = 9.1 × 10−4).
ERAP1 risk alleles reside in a relatively low expressed haplotype
The Birdshot risk variant rs2287987 (349V in ERAP1) is in near complete LD (r2 = 0.99) with rs10050860 (575N in ERAP1, P = 1.46 × 10−6 for association with Birdshot). The SNP rs10050860 was recently suggested to represent a splice interfering variant (rs7063), which was linked to changes in the proportion of expression of two major ERAP1 isoforms (23). We explored the rs7063 variant using rs1057569 (r2 = 1). Indeed, we observed strong association between the rs1057569 genotype and the expression of the two major ERAP1 transcripts in samples of European ancestry (Supplementary Table 4). This variant is also among the top eQTLs for ERAP1 in lymphoid cell lines investigated in the Genotype-Tissue Expression (GTEx) project (Supplementary Fig. 7). The rs2287987 is in moderate LD with rs1057569 (r2 = 0.53 in the combined 2993 controls) and associated with ∼50% change in expression of the major ERAP1 isoform (Supplementary Table 4 and Fig. 2E), which results in ∼5x lower cumulative levels of ERAP1 protein in HLA-A29-positive individuals homozygous for the C allele of rs2287987 (Fig. 2F). However, the eQTL effects of rs2287987 on ERAP1 transcripts were completely abrogated once adjusted for rs1057569 (Supplementary Table 4). Therefore, we tested whether the rs2287987 disease signal was mediated by rs1057569. Conditioning on rs1057569, we found the rs2287987 signal to be modestly affected (pmeta = 4.61 × 10-4), while conditioning on rs2287987 neutralized the signal (pmeta = 0.39). These results indicate that the association signal at rs2287987 represents a signal beyond the association with the splice interfering variants in ERAP1. We note that the A allele of rs1057569 (T allele of rs7063) associated with low expression of ERAP1 protein is present in >90% of ERAP1 haplotypes that encode Hap10 and Hap6, and <10% of the other common ERAP1 haplotypes in controls. In the Nigergian (YRI) population (n = 82), the rs2287987 genotype is not in LD with rs1057569 (r2 = 0.009), and not linked to ERAP1 expression (transcript ENST00000443439.6, IRR2 [95% CI] = 0.93[0.65–1.32], P = 0.68).
Full-length ERAP1 sequencing of the risk haplotype
We cloned the full-length coding sequence of ERAP1 from two patients that were heterozygous or homozygous for Hap10 to reveal the amino acid residues for additional reported amino acid positions in ERAP1 (24). Analysis of the coding sequence revealed that both copies (based upon nine individual clones) of Hap10 encoded the ancestral alleles for additional reported amino acid positions (Supplementary Table 5). All detectable transcripts contained a C-terminus identical to the 19-exon transcript ERAP1-202 (ENST00000443439).
Disease-associated ERAP1 displays distinct enzymatic activity.
Considering the association of rs2287987 exceeded variants that govern ERAP1 expression and that rs2287987 has previously been shown to affect trimming properties of recombinant ERAP1 (25,26), we assessed the relative enzymatic activities of ERAP1 haplotypes in vitro. We compared normalized concentrations of ERAP1 protein precipitated from lymphoid cell lines of patients and controls that carry the Hap10 and Hap2, the latter haplotype showing evidence for protection against Birdshot (Table 1). Total cellular ERAP1 protein appeared to have slower rates for enzymatic trimming of the archetypical fluorogenic substrate L-Leucine-4-methylcoumaryl-7-amide (Leu-AMC) with each sequential increase in Hap10 compared with homozygous carriers of Hap2 (Fig. 2G). These results confirm that changes of enzymatic properties in ERAP1 are mediated by haplotypes that carry the C allele of rs2287987 (i.e. Hap10), thereby governing functional consequences for patients with Birdshot.
Non-random distribution of ERAP1-ERAP2 haplotypes in the European population.
ERAP2 encodes two common haplotypes (9): the A allele of rs2248374 tags Haplotype A [HapA], which encodes a canonical protein, while the G allele tags HapB, which usually lacks functional protein (Supplementary Fig. 8). Since ERAP1 is postulated to be complemented by ERAP2 in trimming peptides (27), we hypothesized that the expression of ERAP2 protein could be dependent on the ERAP1 haplotype and that this relationship should be evident in the population. To explore this hypothesis, we used phased genotype data of three populations of 3353 controls to test whether the frequency of HapA of ERAP2 differed between ERAP1 haplotypes encoded along the same chromosome. We observed a non-random distribution of ERAP2-HapA across the ERAP1 haplotypes for each European population (Fig. 3 and Supplementary Table 6). This reflects the complex pattern of LD (D’) between non-synonymous variants in ERAP1 and rs2248374 in ERAP2 (Supplementary Fig. 9). A similar non-random distribution was observed for ERAP1 haplotypes considering five non-synonymous variants in ERAP1 (12) (Supplementary Fig. 10). This demonstrates that the co-occurrence of functional ERAP2 protein is dependent on the ERAP1 haplotype background.
Figure 3.
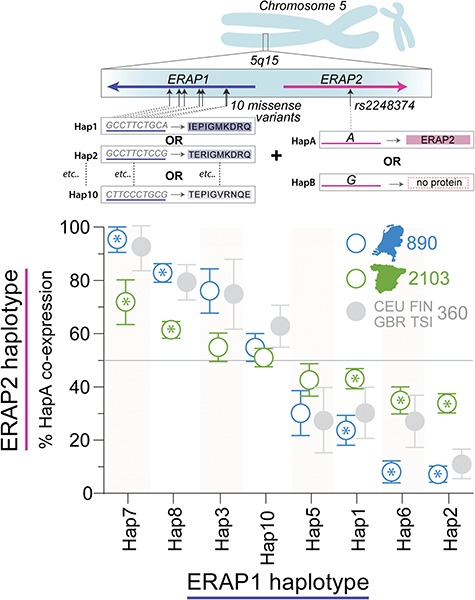
Non-random distribution of the ERAP2-protein coding haplotype across common ERAP1 haplotypes. Ten missense variants in ERAP1 encode eight discrete and common haplotypes, while ERAP2 encodes a common protein-coding haplotype (HapA tagged by the A allele of rs2248374) and non-coding haplotypes (tagged by the G allele). Each chromosome 5 harbors one ERAP1 haplotype and one ERAP2 haplotype. Using phased ERAP1-ERAP2 haplotype data of 890 Dutch controls (1780 phased haplotypes) and 2103 Spanish controls (4206 phased haplotypes), we outlined the percentage of HapA per ERAP1 haplotype on the same chromosome. Error bars indicate the 95% confidence interval based upon 20 000 bootstrap samples. Frequency of HapA of ERAP2 for each of the ERAP1 haplotypes was considered non-random if the observed frequency deviated from an expected 50% using an exact binominal test with [A or G rs2248374 for 8 ERAP1 haplotypes = 0.05/16] P < 2.5 × 10-3 in both the Dutch and Spanish populations with consistent direction of effect. ERAP1 haplotypes that fulfilled these criteria are highlighted by asterisks (for summary statistics see Supplementary Table 6). As a reference, we plotted the ERAP1-ERAP2 haplotype data for the 360 samples from the CEU, FIN, GBR and TSI populations from the 1000 Genomes sample collection used for functional investigation in this study (48).
Discussion
We defined the common ERAP1 and ERAP2 haplotype structure in Birdshot, confirmed the genetic association with rs10044354 (19) and showed that despite strong LD between rs10044354 and splice region variant rs2248374 in ERAP2, the strong eQTL effects of rs10044354 on ERAP2 expression are independent from rs2248374. This finding suggests that a change in expression, and not merely the presence of ERAP2, confers risk for Birdshot and other ERAP2-linked conditions (28). Thus, future treatment strategies to dampen ERAP2 expression may complement current research efforts focused on the pharmacological inhibition of its aminopeptidase activity (29).
We revealed a second independent association for rs2287987 in ERAP1, an SNP that nearly always resides in the common ERAP1 haplotype Hap10 (21). The rs2287987 risk genotype is accompanied by changes in ERAP1 transcript levels and lower protein expression as a result of LD with rs1057569, a strong eQTL for ERAP1 (Supplementary Fig. 6). The functional implications of ERAP1 expression need further investigation, especially since the risk genotype linked to AS results in increased ERAP1 expression (23). However, the association with rs2287987 was independent from rs1057569, suggesting disease modifying effects beyond the expression of ERAP1. Indeed, we ascertained that the Birdshot-associated allele of rs2287987 (349V located near the zinc-binding motif of the enzyme) resides in an haplotype that within cells exhibits distinct enzymatic activity and our data support previous studies indicating that Hap10 can be considered functionally antagonistic to Hap2 and Hap1 (24,26). Also, direct comparison by molecular cloning revealed that Hap10 in Birdshot and a previously investigated 15 non-synonymous variant (i.e. affecting amino acid residues) ERAP1 haplotype, termed *001, represent identical ERAP1 haplotypes (10,24). The *001 (Hap10) was previously shown to provide suboptimal peptide cargo for MHC-I, by reduced trimming of peptides (10,24). Therefore, Hap10 may confer risk by incomplete destruction of immunogenic epitopes that bind HLA-A29. Eye tissues of Birdshot patients are infiltrated by CD8+ T cells, but to date, evidence for their direct involvement is lacking (18). ERAP1 and ERAP2 may perhaps also influence T or NK cells function via Killer immunoglobulin-like receptors, a class of immune receptors that interact with MHC-I that have been genetically linked to Birdshot (30).
Hap10 is also the major risk ERAP1 haplotype for patients with BD and is highly protective against AS (11,12). Hap10 is associated with BD assuming a recessive model (Birdshot, pmeta-recessive = 9.81 × 10−5). Hap1/Hap2 are risk haplotypes for AS and psoriasis (8,12), while being protective for BD and Birdshot. Therefore, Hap10 and Hap2 of ERAP1 play a central role for patients within the MHC-I-opathy cluster: patients with psoriasis have an increased risk of uveitis (31). Patients with AS and BD often have uveitis (11,42), and the ERAP1 associations are much stronger in cases that developed uveitis compared with cases with systemic disease alone (2,32). Although HLA-A29 is also common in some non-European populations (∼7–14% of Caucasian populations is HLA-A29 positive) (18), Hap10 and particularly the Hap10-ERAP2 (rs2287987-rs10044354) haplotype are less common in Asian and African individuals compared with European and, as we show, if present may exhibit different functional effects on ERAP1, which may explain why Birdshot primarily affects patients of European descent (18).
Although 90% of all cases carried either a risk allele for rs2287987 or rs10044354, 50% of all cases were positive for the combined haplotype. A proportion of Birdshot patients may have self-limiting disease, while others will develop irreversible retinal dysfunction and blindness (13). It will be interesting to evaluate the potential relation between ERAP haplotypes and clinical outcome (e.g. electroretinography) in follow-up studies. This was not possible because of the relatively limited sample size, owing to low prevalence of this rare disease (13,14). This limitation also hampered the investigation for association with other SNPs with evidence for more moderate effect size (e.g. rs30187). Regardless, the strongest associations for Birdshot include the rs2287987-rs10044354 haplotype and HLA-A29, a triad of loci that encode functionally tightly related proteins of the class I antigen presentation pathway and warrants further investigation into antigen presentation in human uveitis. Whether ERAP1 and ERAP2 contribute to disease by direct interaction in the endoplasmic reticulum is not known (43–45). In the absence of ERAP2 protein, cells with distinct ERAP1 haplotypes show substantial differences in HLA-A29 peptidome composition, which demonstrates that ERAP1 is able to directly influence HLA-A29 (43). We recently showed that ERAP2 increases the number of 9-mer peptides presented by HLA-A29 (36). This suggests that some peptides bound by ERAP2 may be protected from destruction by ERAP1, which will be further facilitated in a hypoactive ERAP1 background (i.e. Hap10). These results favor our hypothesis that a limited set of uveitogenic peptides drive T cell mediated pathology in Birdshot (18). However, ERAP2 has different peptide preferences compared with ERAP1 (27,29) and MHC-I peptidome studies support separate contributions of Hap10 and ERAP2 in manipulating the peptide composition presented on the cell surface (35). This reflects the lack of LD between the protein-coding ERAP2 haplotype and MHC-I-opathy linked Hap10 and Hap2/Hap1 in the European populations investigated. However, ERAP2 protein expression more often occurs in ERAP1 haplotypes Hap8 and Hap7 (and perhaps Hap3) background, as a result of LD (D’) between SNPs including rs3734016 and rs26618, and rs2248374 (Supplementary Fig. 9). The lack of detailed MHC-I data hampered further investigation of this phenomena, because ERAP2 may not equally contribute to the peptidome of each MHC-I molecule (26,34). However, this would demand a large population study with sufficient power to investigate the ERAP1-ERAP2 haplotypes for distinct MHC-I haplotypes. Such studies will also be key to advance our understanding of ERAP function in inflammation, cancer and less explored fields such as transplantation biology and vaccine efficacy studies.
The original concept of the MHC-I-opathy aims at clustering subsets of clinical phenotypes on the basis of one or multiple unifying molecular fingerprints (1). Here, we explicitly refer to the subgroup of patients that carry risk MHC-I alleles (and ERAP variants in epistasis) when referring to ‘MHC-I-opathy’ patients. Our results in Birdshot further substantiate the existence of such concept. We envision that molecular reclassification based on a patient’s MHC-I and ERAP haplotype may improve clinical decision making as well as drug discovery (e.g. ERAP inhibitors) in the near future. Emerging studies explore this concept, those of which that have linked clinical response to ERAP variants (37–40). Ultimately, understanding how to pharmacologically influence ERAP function (41) may lay the foundation for the design of effective therapeutic strategies to correct ERAP function in accordance with the patient’s molecular fingerprint to tailor the treatment of MHC-I-opathy patients specifically.
Materials and Methods
5q15 variant and haplotype testing in Dutch and Spanish cohorts
This study was performed in compliance with the guidelines of the Declaration of Helsinki and has the approval of the local Institutional Review Board (University Medical Center Utrecht). Genotype data for 13 key variants at 5q15 (Supplementary Table 1) and HLA-A29 status from 84 Dutch cases and 890 Dutch controls from European ancestry were obtained from a previously described discovery collection and the Genome of the Netherlands Project (GoNL) release 4 (19,42). The number of samples is slightly fewer than the Dutch discovery cohort because some samples no longer met the criteria for Birdshot Uveitis (e.g. presence of systemic disease) or failed to pass quality control requirements. Imputation of untyped SNPs was conducted using the GoNL and 1000 Genomes Project Phase 3 data (imputed SNPs with info > 0.95) (43). Disease association testing was performed with PLINK v1.07 (44) using a logistic regression model correcting for the top two principal components and sex (Supplementary Fig. 1). For replication, the genotype data (n = 13) (Supplementary Table 1) and HLA-A29 status were extracted from 46 Spanish cases and 2103 Spanish controls from a previously described non-infectious uveitis collection (45) and tested with a logistic regression model (replication was defined as Bonferroni-corrected P-value for two LD blocks at 5q15 = 0.025). Meta-analysis of Dutch and Spanish cohorts was performed using inverse variance-weighted meta-analysis (46). Common ERAP1 haplotypes (based on 10-non-synonymous variants, which occur in >1% of controls, see Table 1 and Supplementary Fig. 2) were phased using PLINK. The estimated haplotype frequencies for the identified eight common ERAP1 haplotypes matched the frequency of haplotypes estimates of genotype data by SHAPEIT2 (considered the most accurate method available) (47). Therefore, the phasing accuracy of PLINK was considered sufficient for ERAP1 haplotype association testing using logistics regression (Supplementary Fig. 3). The total testing burden for SNP association was estimated at ([13 variants + 8 ERAP1 haplotypes] × 3 models + 4 rs2287987-rs10044354 haplotypes = 67 independent tests) a 1% type I error rate with P < 1.49 × 10−4, which was used as a threshold to prioritize SNP associations calculated in the discovery and meta-analysis. We tested for the rs2287987-rs10044354 haplotype in the 129 HLA-A29-positive Dutch and Spanish cases and 439 HLA-A29 positive Dutch and Spanish controls using a logistic regression model using the glm function n R (v3.3.2) with the command ‘family = binomial’. The OR for each possible genotype was calculated by exponentiating the coefficient from the logistic regression. The low risk rs10044354-CC/rs2287987-TT genotype was set as the baseline (OR = 1) and the other genotype combinations were coded according to a series of dichotomous indicator variables. P-values from the logistic regression below P < 1.49 × 10−4 were considered significant.
Investigation of the distribution of ERAP1 and ERAP2 haplotypes in European populations
The major haplotype of ERAP2 is under balancing selection, where the G allele rs2248374 tags a non-protein coding haplotype (HapB) and the A allele of rs2248374 tags the protein-coding haplotype (HapA) (9). The frequency of the A allele of rs2248374 co-occurring on the same chromosome with each of the eight common ERAP1 haplotypes was determined in the 890 Dutch and 2103 Spanish controls using phased haplotype data using SHAPEIT2 (47), and phased haplotype data were obtained for the British [GBR], Finnish [FIN], Utah residents with Northern and Western European ancestry [CEU] and Tuscany [TSI] populations from the GEUVIDAS consortium (48). The 95% confidence intervals of the observed frequencies were estimated using bootstrap sampling (n = 20 000). We used a two-sided exact binominal test to assess whether the observed frequencies were non-random (deviation from a test probability of 0.5 or the hypothesis that ERAP2 co-occurs in 50% of any particular ERAP1 haplotype). Frequency distribution of ERAP2 for each of the ERAP1 haplotypes was considered non-random at [A or G rs2248374 for eight ERAP1 haplotypes = 0.05/16] P < 2.5 x 10−3 in both the Dutch and Spanish populations (with consistent direction of effect).
Statistical framework for 5q15 eQTL analysis
To investigate the transcriptional regulation of the variants at 5q15, RNA-sequencing data from the GEUVADIS consortium of 360 lymphoid cell lines of European ancestry and 82 African samples (48) were extracted via the recount2 package, a resource for expression data for uniformly processed and quantified RNA-sequencing reads from >70 000 human samples (49). Transcript-level counts of all samples of the GEUVADIS study were scaled using the scale counts function in the recount2 package to take into account differing coverage between samples. Data corresponding to reported transcripts from ERAP1, ERAP2 and LNPEP (n = 25 using Ensembl (22) transcript IDs) were extracted for all samples, of which 4 transcripts were sufficiently expressed and considered for testing the effect of genotype on transcript expression (ERAP1∼201, ENST00000296754.7; ERAP1∼202, ENST00000443439.6; ERAP2∼202, ENST00000437043.7; LNPEP∼201, ENST00000231368.9). The phased genotype data were obtained via the Ensembl portal (22). To interrogate the effect of genotype on local transcript expression, we fitted a generalized linear mixed model (GLMM) using a negative binominal fit of the count data and logarithmic link using the lme4 package in R (v3.3.2) (50), with a fixed effect of patients sex and random effects of laboratory (n = 7) and population (n = 5) of the 1000 Genomes project (formula 1). For pairwise conditioning, the genotype of the studied SNP was added as a covariate into the GLMM (formula 2):
(1): glmer.nb(expression ∼ genotype + sex + (1|Laboratory) + (1|Population))
(2): glmer.nb(expression ∼ genotype + sex + conditional SNP genotype + (1|Laboratory) + (1|Population)).
Genotypes were ranked using their imputed dosage value (a number from 0 to 2, indicating the number of risk alleles for each SNP; Supplementary Table 1) adapted from a previous method (23). Patients homozygous for the G allele of rs2248374 (n = 94) were removed to fit a model for ERAP2 expression in HapA-positive individuals. The incident rate ratio (IRR) is the exponential of the coefficient returned from the GLMM. We used IRR2 as the metric for change in transcript expression (23), which is conceptually similar and close to the fold change in expression (mean count of the transcript) between individuals homozygous for the protective over individuals homozygous for the risk alleles. IRR2 < 0.60 or >1.30 (>30% change in expression) were prioritized for reporting as biologically relevant. Effect size metrics of eQTL analysis from lymphoid cell lines and whole blood from the GTEx project Phase 2 for genes at 5q15 were obtained via the GTEx portal (51).
Western blot analysis of ERAP1 and ERAP2
Human lymphoid cell lines were generated from peripheral blood of Dutch cases recruited at the Departments of Ophthalmology at the University Medical Center Utrecht and Radboud University Medical Center, the Netherlands. Cells were grown in Roswell Park Memorial Institute 1640 medium supplemented with 10% heat-inactivated fetal bovine serum and antibiotics. The following HLA-A29-positive cell lines were obtained from the National Institute of General Medical Sciences Human Genetic Cell Repository at the Coriell Institute for Medical Research: GM19310, GM19397, GM19452, HG00096, HG00113, HG00116, HG01082. Protein lysates (10 μg/lane) of >2 × 106 cells were separated on a 4–20% Mini-PROTEAN TGX gel (Bio-Rad Laboratories, Richmond, CA, USA) and transferred to a Polyvinylidene difluoride membrane and detected using antibodies to ERAP1 and ERAP2 (AF3830, AF2334, both R&D Systems), and α-tubulin (T6199, Sigma), and secondary antibodies conjugated to Horseradish Peroxidase. The ratio of the intensity of the ERAP1 over α-tubulin band was calculated using Image Lab 5.1 (Bio-Rad Laboratories) and grouped according to the distribution of rs2287987 genotype. Details on the Western blot analyses of lymphoid cell lines are outlined under Supplementary Fig. 4.
LNPEP sequencing
We sequenced exon 5–8 of LNPEP from total RNA (cDNA) in steady-state or 24 h stimulated [TLR7/8 (R848) and TLR9 (CpG-B ODN 2006) agonists] lymphoid cell lines of four individuals homozygous for the deletion and four homozygous for the C allele of rs3836862, as determined by sanger sequencing. Detailed methods and used primers are provided in Supplementary Fig. 5.
Full-length ERAP1 cloning and sequencing
RNA was isolated from >2 × 106 cells of two patients with Quick-RNA™ MiniPrep kits (Zymo Research, Irving, CA, USA) and immediately used to generate cDNA with the Transcriptor High Fidelity cDNA synthesis kit (Roche, Mannheim, Germany) using the provided random hexamere primers. ERAP1 was directly amplified from cDNA as described (10). The product was gel-purified, and cloned into the blunt-end cloning vector, pCR-Blunt II, using the Zero Blunt® TOPO® PCR Cloning kit (Life Technologies) or cloned into StarGate® pESG-IBA vectors (IBA Lifesciences), and sequenced.
Cellular ERAP1 enzymatic activity
Total cellular ERAP1 was immunoprecipitated with anti-ERAP1 (5 ug/mL, AF2334, R&D Systems) antibody and protein G-sepharose beads as described (52). Equivalent concentrations of protein samples (normalized by Bradford assay) of purified ERAP1 were incubated with 50 μM Leucine-aminomethyl-coumarin and fluorescent intensity was followed for 30 min with excitation at 365/380 nM and emission at 440/450 nm.
Supplementary Material
Acknowledgements
We would like to thank professor Aniki Rothova for support in evaluation of Birdshot patients.
Conflict of Interest statement. None declared.
Funding
VENI Award from the Netherlands Organization for Scientific Research (NWO; 016.186.006, 016.186.071 to J.J.W.K. and S.L.P.). Miguel Servet fellowship from the Spanish Ministry of Economy, Industry and Competitiveness (CP17/00008 to A.M.). Fischer Stichting, the Landelijke Stichting voor Blinden en Slechtzienden and the Algemene Nederlandse Vereniging ter Voorkoming van Blindheid, which contributed through UitZicht (to J.J.W.K.). The funding organizations had no role in the design or conduct of this research.
References
- 1. McGonagle D., Aydin S.Z., Gül A., Mahr A. and Direskeneli H. (2015) `MHC-I-opathy'-unified concept for spondyloarthritis and Behçet disease. Nat. Rev. Rheumatol., 11, 731–740. [DOI] [PubMed] [Google Scholar]
- 2. Kirino Y., Bertsias G., Ishigatsubo Y., Mizuki N., Tugal-Tutkun I., Seyahi E., Ozyazgan Y., Sacli F.S., Erer B., Inoko H. et al. (2013) Genome-wide association analysis identifies new susceptibility loci for Behçet's disease and epistasis between HLA-B*51 and ERAP1. Nat. Genet., 45, 202–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cortes A., Pulit S.L., Leo P.J., Pointon J.J., Robinson P.C., Weisman M.H., Ward M., Gensler L.S., Zhou X., Garchon H.J. et al. (2015) Major histocompatibility complex associations of ankylosing spondylitis are complex and involve further epistasis with ERAP1. Nat. Commun., 6, 7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans D.M., Spencer C.C., Pointon J.J., Su Z., Harvey D., Kochan G., Oppermann U., Dilthey A., Pirinen M., Stone M.A. et al. (2011) Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet., 43, 761–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tang H., Jin X., Li Y., Jiang H., Tang X., Yang X., Cheng H., Qiu Y., Chen G., Mei J. et al. (2014) A large-scale screen for coding variants predisposing to psoriasis. Nat. Genet., 46, 45–50. [DOI] [PubMed] [Google Scholar]
- 6. Giza M., Koftori D., Chen L. and Bowness P. (2018) Is Behçet's disease a 'class 1-opathy'? The role of HLA-B*51 in the pathogenesis of Behçet's disease. Clin. Exp. Immunol., 191, 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Yazici H., Seyahi E., Hatemi G. and Yazici Y. (2018) Behçet syndrome: a contemporary view. Nat. Rev. Rheumatol., 14, 107–119. [DOI] [PubMed] [Google Scholar]
- 8. López de Castro J.A., Alvarez-Navarro C., Brito A., Guasp P., Martín-Esteban A. and Sanz-Bravo A. (2016) Molecular and pathogenic effects of endoplasmic reticulum aminopeptidases ERAP1 and ERAP2 in MHC-I-associated inflammatory disorders: towards a unifying view. Mol. Immunol., 77, 193–204. [DOI] [PubMed] [Google Scholar]
- 9. Andrés A.M., Dennis M.Y., Kretzschmar W.W., Cannons J.L., Lee-Lin S.Q., Hurle B., NISC Comparative Sequencing Program, Schwartzberg P.L., Williamson S.H., Bustamante C.D. et al. (2010) Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet., 6, e1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reeves E., Edwards C.J., Elliott T. and James E. (2013) Naturally occurring ERAP1 haplotypes encode functionally distinct alleles with fine substrate specificity. J. Immunol., 191, 35–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Takeuchi M., Ombrello M.J., Kirino Y., Erer B., Tugal-Tutkun I., Seyahi E., Özyazgan Y., Watts N.R., Gül A., Kastner D.L. and Remmers E.F. (2016) A single endoplasmic reticulum aminopeptidase-1 protein haplotype is a strong risk factor for Behçet's disease in HLA-B*51 carriers. Ann. Rheum. Dis., 75, 2208–2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Roberts A.R., Appleton L.H., Cortes A., Vecellio M., Lau J., Watts L., Brown M.A. and Wordsworth P. (2017) ERAP1 association with ankylosing spondylitis is attributable to common genotypes rather than rare haplotype combinations. Proc. Natl. Acad. Sci. U. S. A., 114, 558–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Minos E., Barry R.J., Southworth S., Folkard A., Murray P.I., Duker J.S., Keane P.A. and Denniston A.K. (2016) Birdshot chorioretinopathy: current knowledge and new concepts in pathophysiology, diagnosis, monitoring and treatment. Orphanet. J. Rare. Dis., 11, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shah K.H., Levinson R.D., Yu F., Goldhardt R., Gordon L.K., Gonzales C.R., Heckenlively J.R., Kappel P.J. and Holland G.N. (2005) Birdshot chorioretinopathy. Surv. Ophthalmol., 50, 519–541. [DOI] [PubMed] [Google Scholar]
- 15. Yang P. and Foster C.S. (2013) Interleukin 21, interleukin 23, and transforming growth factor β1 in HLA-A29-associated birdshot retinochoroidopathy. Am. J. Ophthalmol., 156, 400–406.e2. [DOI] [PubMed] [Google Scholar]
- 16. Molins B., Mesquida M., Llorenç V., Sainz de la Maza M. and Adán A. (2016) Elevated serum immune mediators and subclinical inflammation in HLA-A29-associated birdshot chorioretinopathy. Ocul. Immunol. Inflamm., 24, 647–652. [DOI] [PubMed] [Google Scholar]
- 17. Kuiper J.J., Mutis T., Jager W., Groot-Mijnes J.D. and Rothova A. (2011) Intraocular interleukin-17 and proinflammatory cytokines in HLA-A29-associated birdshot chorioretinopathy. Am. J. Ophthalmol., 152, 177–182.e1. [DOI] [PubMed] [Google Scholar]
- 18. Kuiper J., Rothova A., Boer J. and Radstake T. (2015) The immunopathogenesis of birdshot chorioretinopathy; a bird of many feathers. Prog. Retin. Eye. Res., 44, 99–110. [DOI] [PubMed] [Google Scholar]
- 19. Kuiper J.J., Van Setten J., Ripke S., Van 'T Slot R., Mulder F., Missotten T., Baarsma G.S., Francioli L.C., Pulit S.L., De Kovel C.G. et al. (2014) A genome-wide association study identifies a functional ERAP2 haplotype associated with birdshot chorioretinopathy. Hum. Mol. Genet., 23, 6081–6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Herbort C.P. Jr., Pavésio C., LeHoang P., Bodaghi B., Fardeau C., Kestelyn P., Neri P., Papadia M. et al. (2017) Why birdshot retinochoroiditis should rather be called 'HLA-A29 uveitis'? Br. J. Ophthalmol., 101, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ombrello M.J., Kastner D.L. and Remmers E.F. (2015) Endoplasmic reticulum-associated amino-peptidase 1 and rheumatic disease: genetics. Curr. Opin. Rheumatol., 27, 349–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G. et al. (2018) Ensembl 2018. Nucleic. Acids. Res., 46, D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanson A.L., Cuddihy T., Haynes K., Loo D., Morton C.J., Oppermann U., Leo P., Thomas G.P., Lê Cao K.A., Kenna T.J. et al. (2018) Genetic variants in ERAP1 and ERAP2 associated with immune-mediated diseases influence protein expression and the isoform profile. Arthritis Rheumatol., 70, 255–265. [DOI] [PubMed] [Google Scholar]
- 24. Reeves E., Colebatch-Bourn A., Elliott T., Edwards C.J. and James E. (2014) Functionally distinct ERAP1 haplotype combinations distinguish individuals with ankylosing spondylitis. Proc. Natl. Acad. Sci. U. S. A., 111, 17594–17599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Seregin S.S., Rastall D.P., Evnouchidou I., Aylsworth C.F., Quiroga D., Kamal R.P., Godbehere-Roosa S., Blum C.F., York I.A., Stratikos E. et al. (2013) Endoplasmic reticulum aminopeptidase-1 alleles associated with increased risk of ankylosing spondylitis reduce HLA-B27 mediated presentation of multiple antigens. Autoimmunity, 46, 497–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guasp P., Barnea E., González-Escribano M.F., Jiménez-Reinoso A., Regueiro J.R., Admon A. and López de Castro J.A. (2017) The Behçet's disease-associated variant of the aminopeptidase ERAP1 shapes a low-affinity HLA-B*51 peptidome by differential subpeptidome processing. J. Biol. Chem., 292, 9680–9689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Saveanu L., Carroll O., Lindo V., Del Val M., Lopez D., Lepelletier Y., Greer F., Schomburg L., Fruci D., Niedermann G. et al. (2005) Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat. Immunol., 6, 689–697. [DOI] [PubMed] [Google Scholar]
- 28. Robinson P.C., Costello M.E., Leo P., Bradbury L.A., Hollis K., Cortes A., Lee S., Joo K.B., Shim S.C., Weisman M. et al. (2015) ERAP2 is associated with ankylosing spondylitis in HLA-B27-positive and HLA-B27-negative patients. Ann. Rheum. Dis., 74, 1627–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mpakali A., Giastas P., Deprez-Poulain R., Papakyriakou A., Koumantou D., Gealageas R., Tsoukalidou S., Vourloumis D., Mavridis I.M., Stratikos E. et al. (2017) Crystal structures of ERAP2 complexed with inhibitors reveal pharmacophore requirements for optimizing inhibitor potency. A.C.S. Med. Chem. Lett., 8, 333–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Levinson R.D., Du Z., Luo L., Monnet D., Tabary T., Brezin A.P., Zhao L., Gjertson D.W., Holland G.N., Reed E.F. et al. (2008) Combination of KIR and HLA gene variants augments the risk of developing birdshot chorioretinopathy in HLA-A*29-positive individuals. Genes. Immun., 9, 249–258. [DOI] [PubMed] [Google Scholar]
- 31. Chi C.C., Tung T.H., Wang J., Lin Y.S., Chen Y.F., Hsu T.K. and Wang S.H. (2017) Risk of uveitis among people with psoriasis: a nationwide cohort study. JAMA. Ophthalmol., 135, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Robinson P.C., Leo P.J., Pointon J.J., Harris J., Cremin K., Bradbury L.A., Wellcome Trust Case Control Consortium, Australasian Osteoporosis Genetics Consortium (AOGC), Stebbings S., Harrison A.A. et al. (2016) The genetic associations of acute anterior uveitis and their overlap with the genetics of ankylosing spondylitis. Genes. Immun., 17, 46–51. [DOI] [PubMed] [Google Scholar]
- 33. Alvarez-Navarro C., Martín-Esteban A., Barnea E., Admon A. and López de Castro J.A. (2015) Endoplasmic reticulum aminopeptidase 1 (ERAP1) polymorphism relevant to inflammatory disease shapes the peptidome of the birdshot chorioretinopathy-associated HLA-A*29:02 antigen. Mol. Cell. Proteomics., 14, 1770–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen H., Li L., Weimershaus M., Evnouchidou I., Endert P. and Bouvier M. (2016) ERAP1-ERAP2 dimers trim MHC I-bound precursor peptides; implications for understanding peptide editing. Sci. Rep., 12, 28902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Martín-Esteban A., Sanz-Bravo A., Guasp P., Barnea E., Admon A. and López de Castro J.A. (2017) Separate effects of the ankylosing spondylitis associated ERAP1 and ERAP2 aminopeptidases determine the influence of their combined phenotype on the HLA-B*27 peptidome. J. Autoimmun., 79, 28–38. [DOI] [PubMed] [Google Scholar]
- 36. Sanz-Bravo A., Martin-Esteban A., Kuiper J.J.W., García-Peydró M., Barnea E., Admon A. and López de Castro J.A. (2018) Allele-specific alterations in the peptidome underlie the joint association of HLA-A*29:02 and endoplasmic reticulum aminopeptidase 2 (ERAP2) with birdshot chorioretinopathy. Mol. Cell. Proteomics., 8, 1564–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Nossent J.C., Johnsen S. and Bakland G. (2016) The influence of ERAP1 gene variants on clinical phenotype in ankylosing spondylitis. Scand. J. Rheumatol., 45, 474–479. [DOI] [PubMed] [Google Scholar]
- 38. Masouri S., Stefanaki I., Ntritsos G., Kypreou K.P., Drakaki E., Evangelou E., Nicolaidou E., Stratigos A.J. and Antoniou C. (2016) A pharmacogenetic study of psoriasis risk variants in a greek population and prediction of responses to anti-TNF-α and anti-IL-12/23 agents. Mol. Diagn. Ther., 20, 221–225. [DOI] [PubMed] [Google Scholar]
- 39. Ozen G., Deniz R., Eren F., Erzik C., Unal A.U., Yavuz S., Aydin S.Z., Inanc N., Direskeneli H. and Atagunduz P. (2017) Association of ERAP1, IL23R and PTGER4 polymorphisms with radiographic severity of ankylosing spondylitis. Open. Rheumatol. J., 16, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Baeten D., Baraliakos X., Braun J., Sieper J., Emery P., Heijde D., McInnes I., Laar J.M., Landewé R., Wordsworth P. et al. (2013) Anti-interleukin-17A monoclonal antibody secukinumab in treatment of ankylosing spondylitis: a randomised, double-blind, placebo-controlled trial. Lancet, 382, 1705–1713. [DOI] [PubMed] [Google Scholar]
- 41. Georgiadis D., Mpakali A., Koumantou D. and Stratikos E. (2018) Inhibitors of ER aminopeptidase 1 and 2: from design to clinical application. Curr. Med. Chem., epub. [DOI] [PubMed] [Google Scholar]
- 42. Genome of the Netherlands Consortium (2014) Whole-genome sequence variation, population structure and demographic history of the Dutch population. Nat. Genet., 46, 818–825. [DOI] [PubMed] [Google Scholar]
- 43. 1000 Genomes Project Consortium (2015) A global reference for human genetic variation. Nature, 526, 68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., Bakker P.I., Daly M.J. et al. (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet., 81, 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Márquez A., Cordero-Coma M., Martín-Villa J.M., Gorroño-Echebarría M.B., Blanco R., Díaz V.D., Del Rio M.J., Blanco A., Olea J.L., Cordero Y. et al. (2017) New insights into the genetic component of non-infectious uveitis through an Immunochip strategy. J. Med. Genet., 54, 38–46. [DOI] [PubMed] [Google Scholar]
- 46. Evangelou E. and Ioannidis J.P. (2013) Meta-analysis methods for genome-wide association studies and beyond. Nat. Rev. Genet., 14, 379–389. [DOI] [PubMed] [Google Scholar]
- 47. Delaneau O., Marchini J. and Zagury J.F. (2011) A linear complexity phasing method for thousands of genomes. Nat. Methods, 9, 179–181. [DOI] [PubMed] [Google Scholar]
- 48. Lappalainen T., Sammeth M., Friedländer M.R., 't Hoen P.A., Monlong J., Rivas M.A., Gonzàlez-Porta M., Kurbatova N., Griebel T., Ferreira P.G. et al. (2013) Transcriptome and genome sequencing uncovers functional variation in humans. Nature, 501, 506–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Collado-Torres L., Nellore A., Kammers K., Ellis S.E., Taub M.A., Hansen K.D., Jaffe A.E., Langmead B. and Leek J.T. (2017) Reproducible RNA-seq analysis using recount 2. Nat. Biotechnol., 35, 319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bates D., Mächler M., Bolker B. and Walker S. (2015) Fitting linear mixed-effects models using lme4. J. Stat. Softw., 67, https://www.jstatsoft.org/article/view/v067i01/0. [Google Scholar]
- 51. GTEx Consortium (2017) Genetic effects on gene expression across human tissues. Nature, 550, 204–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fruci D., Ferracuti S., Limongi M.Z., Cunsolo V., Giorda E., Fraioli R., Sibilio L., Carroll O., Hattori A., Endert P.M. et al. (2006) Expression of endoplasmic reticulum aminopeptidases in EBV-B cell lines from healthy donors and in leukemia/lymphoma, carcinoma, and melanoma cell lines. J. Immunol., 176, 4869–4879. [DOI] [PubMed] [Google Scholar]
- 53. Pruim R.J., Welch R.P., Sanna S., Teslovich T.M., Chines P.S., Gliedt T.P., Boehnke M., Abecasis G.R. and Willer C.J. (2010) LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics, 26, 2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


