Abstract
Summary: LS-SNP/PDB is a new WWW resource for genome-wide annotation of human non-synonymous (amino acid changing) SNPs. It serves high-quality protein graphics rendered with UCSF Chimera molecular visualization software. The system is kept up-to-date by an automated, high-throughput build pipeline that systematically maps human nsSNPs onto Protein Data Bank structures and annotates several biologically relevant features.
Availability: LS-SNP/PDB is available at http://ls-snp.icm.jhu.edu/ls-snp-pdb and via links from protein data bank (PDB) biology and chemistry tabs, UCSC Genome Browser Gene Details and SNP Details pages and PharmGKB Gene Variants Downloads/Cross-References pages.
Contact: karchin@jhu.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Single nucleotide polymorhpisms (SNPs) are the most common type of inter-individual human genetic variation (Goodstadt and Ponting, 2001), and the number of human SNPs catalogued in NCBI's dbSNP database (Sherry et al., 2001) has grown exponentially over the past few years. Build 129 of dbSNP (June 10, 2008) contains over 1.4 million uniquely mapped human SNPs, of which over 90 000 are non-synonymous. Because SNPs can impact an individual's susceptibility to disease and sensitivity to drugs (Evans and Relling, 1999; Sunyaev et al., 2001), there is an increasing interest in computational tools to identify SNPs that may affect molecular function. We have developed a resource to contribute to this effort, using genome-wide mapping of nsSNPs onto experimentally determined protein structures.
LS-SNP/PDB annotates all human SNPs that produce an amino acid change in a protein structure in PDB (Deshpande et al., 2005), using features of their local structural environment, putative binding interactions and evolutionary conservation. For nsSNPs that are accessible to solvent, we provide two colorized views of the protein surface to represent: (i) evolutionary conservation among homologous proteins and (ii) electrostatic potential. The presence of a nsSNP in a highly conserved surface patch or a charged surface patch suggests possible biological importance. These annotations allow users to quickly scan a large number of nsSNPs of interest and prioritize those with higher likelihood of impacting normal protein activities.
This work builds on an earlier version of LS-SNP (Karchin et al., 2005) that mapped human nsSNPs onto protein homology models. It features completely redesigned pipeline software, fully automatic builds, weekly incremental updates, new annotations, high-quality protein images for molecular visualization and a new web interface.
2 SYSTEMS AND METHODS
The pipeline integrates data from a variety of sources and a new build is run for each new release of the UCSC Genome Browser (Karolchik et al., 2008) SNP track as follows: Users can access the database with a query form and select nsSNPs by dbSNP, HUGO gene, UniProtKB, KEGG Pathway ID, or PDB ID(s), genomic region or cytogenic band. Queries can be further restricted by: solvent accessibility (exposed, intermediate, buried); secondary structure (helix, strand, coil), proximity to ligands or domain interfaces; severity of amino acid change (radical, moderate, mild, conservative); evolutionary conservation (low, medium, high); and PDB structure type (X-ray, NMR).
mapping of all human UniProtKB (Wu et al., 2006) protein sequences onto genomic DNA to identify the (unknown) codon that yielded each amino acid residue. Mapping is done by aligning the UniProtKB proteins to all human mRNAs in the Genome Browser and selecting the best match;
taking the genomic address of each SNP from dbSNP (currently from NCBI Build 36.1/hg18) and finding SNPs that alter an amino acid residue;
using alignments of UniProtKB and PDB protein sequences to map each nsSNP onto a PDB structure;
annotating nsSNPs by structural location, secondary structure, solvent accessibliity, evolutionary conservation, electrostatic potential and proximity to small molecule ligands and domain interfaces;
creating high-quality protein images for each nsSNP.
3 IMPLEMENTATION
The LS-SNP/PDB build controller is written in Python and runs on a Fedora Core Linux server and SGE-managed cluster of compute nodes. Data are stored in a mySQL relational database. The web site is implemented in Java/J2EE (JSP and servlets) and run on a tomcat webserver. Details are provided in Supplementary Material.
4 DISCUSSION
We illustrate how LS-SNP/PDB can identify biologically interesting nsSNPs with two examples.
Filtered searching: LS-SNP/PDB query search for SNPs in the q13 band of chromosome 11, filtered by moderate/radical amino acid change, medium/high conservation and domain interface proximity yielded rs116049121 (aspartic acid to alanine), a SNP in the barrier-to-autointegration factor (BAF) protein, which protects DNA from retrovirus integration (Zheng et al., 2000) (Fig. 1). It occurs at an evolutionarily conserved region and is near an acidic surface patch. Functional studies have shown that the aspartic acid is a critical residue in a network of salt bridges, which connect two halves of the BAF homodimer (Umland et al., 2000). Alanine likely decreases the stability of functional BAF, possibly improving resistance of indviduals with this nsSNP to viral infection.
Fig. 1.
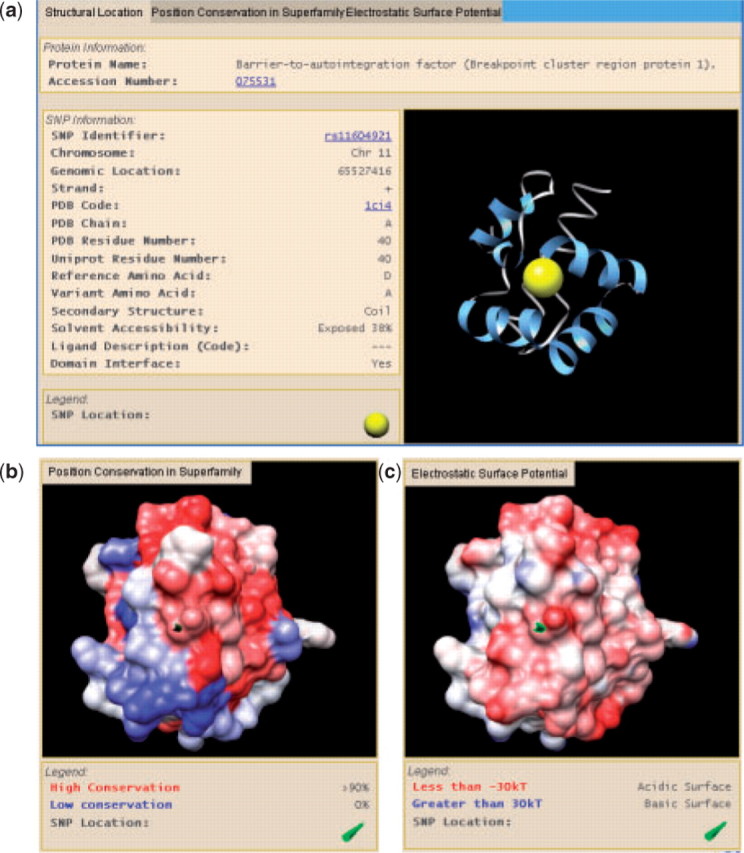
Screenshot of rs11604921. (a) Location in PDB 1ci4A. (b) Protein surface rendering colored by conservation; and (c) colored by electrostatic potential.
GWAS SNPs: a recent genome-wide association study found several SNPs in intercellular adhesion molecule 1 to be significantly associated with lower soluble protein concentration in plasma (a vascular disease biomarker) (Pare et al., 2008). The rs1799969 (glycine to arginine) is in an ICAM-1 domain that binds the integrin MAC-1 (Diamond et al., 1991). This domain has been shown to bind a charged surface on MAC-1 (Yang et al., 2007), and one study suggested that rs1799969 weakens this interaction (Ponthieux et al., 2003). LS-SNP/PDB electrostatic potential view of PDB ID 1p53 shows that it is located on an acidic surface patch, thus partial neutralization of the charged region by arginine could explain weakened binding. Alternatively, there may be a steric requirement for glycine at this position (a turn in the protein backbone). Relative importance of electrostatics and sterics for MAC-1 binding could be experimentally tested by comparing stability and MAC-1 binding of wild-type ICAM-1 with R241 and E241 mutants.
5 CONCLUSION
Plans for LS-SNP include annotation of small deletions, insertions and inversions, integration of high-quality homology models, machine learning classification of variants and handling of custom inputs, such as rare variants not in dbSNP. We will incorporate more detailed information about evolutionary conservation and electrostatic potential at each SNP site. In the near future, we will be applying the pipeline to somatic DNA sequence alterations found in the exons of tumor genomes.
Conflict of Interest: none declared.
Supplementary Material
REFERENCES
- Deshpande N, et al. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond MS, et al. Binding of the integrin Mac-1 (CD11b/CD18) to the third immunoglobulin-like domain of ICAM-1 (CD54) and its regulation by glycosylation. Cell. 1991;65:961–971. doi: 10.1016/0092-8674(91)90548-d. [DOI] [PubMed] [Google Scholar]
- Evans WE, Relling MV. Pharmacogenomics: translating functional genomics into rational therapeutics. Science. 1999;286:487–491. doi: 10.1126/science.286.5439.487. [DOI] [PubMed] [Google Scholar]
- Goodstadt L, Ponting CP. Sequence variation and disease in the wake of the draft human genome. Hum. Mol. Genet. 2001;10:2209–2214. doi: 10.1093/hmg/10.20.2209. [DOI] [PubMed] [Google Scholar]
- Karchin R, et al. LS-SNP: large-scale annotation of coding non-synonymous SNPs based on multiple information sources. Bioinformatics. 2005;21:2814–2820. doi: 10.1093/bioinformatics/bti442. [DOI] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC genome browser database: update 2008. Nucleic Acids Res. 2008;36:D773–D779. doi: 10.1093/nar/gkm966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pare G, et al. Novel association of ABO histo-blood group antigen with soluble ICAM-1: results of a genome-wide association study of 6,578 women. PLoS Genet. 2008;4:e1000118. doi: 10.1371/journal.pgen.1000118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ponthieux A, et al. Association between Gly241Arg ICAM-1 gene polymorphism and serum sICAM-1 concentration in the Stanislas cohort. Eur. J. Hum. Genet. 2003;11:679–686. doi: 10.1038/sj.ejhg.5201033. [DOI] [PubMed] [Google Scholar]
- Sherry ST, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunvaev S, et al. Integration of genome data and protein structures:prediction of protein folds, protein interactions and “molecular phenotypes” of single nucleotide polymorphisms. Curr. Opin. Struct. Biol. 2001;11:123–130. doi: 10.1016/s0959-440x(00)00175-5. [DOI] [PubMed] [Google Scholar]
- Umland TC, et al. Structural basis of DNA bridging by barrier-to-autointegration factor. Biochemistry. 2000;39:9130–9138. doi: 10.1021/bi000572w. [DOI] [PubMed] [Google Scholar]
- Wu CH, et al. The Universal Protein Resource (UniProt): an expanding universe of protein information. Nucleic Acids Res. 2006;34:D187–D191. doi: 10.1093/nar/gkj161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, et al. Interaction between single molecules of Mac-1 and ICAM-1 in living cells: an atomic force microscopy study. Exp. Cell Res. 2007;313:3497–3504. doi: 10.1016/j.yexcr.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Zheng R, et al. Barrier-to-autointegration factor (BAF) bridges DNA in a discrete, higher-order nucleoprotein complex. Proc. Natl Acad. Sci. USA. 2000;97:8997–9002. doi: 10.1073/pnas.150240197. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


