ABSTRACT
The 19q13 allele rs11672691 has been reproducibly found in association with aggressive form of prostate cancer, yet the underlying mechanism remains totally unknown. We have recently uncovered a mechanism by which rs11672691 influenced a novel oncogenic regulatory circuit, including HOXA2, PCAT19 and CEACAM21, thereby contributing to prostate cancer aggressiveness.
KEYWORDS: 19q13 locus, rs11672691, HOXA2-PCAT19-CEACAM21 regulatory circuit, aggressive prostate cancer, integrated genomic analysis
Prostate cancer remains the most common noncutaneous malignancy, and the second most common cancer-related death among men in the Western world1. Among the risk factors for prostate cancer, the genetic heritability estimates were 57%2. Genome-wide association studies (GWAS) have thus far identified 150 susceptibility single nucleotide polymorphisms (SNPs), together captured 28.4% of the familial relative risk in prostate cancer3. While the vast majority of these SNPs fall within noncoding genomic regions, making it a daunting challenge to interpret, ongoing efforts have sought to uncover the underlying molecular mechanism for the SNPs residing in gene regulatory elements4-6.
Management of early-stage prostate cancer is usually effective, whereas the advanced stage aggressive forms are difficult to treat. Variants associated with prostate cancer susceptibility have been relatively well studied, however, few loci linked to aggressive disease are investigated. The 19q13 allele rs11672691 within the intronic region of a long non-coding RNA (lncRNA) gene, prostate cancer associated transcript 19 (PCAT19) was discovered to be associated with aggressive prostate cancer in two independent large case-only stidues7,8. More recently9, we independently validated this association and defined an elegant biological mechanism underlying the 19q13 locus, therefore likely informing aggressive prostate cancer poor prognosis and treatment (see Figure 1).
Figure 1.
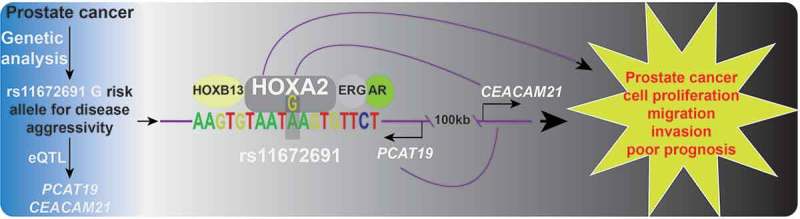
Molecular and clinical underpinnings of the aggressive prostate cancer risk 19q13 locus. Previous GWASs and our large-scale independent genetic analysis revealed an association of the 19q13 allele rs11672691 G with prostate cancer aggressiveness. The rs11672691 G allele is strongly correlated with elevated expression of CEACAM21 and the lncRNA PCAT19 in an eQTL analysis. Subsequently, HOXA2 was found to preferentially occupy a PCAT19 intronic enhancer carrying the G allele of rs11672691, which together with other transcription factors AR, HOXB13, and ERG contributed to enhanced expression of PCAT19 and CEACAM21, thereby promote prostate cancer cell proliferation and aggressiveness. In the clinical setting, the rs11672691 genotype, HOXA2, PCAT19, and CEACAM21 expression were discovered as a potential biomarker in prostate cancer prognosis.
To get more insights into how the 19q13 allele impacts aggressive prostate cancer, we first performed an expression quantitative trait locus (eQTL) analysis in Swedish, TCGA, and Wisconsin cohorts, leading to the discovery that the rs11672691 G allele is significantly associated with the elevated expression levels of carcinoembryonic antigen related cell adhesion molecule 21 (CEACAM21) and PCAT19 (see Figure 1). Both genes are new to prostate cancer. The identification of novel genes expands a possible mechanism by which these genes account for prostate cancer. We thus knocked down CEACAM21 or PCAT19 in multiple PCa cell lines, and observed that the attenuated levels of CEACAM21 or PCAT19 expression markedly reduce cell proliferation, migration, and invasion. Accordingly, PCAT19 or CEACAM21 overexpression promote prostate cancer cell growth, and metastatic capacity9,10. Moreover, PCAT19 and CEACAM21 highly expressed in PCa tumor specimens as compared to normal tissues, and their high expression levels positively correlated with shortened disease-free survival of prostate cancer patients9,10, demonstrating that PCAT19 and CEACAM21 are two plausible causal genes explaining the association of the 19q13 locus with aggressive prostate cancer.
These findings also raise the question if the noncoding genomic variant rs11672691 contributes to the regulation of its eQTL genes. We thereby conducted a genome-wide analysis of epigenome and transcription factor binding data determined by chromatin immunoprecipitation sequencing (ChIP-seq). This analysis in combination with computational prediction using transcription factor DNA-binding position weight matrix data, led to the finding of the rs11672691 region as an active enhancer with epigenetic marks, H3K4me1/2 and H3K27ac, and occupancy of the transcription factors androgen receptor (AR), homeobox B13 (HOXB13), ETS-related gene (ERG), and homeobox A2 (HOXA2). Intriguingly, rs11672691 was mapped within a HOXA2 DNA-binding motif where the aggressive G allele is likely to increase the binding affinity of HOXA2 as compared to the A allele (see Figure 1). We further confirmed this enhanced DNA-binding of HOXA2 to the rs11672691 G risk allele containing sequence in vitro and in vivo.
Thus, the rs11672691 enhancer is a highly occupied target region bound with several transcription factors. In contrast to the well-studied regulators AR, HOXB13, and ERG in prostate cancer, HOXA2 is brand new. We thus sought to explore the function of HOXA2 in prostate cancer. This analysis revealed that HOXA2 is an androgen-responsive gene, and essential for prostate cancer cell growth and invasiveness. Furthermore, clinical data showed that HOXA2 mRNA levels greatly increased in primary and metastatic specimens of prostate cancer patients, and high HOXA2 levels served as an independent predictor of prostate cancer relapse and overall survival (see Figure 1). Surprisingly, we found that HOXA2 levels were significantly predictive of disease relapse in prostate cancer cases with low intermediate risk (Gleason score 7), a subcohort with the most uncertainty in deciding the right balance between active surveillance and immediate treatment. Given that the rs11672691 region is a targeted enhancer and a motif disruptor of HOXA2, we further evaluate if HOXA2 regulates the expression of the rs11672691-associated genes. We thus performed a series of chromatin and gene knockdown assays, and concluded that both PCAT19 and CEACAM21 are the direct target genes of HOXA2. In addition, the lncRNA PCAT19 possesses enhancer-like function in regulating CEACAM21 expression. To prove how the rs11672691 enhancer or PCAT19 regulate CEACAM21 over a 100kb interval, we applied quantitative chromosome conformation capture assays (3C-qPCR), and revealed a direct chromatin loop formation between PCAT19 and CEACAM21 loci (see Figure 1).
These findings suggest a likely model of rs11672691-mediated HOXA2 in regulating the expression of PCAT19 and CEACAM21 through a long-range chromatin interaction, raising the possibility if rs11672691 plays a direct role in the process. We therefore applied the CRISPR/Cas9 genome-editing tool to convert the genotype of rs11672691 G/A in 22Rv1 cells into G/G or A/A. Our follow-up analyses show that, among the three types of cells, the rs11672691 G/G cell line indicates the highest mRNA levels of PCAT19 and CEACAM21. Consistently, HOXA2 shows the most strong chromatin occupancy at rs11672691 enhancer in the G/G cell line. Unexpectedly, we observed that the G/G cells phenotypically appear to be aggressive, and indicate higher levels of proliferation and migration potential than that of the other two types of cell lines. Interestingly, in the clinical setting, the prostate cancer patients carrying rs11672691 G allele indicate increased risk of biochemical recurrence. Furthermore, the G genotype of rs11672691 can synergize with PCAT19 or CEACAM21 expression data to improve the predictive values in prostate cancer prognosis.
Thus, our combination of intense genetic, functional genomic and clinical data analyses give insights into the biological mechanisms underlying the 19q13 aggressive prostate cancer risk locus, highlighting value for potential clinical translation and specifically a rs11672691-orchestrated oncogenic regulatory circuit, including HOXA2, PCAT19 and CEACAM21 as potential biomarkers to improve patient risk stratification and management. In the future, it would be interesting to identify drugs for oncogene such as HOXA2, CEACAM21, and PCAT19, and to test their efficacy on the treatment of aggressive prostate cancer. We shall further validate our findings of the 19q13 allele rs11672691 mediated oncogenic regulatory circuit in the genetically modified mouse and prostate cancer patient-derived tumor graft models.
Funding Statement
This work was supported by the Academy of Finland [284618].
Acknowledgments
This work was supported by Academy of Finland (284618 and 279760), Jane and Aatos Erkko Foundation, and Finnish Cancer Foundation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
References
- 1.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F.. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2012;136:E359–E386. PMID: 25220842. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Mucci LA, Hjelmborg JB, Harris JR, Czene K, Havelick DJ, Scheike T, Graff RE, Holst K, Möller S, Unger RH, et al. Familial risk and heritability of cancer among twins in nordic countries. JAMA. 2016; 315(1): 68–76. doi: 10.1001/jama.2015.17703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schumacher FR, Al Olama AA, Berndt SI, Benlloch S, Ahmed M, Saunders EJ, Dadaev T, Leongamornlert D, Anokian E, Cieza-Borrella C, et al. Association analyses of more than 140,000 men identify 63 new prostate cancer susceptibility loci. Nat Genet. 2018;50::928–936. PMID:29892016. doi: 10.1038/s41588-018-0142-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Q, Whitington T, Gao P, Lindberg JF, Yang Y, Sun J, Väisänen MR, Szulkin R, Annala M, Yan J, et al. A prostate cancer susceptibility allele at 6q22 increases RFX6 expression by modulating HOXB13 chromatin binding. Nat Genet. 2014;46(2):126–135. PMID: 24390282. doi: 10.1038/ng.2862. [DOI] [PubMed] [Google Scholar]
- 5.Whitington T, Gao P, Song W, Ross-Adams H, Lamb AD4, Yang Y, Svezia I, Klevebring D, Mills IG, Karlsson R, et al. Gene regulatory mechanisms underpinning prostate cancer susceptibility. Nat Genet. 2016;48(4):387–397. PMID: 26950096. doi: 10.1038/ng.3523. [DOI] [PubMed] [Google Scholar]
- 6.Zhang P, Xia JH, Zhu J, Gao P, Tian YJ, Du M, Guo YC, Suleman S, Zhang Q, Kohli M, et al. High-throughput screening of prostate cancer risk loci by single nucleotide polymorphisms sequencing. Nat Commun. 2018;22;9(1):2022 PMID: 29789573. doi: 10.1038/s41467-018-04451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amin A, Olama A, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, Giles GG, Severi G, Neal DE, et al. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;15;22(2):408–415. PMID: 23065704. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shui IM, Lindström S, Kibel AS, Berndt SI, Campa D, Gerke T, Penney KL, Albanes D, Berg C, Bueno-de-Mesquita HB, et al. Prostate cancer (PCa) risk variants and risk of fatal PCa in the National Cancer Institute breast and prostate cancer cohort consortium. Eur Urol. 2014. June;65(6):1069–1075. doi: 10.1016/j.eururo.2013.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao P, Xia JH, Sipeky C, Dong XM, Zhang Q, Yang Y, Zhang P, Cruz SP, Zhang K, Zhu J, et al. Biology and clinical implications of the 19q13 aggressive prostate cancer susceptibility locus. Cell. 2018; 174(3): 576–589. doi: 10.1016/j.cell.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua JT, Ahmed M, Guo H, Zhang Y, Chen S, Soares F, Lu J, Zhou S, Wang M, Li H, et al. Risk SNP-mediated promoter-enhancer switching drives prostate cancer through lncRNA PCAT19. Cell. 2018; 26;174(3): 564–575. doi: 10.1016/j.cell.2018.06.014. [DOI] [PubMed] [Google Scholar]


