Abstract
The field of chemical senses has made major progress in understanding the cellular mechanisms of olfaction and taste in the past 2 decades. However, the molecular understanding of odor and taste recognition is still lagging far behind and will require solving multiple structures of the relevant full-length receptors in complex with native ligands to achieve this goal. However, the development of multiple complimentary strategies for the structure determination of G protein-coupled receptors (GPCRs) makes this goal realistic. The common conundrum of how multi-specific receptors that recognize a large number of different ligands results in a sensory perception in the brain will only be fully understood by a combination of high-resolution receptor structures and functional studies. This review discusses the first steps on this pathway, including biochemical and physiological assays, forward genetics approaches, molecular modeling, and the first steps towards the structural biology of olfactory and taste receptors.
Keywords: biochemistry, functional assays, molecular modeling, olfaction, structural biology, taste
Towards the high-resolution structures of chemosensory receptors
In recent years, there has been increased success in the determination of membrane protein structures due to the development of a range of novel and improved methods. In particular, there have been a series of protein engineering breakthroughs that have facilitated the structure determination of G protein-coupled receptors (GPCRs) (Tate and Schertler 2009). This has transformed the field. The structures have been instrumental in developing a unified theory of how the conformation change in the receptor that occurs upon agonist binding results in G protein coupling on the intracellular face of the receptor (Rosenbaum et al. 2009; Katritch et al. 2013; Venkatakrishnan et al. 2013, 2016). In addition, structures of GPCRs bound to G proteins have illuminated how the subsequent signaling pathway is activated (Flock et al. 2015). High-resolution structures of specific GPCRs, such as the β1-adrenoceptor, bound to ligands of different efficacy and potency have resulted in a detailed understanding of the relationship between the chemical structure of a ligand and how it affects the conformation of the receptor (Warne et al. 2011). It is now possible to use structure-based drug design to develop novel highly potent and subtype specific drug candidates, that will result in better drugs being available in the future for a wide range of diseases such as diabetes, neurodegeneration, and cancer (Congreve et al. 2011; Mason et al. 2012). The structural biology of GPCRs is a vast field and the interested reader is guided towards the excellent reviews cited above.
Despite intense efforts in GPCR structural biology over the past decades and the crystallographic analysis of the extracellular region of a taste receptor (Nuemket et al. 2017), there is no high-resolution structure of any full-length taste receptor or olfactory receptor (OR), which has hampered an in-depth understanding into their molecular mechanism. Problems in obtaining high-resolution structures include poor functional heterologous expression, post-translational modifications, high flexibility, and low stability in detergent. The latter problem is especially important to resolve in order to allow the formation of well-diffracting crystals, which has been achieved by engineering thermostable mutants, protein fusion strategies or the formation of GPCR-G protein complexes with mini-G proteins and the addition of nanobodies (Cherezov et al. 2007; Ghosh et al. 2015; Carpenter et al. 2016; Magnani et al. 2016). Once it has been decided which strategy to use, the receptor needs to be expressed purified and crystallised, and each stage needs careful optimization. Overexpression systems using either insect cells or mammalian cells are the most successful, such as the Baculovirus expression system (Saarenpää et al. 2015), the use of stable inducible cell lines (Andréll et al. 2016) or the BacMam expression system (Goehring et al. 2014). Green fluorescent protein fused at the C-terminus of GPCRs is a well-established method for rapid and efficient screening of expression levels, and for distinguishing folded versus unfolded receptor by a combination of fluorescence size exclusion chromatography (FSEC), differential solubilisation and confocal microscopy (Thomas and Tate 2014). N-glycosylation may be problematic, as it may be necessary for efficient folding of the chemosensory receptors, but it interferes with the formation of well-diffracting crystals. The use of a specific mammalian cell line HEK293-GnTI-, which produces truncated N-glycosylation or the treatment with the enzymes EndoH or PNGaseF can address this problem (Thomas and Tate 2014). Once large amounts of fully functional receptor have been expressed, then it needs to be purified in a functional state, and the recent development of novel mild detergents has been critical for increased successes of this step (Bae et al. 2016; Hussain et al. 2016). Crystallisation remains a real challenge, but the revival and further development of the lipidic cubic phase technique has been pivotal in the crystallisation success of GPCRs (Huang et al. 2016). Once high quality and well diffracting crystals have been obtained then the exacting procedures of crystal mounting, data collection and processing have to be performed carefully to get the maximum information from them. Usually hundreds of crystals have to be grown and screened by X-ray diffraction to determine a high-resolution structure. However, the recent rapid developments of the resolution revolution in the field of electron cryo-microscopy (cryo-EM) will undoubtedly start to have an impact on the structure determination of GPCRs, initially on the largest receptors in complex with G proteins, which will have a molecular weight of about 150–250 kDa. However, as the techniques improve, the routine structure determination of GPCRs with molecular weights less than 100 kDa will become a reality (Liang et al. 2017). The tremendous efforts accomplishing a high-resolution structure of a GPCR such as in the case of the human histamine H1 receptor in complex with the antihistaminic compound doxepin revealed the biochemical action of differences between a first and a second generation antihistamine (Shimamura et al. 2011). A phosphate ion from the buffer used during purification, was bound in the ligand binding site together with doxepin, a first generation anti-histamine drug. The negatively charged phosphate ion binds exactly in the region of the binding pocket that provides receptor specificity for second generation anti-histamines. Only due to the structural insights at atomic detail it is now possible to rationally design optimized tailor made antihistaminic compounds, with maximum pharmacological potential and minimum side effects. Another example of where a GPCR structure has led to a molecular understanding of a functional phenomenon is the inactivation of the adenosine A2A receptor (A2AR) by the antibody Fab fragment Fab2838. A2AR is a major drug target due to its role in regulating blood flow to the cardiac muscle and the regulation of glutamate and dopamine release in the brain. The high resolution structure of this receptor in complex with an allosteric antibody revealed the molecular mechanism of the underlying receptor inactivation, which is based on the prevention of agonist but not antagonist binding to the extracellular ligand-binding pocket. The structure showed that Fab2838 recognizes the intracellular surface of A2AR and that its complementarity-determining region, CDR-H3, penetrates into the receptor (Hino et al. 2012). A2AR is also a good example showing how structure-based drug design can result in the development of novel potent and specific inhibitors that have the potential for the treatment of Parkinson’s disease (Congreve et al. 2012). These examples highlight the synergism of combining structural and functional studies. The initial steps for the structure determination of chemosensory receptors are still ongoing, and there is a long and rocky road to achieve the ultimate goal of determining their structures. As the 17th International Symposium on Olfaction and Taste, Yokohama, Japan, 2016 showed, there is a wide range of structure–function studies ongoing in several different laboratories around the world. The symposium “structure–function relationships of olfactory and taste receptors” brought both sides closer together and increased mutual understanding of the problems facing the field. Only a combined approach and a team effort on structural and functional studies will ultimately reveal the underlying molecular mechanisms of chemosensory receptor function.
Structure–function relationship of ORs
Over 40 different GPCRs have been crystallised and their structures determined in multiple different conformations by X-ray crystallography and, more recently, by electron cryomicroscopy. However, experimental structure elucidation of the largest family of GPCRs, the ORs, has remained elusive (Figure 1) (Buck and Axel 1991; Probst et al. 1992; Pilpel and Lancet 1999).
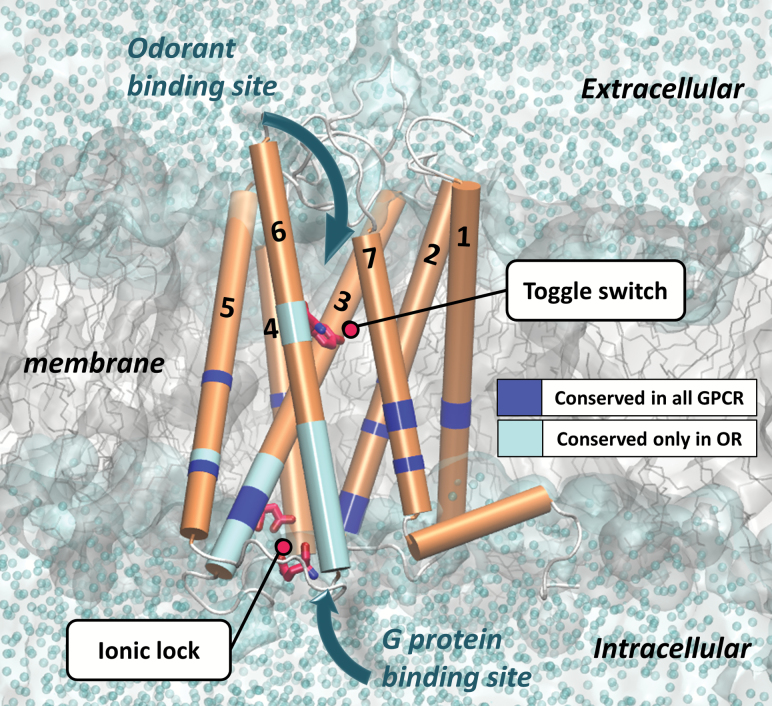
Figure 1.
Homology model of MOR256-3 embedded in a membrane and solvated in water. The backbone of the receptor is represented in orange and the toggle switch and the ionic lock are detailed in pink. The conserved residues locations are highlighted in dark blue for those common to all the class A GPCR and in light blue for those specific to the OR family. The predicted odorant binding site is located at the extracellular part of the receptor close to the toggle switch. The binding site of the G protein is at the opposite side of the bundle, in its intracellular part.
OR sequences have characteristic motifs of conserved amino acid residues present in each of their 7 transmembrane helices (TM) numbered from TM1 to TM7. Some of these motifs are also shared with non-olfactory GPCRs (indicated by * in the following listing). For each TM, the motifs are: (de March et al. 2015a)
- GN* in TM1,
- LHxPMY and LxxxD* in TM2,
- MAY, DRY* and VAICxPLxY in TM3,
- W* in TM4,
- P* and SY* in TM5,
- KAFSTCASH and FYG in TM6,
- NPxxY* in TM7.
Though hindered by the lack of experimentally verified structures, advances in computational techniques in combination with functional assessments of OR mutants have enabled investigation of ligand–receptor interactions at the atomic level. Application of these methods to ORs, guided by their similarities with non-olfactory GPCRs and experimental ligand-mediated receptor activation data, has been demonstrated to be a powerful tool for the study of their dynamics and their interactions with odorant molecules (de March and Golebiowski 2014).
Olfactory sensation begins with the detection of odor ligands by ORs. Since the human genome encodes over ~400 ORs, it has been thought a combinatorial code is utilized, whereby one OR can be activated by a set of odorants and one odorant can activate a combination of ORs (Malnic et al. 1999). To accommodate structurally diverse odorants, the ligand-binding cavity is supposed to be highly variable between the ORs (Figure 1) (Pilpel and Lancet 1999; de March et al. 2015a). The chemical space associated with ORs response cover a large spectrum, leading to the denomination of narrowly and broadly tuned receptors for OR responding to a small or large number of odorants, respectively. This characteristic is influenced by the permissiveness of the OR cavity and its ability to be activated (Yu et al. 2015).
The activation mechanism of GPCRs (including ORs) is based on favoring an active state with respect to an inactive state when an agonist molecule is bound. Homology models of ORs based on conserved residues with other GPCR has successfully predicted ligand-mediated activation in cell-culture based assays, providing evidence for its accuracy in ligand-binding prediction. (Floriano et al. 2000; Gelis et al. 2012; Topin et al. 2014) The calculation of affinity between an odorant molecule and the ligand cavity of an OR is an efficient parameter for discriminating agonists and antagonists, but high affinity is not necessarily translated into activation. Indeed, inverse agonists bind to the cavity of a receptor, but do not trigger activation (de March et al. 2015b). Once the receptor is activated, a binding site is opened in its intracellular part allowing the binding of a G protein, triggering neuronal activation (Nakamura et al. 2013; Flock et al. 2015). Assuming that all ORs share G-protein coupling as a common activation mechanism, it is reasonable to hypothesize that some residues conserved in this family control the activation (de March et al. 2015c) (Figure 1). The structures of GPCRs are highly conserved and the mechanism of receptor activation is also highly conserved, where agonist binding causes a conformational change that results in the opening of a cleft on the intracellular surface where the G protein binds (Venkatakrishnan et al. 2013).
Despite the continued absence of an experimentally verified structure, our understanding of the structure–function relationship of ORs has dramatically increased in recent years through modeling in silico. However, the structure of the extracellular and intracellular loops remains unclear, because only the disulphide bridge between 2 Cys residues from the extracellular part of TM3 and extracellular loop 2 is conserved between ORs and non-olfactory GPCRs. The existence of a second disulphide bridge in extracellular loop 2 has also been proposed for 1 human OR (Cook et al. 2009).
One of the reasons why OR structures have not been determined is that heterologous expression of ORs for large-scale production is notoriously difficult. ORs are not efficiently transported to the cell surface when expressed in cells other than olfactory sensory neurons (Gimelbrant et al. 1999; Lu et al. 2004; Wu et al. 2012; Sharma et al. 2017). The features controlling this surface expression are thought to be dictated by certain amino acids within the ORs sequences. It could rely as well as on the interaction of the OR with the RTP1S (facilitator of OR trafficking) (Wu et al. 2012) or with the membrane composition. Understanding the mechanisms underlying the surface expression of ORs could facilitate the development of new ways to produce large quantities of functional ORs for structural studies.
Structure–function relationship of sweet and umami taste receptors
The vertebrate gustatory system is an indispensable tool for the selection of edible food items. Five taste qualities [sweet, sour, salty, umami (typically elicited by the L-amino acid, glutamate), and bitter] are generally accepted as the basic tastes. Each taste has a specific role to evaluate the caloric content, the electrolyte concentration and the presence of carbohydrate- and protein-rich foods or potentially harmful substances. Detection of the corresponding food components is mediated by taste receptors expressed in sensory cells residing in the oral cavity (Behrens and Meyerhof 2015). Sweet-tasting compounds are perceived through the activation of a heterodimeric GPCR composed of, TAS1R2 (taste receptor type 1, member 2) and TAS1R3 (taste receptor type 1, member 3) (Figure 2). Umami tasting compounds are perceived through a GPCR heterodimer composed of TAS1R1 (taste receptor type 1, member 1) and TAS1R3 (taste receptor type 1, member 3). The sweet and umami taste receptors (TAS1Rs) are members of the small family of class C GPCRs that includes the metabotropic glutamate receptors (mGluRs), the γ-aminobutyric acid receptor B (GABABR), and the calcium-sensing receptor (CaSR). In addition to the characteristic heptahelical transmembrane domain (TMD), TAS1Rs possess a large extra-cellular domain, composed of the Venus Flytrap (VFT) domain followed by a short cysteine-rich region. Cellular assays, molecular docking, and site-directed mutagenesis studies have revealed that the VFT domain of TAS1R2 (TAS1R2-VFT) or TAS1R1 (TAS1R-VFT) contains the primary binding site for most of the sweet ligands including natural sugars artificial sweeteners and natural sweeteners or umami ligands including amino acids and purine 5ʹ nucleotides (Nelson et al. 2001; Li et al. 2002; Nelson et al. 2002; Xu et al. 2004). However, some studies have revealed that the VFT, the cysteine-rich region and the TMD of T1R3 interact with some sweeteners including sweet-tasting proteins (Jiang et al. 2004, 2005a, 2005b; Winnig et al. 2007; Maîtrepierre et al. 2012). To elucidate the contribution of TAS1R2-VFT to sweet tastant binding, Briand and co-workers heterologously expressed human TAS1R2-VFT in Escherichia coli (paper in preparation). In agreement with previously published data (Nie et al. 2005), ligand-binding studies using intrinsic tryptophan fluorescence revealed micromolar and millimolar range affinities of TAS1R2-VFT with natural sugars and various sweeteners, confirming the functional state of the recombinant protein. Site-directed mutagenesis combined with fluorescent binding assays demonstrated that TAS1R2-VFT binding is specific. The interaction of TAS1R2-VFT with the 2 recombinant sweet-tasting proteins, brazzein and monellin, was measured using Bio-Layer Interferometry (BLI). This optical technique analyses the signal variations in the interference pattern generated from visible light reflected from an optical layer and a biolayer containing the immobilized protein of interest. BLI experiments demonstrated that TAS1R2-VFT binds these 2 sweet-tasting proteins with affinities in agreement with the physiological range. The strategy for the large-scale production of functional VFT domains of TAS1Rs will be used in future work to investigate the structure–function relationships of these taste receptors using NMR or crystallographic approaches.
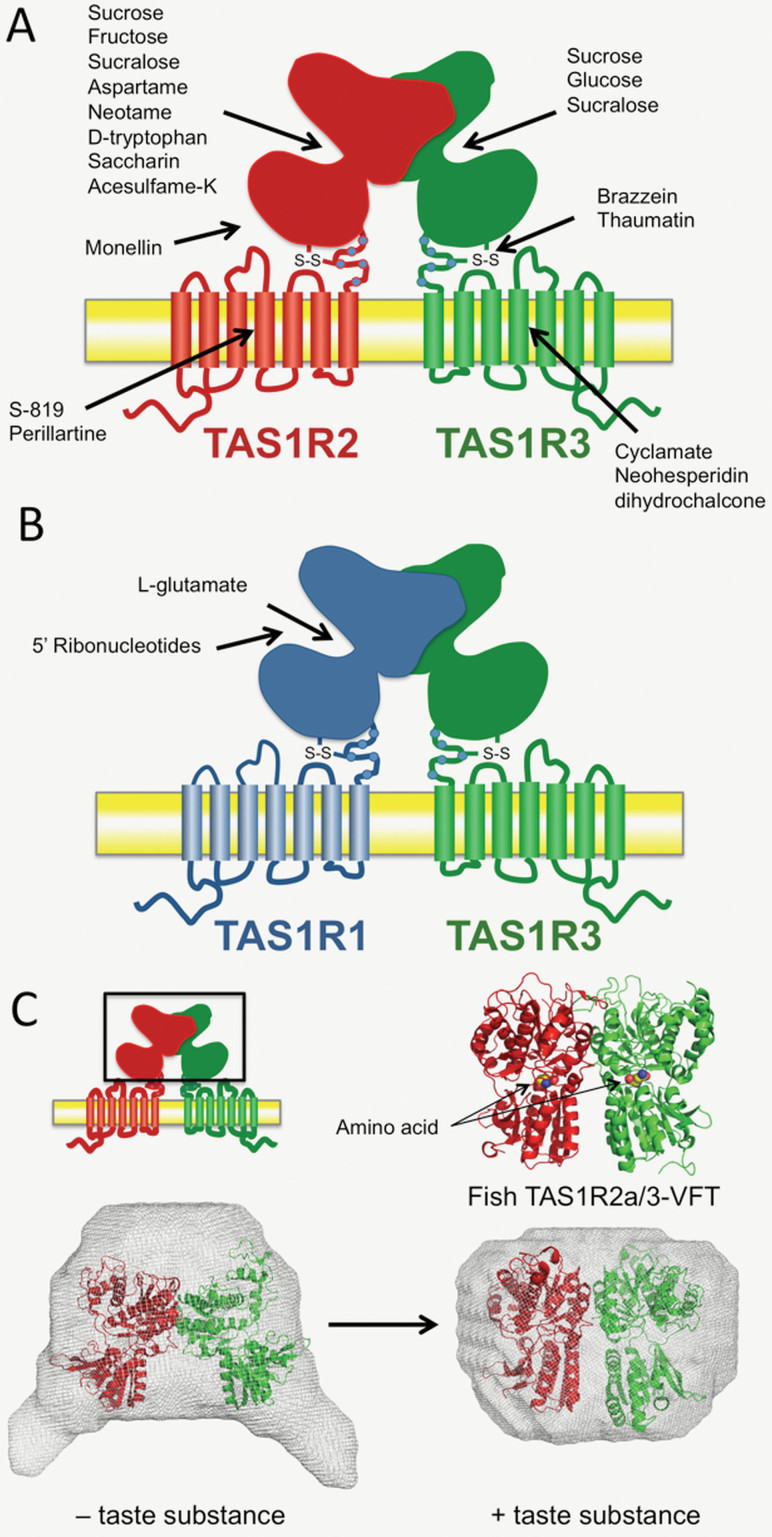
Figure 2.
Sweet and umami taste receptors (TAS1Rs) structure. (A) The sweet taste receptor possesses multiples binding sites. The sweet taste receptor is composed of 2 subunits named TAS1R2 and TAS1R3. The VFT domain of TAS1R2 contains the primary binding site for most of the sweet ligands including natural sugars and, artificial and natural sweeteners, including the sweet tasting protein monellin. Four additional binding loci are indicated. (B) The umami taste receptor is composed of 2 subunits named TAS1R1 and TAS1R3. The VFT domain of TAS1R1 contains the primary binding site for umami ligands including L-glutamate and 5ʹ-ribonucleotide. (C) Conformational change of the VFT domains upon taste substance binding. Molecular envelopes displayed representative low-resolution models reconstructed from the SAXS data of fish TAS1R2a/TAS1R3-VFT heterodimer. The crystal structure of mGluR1-VFT dimer in a resting (PDB: 1EWT) and an active (PDB: 1EWK) state was superimposed on the restored model in the absence (left) and presence (right) of a taste substance, respectively (Adapted from Nango et al. 2016).
Another important factor regarding the structure–function relationship of TAS1Rs is dimerization. Class C GPCRs constitutively exist as homo- or heterodimers, and heterodimerization of TAS1Rs are indeed essential for receptor functions for umami and sweet taste perception (Nelson et al. 2001; Li et al. 2002; Nelson et al. 2002; Zhao et al. 2003). The previous crystallographic structure analyses of mGluR1 VFT domains revealed dimer structures with 2 different arrangements. One is a compact arrangement like an inverted “U”-shape with interprotomer interactions at each VFT region. The other arrangement is a more open inverted “V”-shape resulting from a scissoring rearrangement of each protomer (Kunishima et al. 2000). The “U” and “V” conformations were considered to be an active state and a resting state, respectively, and the observed dimer rearrangement has been presumed to underlie the receptor activation. In theory, these conformation changes could induce rearrangements of the transmembrane regions, thus resulting in receptor activation. It should be noted, however, that the crystal structures of the other class C GPCR VFTs exhibited various types of conformational changes (Muto et al. 2007; Geng et al. 2013, 2016).
So far, purification of a full length TAS1R heterodimer has been unsuccessful. However, Nango et al. (2016) succeeded in purifying the recombinant heterodimeric TAS1R VFT domains, composed of TAS1R2 and TAS1R3 from the medaka fish (Oryzias latipes), and revealed the manner of heterodimerization and conformational change by integrating several biophysical analyses such as electron microscopy, Förster resonance energy transfer, and small-angle X-ray scattering. They found that TAS1R2/3-VFT heterodimer has a compact structure in the presence of compounds that induce TAS1R2/3 receptor responses in fish, e.g. amino acids. In contrast, the dimer becomes more open in the absence of amino acids, presumably through a rearrangement of the dimer interface (Figure 2B). They also confirmed that the conformational change was accompanied by binding of amino acids to the protein. The results suggested that the mechanism of taste signal transduction by TAS1R taste receptor is similar to that proposed for the other class C GPCRs. Recently, the same group succeeded in determining the crystal structures of the fish TAS1R2/3-VFT in the presence of amino acids, and provided the structural basis for taste substance recognition (Nuemket et al. 2017). The TAS1R2/3-VFT was found to share a similar overall architecture, including the primary ligand-binding pocket, with those of VFTs in other class C GPCR members such as mGluR (Figure 2B). The arrangement of the dimer with any amino acid bound was always compact and very similar to the conformation observed in the active state of mGluR. This reflects both the conserved mechanism of receptor activation in Class C GPCRs and also the broad ligand specificity of fish TAS1R2/3. Further structural studies are required to elucidate detailed molecular mechanisms of TAS1Rs, such as receptor activation induced by the binding of taste substances.
Structure–function relationship of bitter taste receptors
The taste receptor 2 (TAS2R) family are GPCRs and facilitate the detection of countless structurally diverse bitter substances. The human TAS2R gene family encompasses ~25 members which, like odorant receptors, possess different ligand profiles. Whereas the majority of the TAS2Rs responds to only a moderate number of agonists, 3 receptors (TAS2R10, TAS2R14, and TAS2R46) are exceptional because they are sensitive to an extraordinarily large array of bitter substances (Behrens and Meyerhof 2015). Somewhat surprisingly, structure–function analyses of TAS2R46 revealed the existence of a single orthosteric ligand-binding pocket that accommodates all the different agonists (Brockhoff et al. 2010). Recently, an additional vestibular binding site in TAS2R46 was proposed (Sandal et al. 2015). Here, ligands bind only transiently en route to the orthosteric site, which may help to pre-filter ligands from the bulk of irrelevant compounds present in complex food matrices. Focusing on the TAS2R10, Born et al. (2013) concluded that this receptor evolved to interact with many agonists weakly at the expense of binding only a few compounds with high affinity. Amino acid residues in at least 3 positions within the binding pocket exert ambivalent contributions to the receptor’s activation by different agonists. Whereas these residues support binding of some compounds, they decrease the affinity of binding of other agonists. Experimental studies have involved generating inter-receptor chimeras and using functional assays to identify regions within TAS2Rs that are involved in agonist interactions (Brockhoff et al. 2010). In efforts to expand on these initial data, more recent studies in silico have relied on molecular modeling to suggest amino acid residues located in the receptor binding pocket [e.g., (Biarnés et al. 2010; Born et al. 2013; Marchiori et al. 2013; Sandal et al. 2015)]. However, since TAS2Rs are only remotely related to the other GPCR-families (Fredriksson et al. 2003), the current homology models are all based on crystal structures of distantly related receptors. Hence, TAS2R structure–function research would benefit more than any other chemoreceptor-family from the successful crystallization of at least one member of this highly interesting group of receptors. Initially efforts to characterize TAS2Rs were focused on the human receptors (Meyerhof et al. 2010), but in recent years numerous receptors from other species have been characterized and their ligands identified (Behrens and Meyerhof 2016). Most recently, Lossow et al. (2016) added the functional characterization of the majority of mouse TAS2Rs. Intriguingly, it was found that the TAS2R repertoire of mice contains fewer generalist receptors and more narrowly tuned receptors. Despite general assumptions anticipating functional conservation among human and mouse orthologues, Lossow et al. demonstrated that such orthologues rarely share common ligands. The combined data from functional experiments performed with TAS2R repertoires of different species and careful structure–function analyses enables insights into the evolutionary history and the selective forces acting on TAS2R gene repertoires of human and mouse. It is believed that species-specific expansion of TAS2R genes serves an important role in the adaptation of species to the presence of specific bitter substances encountered by each species in their environment (Shi et al. 2003; Di Pizio and Niv 2015). One of these gene expansions occurred after the separation of the rodent and primate lineages, and led to muroid cluster I; this contains 5 homologues in mice and only one (TAS2R10) in humans. The amino acid residues identified previously as crucial for TAS2R10 agonist interactions (Born et al. 2013) were different in all the mouse muroid cluster I receptors, suggesting that the mouse receptors had different selectivity and bound different agonists. Hence, permutation of these positions along with gene expansion appears to be an effective evolutionary strategy to rapidly diversify pharmacological properties. In contrast, the human TAS2R38 receptor that binds phenylthiocarbamide (PTC) (Biarnes et al. 2010; Marchiori et al. 2013) has not been duplicated during mammalian evolution. It was hypothesized that the exquisite PTC-sensitivity of primate TAS2R38 developed after separation of rodent and primate lineages, whereas the orthologous mouse TAS2R138 lost PTC-responsiveness and developed other binding specificities (Lossow et al. 2016).
Remarkably, despite pronounced amino acid sequence divergence, striking functional similarities between bitter taste receptors and other class A GPCRs exist (Di Pizio et al. 2016) suggesting that they activate G proteins in a similar manner.
Future perspectives
The ISOT symposium with the title “Structure–function relationships of olfactory and taste receptors”, which took place on the 7 June 2016 in Yokohama, Japan, demonstrated the importance and necessity of high-resolution structures of chemosensory receptors. Moreover, it clearly showed that the joint effort of structural and functional studies is essential and necessary to achieve this goal. Given the enormous efforts in all aspects of pharmacological, cell and molecular biological, neurobiological, and modeling studies, the structural studies can now be performed in a more rational way by extracting and combining the crucial information of each of the fields presented and letting the structural work being guided by this prior knowledge. Eventually any chemosensory high-resolution structure will shed light on the molecular mechanism of any receptor within this class of proteins.
References
- Andréll J, Edwards PC, Zhang F, Daly M, Tate CG. 2016. Generation of tetracycline-inducible mammalian cell lines by flow cytometry for improved overproduction of membrane proteins. Methods Mol Biol. 1432:63–78. [DOI] [PubMed] [Google Scholar]
- Bae HE, Mortensen JS, Ribeiro O, Du Y, Ehsan M, Kobilka BK, Loland CJ, Byrne B, Chae PS. 2016. Tandem neopentyl glycol maltosides (TNMs) for membrane protein stabilisation. Chem Commun (Camb). 52:12104–12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M, Meyerhof W. 2015. Taste receptors. In: Parker JK, Elmore JS, Methven L, editors. Flavour development, analysis and perception in food and beverages. Cambridge, UK: Woodhead Publishing; p. 297–329. [Google Scholar]
- Behrens M, Meyerhof W. 2016. G protein-coupled taste receptors. In: Zufall F, Munger SD, editors. Chemosensory transduction. London, UK: Elsevier Inc; p. 227–244. [Google Scholar]
- Biarnés X, Marchiori A, Giorgetti A, Lanzara C, Gasparini P, Carloni P, Born S, Brockhoff A, Behrens M, Meyerhof W. 2010. Insights into the binding of Phenyltiocarbamide (PTC) agonist to its target human TAS2R38 bitter receptor. PLoS One. 5:e12394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born S, Levit A, Niv MY, Meyerhof W, Behrens M. 2013. The human bitter taste receptor TAS2R10 is tailored to accommodate numerous diverse ligands. J Neurosci. 33:201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockhoff A, Behrens M, Niv MY, Meyerhof W. 2010. Structural requirements of bitter taste receptor activation. Proc Natl Acad Sci USA. 107:11110–11115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck L, Axel R. 1991. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 65:175–187. [DOI] [PubMed] [Google Scholar]
- Carpenter B, Nehmé R, Warne T, Leslie AG, Tate CG. 2016. Structure of the adenosine A(2A) receptor bound to an engineered G protein. Nature. 536:104–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK et al. . 2007. High-resolution crystal structure of an engineered human beta2-adrenergic G protein-coupled receptor. Science. 318:1258–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, Andrews SP, Doré AS, Hollenstein K, Hurrell E, Langmead CJ, Mason JS, Ng IW, Tehan B et al. . 2012. Discovery of 1,2,4-triazine derivatives as adenosine A(2A) antagonists using structure based drug design. J Med Chem. 55:1898–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congreve M, Langmead CJ, Mason JS, Marshall FH. 2011. Progress in structure based drug design for G protein-coupled receptors. J Med Chem. 54:4283–4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook BL, Steuerwald D, Kaiser L, Graveland-Bikker J, Vanberghem M, Berke AP, Herlihy K, Pick H, Vogel H, Zhang S. 2009. Large-scale production and study of a synthetic G protein-coupled receptor: human olfactory receptor 17-4. Proc Natl Acad Sci USA. 106:11925–11930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de March CA, Golebiowski J. 2014. A computational microscope focused on the sense of smell. Biochimie. 107 Pt A:3–10. [DOI] [PubMed] [Google Scholar]
- de March CA, Kim SK, Antonczak S, Goddard WA 3rd, Golebiowski J. 2015a. G protein-coupled odorant receptors: from sequence to structure. Protein Sci. 24:1543–1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de March CA, Ryu S, Sicard G, Moon C, Golebiowski J. 2015b. Structure–odour relationships reviewed in the postgenomic era. Flavour Frag J. 30:342–361. [Google Scholar]
- de March CA, Yu Y, Ni MJ, Adipietro KA, Matsunami H, Ma M, Golebiowski J. 2015c. Conserved residues control activation of mammalian G protein-coupled odorant receptors. J Am Chem Soc. 137:8611–8616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pizio A, Levit A, Slutzki M, Behrens M, Karaman R, Niv MY. 2016. Chapter 18 - Comparing Class A GPCRs to bitter taste receptors: structural motifs, ligand interactions and agonist-to-antagonist ratios. In: Arun KS, editor. Methods in cell biology. Academic Press; p. 401–427. [DOI] [PubMed] [Google Scholar]
- Di Pizio A, Niv MY. 2015. Promiscuity and selectivity of bitter molecules and their receptors. Bioorg Med Chem. 23:4082–4091. [DOI] [PubMed] [Google Scholar]
- Flock T, Ravarani CNJ, Sun D, Venkatakrishnan AJ, Kayikci M, Tate CG, Veprintsev DB, Babu MM. 2015. Universal allosteric mechanism for Gα activation by GPCRs. Nature. 524:173–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floriano WB, Vaidehi N, Goddard WA 3rd, Singer MS, Shepherd GM. 2000. Molecular mechanisms underlying differential odor responses of a mouse olfactory receptor. Proc Natl Acad Sci USA. 97:10712–10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredriksson R, Lagerström MC, Lundin LG, Schiöth HB. 2003. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 63:1256–1272. [DOI] [PubMed] [Google Scholar]
- Gelis L, Wolf S, Hatt H, Neuhaus EM, Gerwert K. 2012. Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angewandte Chemie International Edition. 51:1274–1278. [DOI] [PubMed] [Google Scholar]
- Geng Y, Bush M, Mosyak L, Wang F, Fan QR. 2013. Structural mechanism of ligand activation in human GABA(B) receptor. Nature. 504:254–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Mosyak L, Kurinov I, Zuo H, Sturchler E, Cheng TC, Subramanyam P, Brown AP, Brennan SC, Mun HC et al. . 2016. Structural mechanism of ligand activation in human calcium-sensing receptor. Elife. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh E, Kumari P, Jaiman D, Shukla AK. 2015. Methodological advances: the unsung heroes of the GPCR structural revolution. Nat Rev Mol Cell Biol. 16:69–81. [DOI] [PubMed] [Google Scholar]
- Gimelbrant AA, Stoss TD, Landers TM, McClintock TS. 1999. Truncation releases olfactory receptors from the endoplasmic reticulum of heterologous cells. J Neurochem. 72:2301–2311. [DOI] [PubMed] [Google Scholar]
- Goehring A, Lee CH, Wang KH, Michel JC, Claxton DP, Baconguis I, Althoff T, Fischer S, Garcia KC, Gouaux E. 2014. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc. 9:2574–2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino T, Arakawa T, Iwanari H, Yurugi-Kobayashi T, Ikeda-Suno C, Nakada-Nakura Y, Kusano-Arai O, Weyand S, Shimamura T, Nomura N et al. . 2012. G-protein-coupled receptor inactivation by an allosteric inverse-agonist antibody. Nature. 482:237–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CY, Olieric V, Ma P, Howe N, Vogeley L, Liu X, Warshamanage R, Weinert T, Panepucci E, Kobilka B et al. . 2016. In meso in situ serial X-ray crystallography of soluble and membrane proteins at cryogenic temperatures. Acta Crystallogr D Struct Biol. 72:93–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain H, Du Y, Scull NJ, Mortensen JS, Tarrasch J, Bae HE, Loland CJ, Byrne B, Kobilka BK, Chae PS. 2016. Accessible mannitol-based amphiphiles (MNAs) for membrane protein solubilisation and stabilisation. Chemistry. 22:7068–7073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang P, Cui M, Zhao B, Liu Z, Snyder LA, Benard LM, Osman R, Margolskee RF, Max M. 2005a. Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J Biol Chem. 280:15238–15246. [DOI] [PubMed] [Google Scholar]
- Jiang P, Cui M, Zhao B, Snyder LA, Benard LM, Osman R, Max M, Margolskee RF. 2005b. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J Biol Chem. 280:34296–34305. [DOI] [PubMed] [Google Scholar]
- Jiang P, Ji Q, Liu Z, Snyder LA, Benard LM, Margolskee RF, Max M. 2004. The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J Biol Chem. 279:45068–45075. [DOI] [PubMed] [Google Scholar]
- Katritch V, Cherezov V, Stevens RC. 2013. Structure-function of the G protein–coupled receptor superfamily. Annu Rev Pharmacol Toxicol. 53:531–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. 2000. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 407:971–977. [DOI] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. 2002. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 99:4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T et al. . 2017. Phase-plate cryo-EM structure of a class B GPCR-G-protein complex. Nature. 546:118–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lossow K, Hübner S, Roudnitzky N, Slack JP, Pollastro F, Behrens M, Meyerhof W. 2016. Comprehensive analysis of mouse bitter taste receptors reveals different molecular receptive ranges for orthologous receptors in mice and humans. J Biol Chem. 291:15358–15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu M, Staszewski L, Echeverri F, Xu H, Moyer BD. 2004. Endoplasmic reticulum degradation impedes olfactory G-protein coupled receptor functional expression. BMC Cell Biol. 5:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnani F, Serrano-Vega MJ, Shibata Y, Abdul-Hussein S, Lebon G, Miller-Gallacher J, Singhal A, Strege A, Thomas JA, Tate CG. 2016. A mutagenesis and screening strategy to generate optimally thermostabilized membrane proteins for structural studies. Nat Protoc. 11:1554–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maîtrepierre E, Sigoillot M, Le Pessot L, Briand L. 2012. Recombinant expression, in vitro refolding, and biophysical characterization of the N-terminal domain of T1R3 taste receptor. Protein Expr Purif. 83:75–83. [DOI] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell. 96:713–723. [DOI] [PubMed] [Google Scholar]
- Marchiori A, Capece L, Giorgetti A, Gasparini P, Behrens M, Carloni P, Meyerhof W. 2013. Coarse-grained/molecular mechanics of the TAS2R38 bitter taste receptor: experimentally-validated detailed structural prediction of agonist binding. PLoS One. 8:e64675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason JS, Bortolato A, Congreve M, Marshall FH. 2012. New insights from structural biology into the druggability of G protein-coupled receptors. Trends Pharmacol Sci. 33:249–260. [DOI] [PubMed] [Google Scholar]
- Meyerhof W, Batram C, Kuhn C, Brockhoff A, Chudoba E, Bufe B, Appendino G, Behrens M. 2010. The molecular receptive ranges of human TAS2R bitter taste receptors. Chem Senses. 35:157–170. [DOI] [PubMed] [Google Scholar]
- Muto T, Tsuchiya D, Morikawa K, Jingami H. 2007. Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc Natl Acad Sci USA. 104:3759–3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura S, Itabashi T, Ogawa D, Okada T. 2013. Common and distinct mechanisms of activation of rhodopsin and other G protein-coupled receptors. Sci Rep. 3:1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nango E, Akiyama S, Maki-Yonekura S, Ashikawa Y, Kusakabe Y, Krayukhina E, Maruno T, Uchiyama S, Nuemket N, Yonekura K et al. . 2016. Taste substance binding elicits conformational change of taste receptor T1r heterodimer extracellular domains. Sci Rep. 6:25745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. 2002. An amino-acid taste receptor. Nature. 416:199–202. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. 2001. Mammalian sweet taste receptors. Cell. 106:381–390. [DOI] [PubMed] [Google Scholar]
- Nie Y, Vigues S, Hobbs JR, Conn GL, Munger SD. 2005. Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr Biol. 15:1948–1952. [DOI] [PubMed] [Google Scholar]
- Nuemket N, Yasui N, Kusakabe Y, Nomura Y, Atsumi N, Akiyama S, Nango E, Kato Y, Kaneko MK, Takagi J et al. . 2017. Structural basis for perception of diverse chemical substances by T1r taste receptors. Nat Commun. 8:15530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilpel Y, Lancet D. 1999. The variable and conserved interfaces of modeled olfactory receptor proteins. Protein Sci. 8:969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst WC, Snyder LA, Schuster DI, Brosius J, Sealfon SC. 1992. Sequence alignment of the G-protein coupled receptor superfamily. DNA Cell Biol. 11:1–20. [DOI] [PubMed] [Google Scholar]
- Rosenbaum DM, Rasmussen SG, Kobilka BK. 2009. The structure and function of G-protein-coupled receptors. Nature. 459:356–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saarenpää T, Jaakola VP, Goldman A. 2015. Baculovirus-mediated expression of GPCRs in insect cells. Methods Enzymol. 556:185–218. [DOI] [PubMed] [Google Scholar]
- Sandal M, Behrens M, Brockhoff A, Musiani F, Giorgetti A, Carloni P, Meyerhof W. 2015. Evidence for a transient additional ligand binding site in the TAS2R46 bitter taste receptor. J Chem Theory Comput. 11:4439–4449. [DOI] [PubMed] [Google Scholar]
- Sharma R, Ishimaru Y, Davison I, Ikegami K, Chien MS, You H, Chi Q, Kubota M, Yohda M, Ehlers M et al. . 2017. Olfactory receptor accessory proteins play crucial roles in receptor function and gene choice. Elife. 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi P, Zhang J, Yang H, Zhang YP. 2003. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 20:805–814. [DOI] [PubMed] [Google Scholar]
- Shimamura T, Shiroishi M, Weyand S, Tsujimoto H, Winter G, Katritch V, Abagyan R, Cherezov V, Liu W, Han GW et al. . 2011. Structure of the human histamine H1 receptor complex with doxepin. Nature. 475:65–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CG, Schertler GF. 2009. Engineering G protein-coupled receptors to facilitate their structure determination. Curr Opin Struct Biol. 19:386–395. [DOI] [PubMed] [Google Scholar]
- Thomas J, Tate CG. 2014. Quality control in eukaryotic membrane protein overproduction. J Mol Biol. 426:4139–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topin J, de March CA, Charlier L, Ronin C, Antonczak S, Golebiowski J. 2014. Discrimination between olfactory receptor agonists and non-agonists. Chemistry. 20:10227–10230. [DOI] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Heydenreich FM, Flock T, Miljus T, Balaji S, Bouvier M, Veprintsev DB, Tate CG et al. . 2016. Diverse activation pathways in class A GPCRs converge near the G-protein-coupling region. Nature. 536:484–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. 2013. Molecular signatures of G-protein-coupled receptors. Nature. 494:185–194. [DOI] [PubMed] [Google Scholar]
- Warne T, Moukhametzianov R, Baker JG, Nehmé R, Edwards PC, Leslie AG, Schertler GF, Tate CG. 2011. The structural basis for agonist and partial agonist action on a β(1)-adrenergic receptor. Nature. 469:241–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winnig M, Bufe B, Kratochwil NA, Slack JP, Meyerhof W. 2007. The binding site for neohesperidin dihydrochalcone at the human sweet taste receptor. BMC Struct Biol. 7:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Pan Y, Chen GQ, Matsunami H, Zhuang H. 2012. Receptor-transporting protein 1 short (RTP1S) mediates translocation and activation of odorant receptors by acting through multiple steps. J Biol Chem. 287:22287–22294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. 2004. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc Natl Acad Sci USA. 101:14258–14263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, de March CA, Ni MJ, Adipietro KA, Golebiowski J, Matsunami H, Ma M. 2015. Responsiveness of G protein-coupled odorant receptors is partially attributed to the activation mechanism. Proc Natl Acad Sci USA. 112:14966–14971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. 2003. The receptors for mammalian sweet and umami taste. Cell. 115:255–266. [DOI] [PubMed] [Google Scholar]