Abstract
The mammalian tastes of sweet, umami, and bitter are initiated by activation of G protein-coupled receptors (GPCRs) of the T1R and T2R families on taste receptor cells. GPCRs signal via nucleotide exchange and hydrolysis, the latter hastened by GTPase-accelerating proteins (GAPs) that include the Regulators of G protein Signaling (RGS) protein family. We previously reported that RGS21, uniquely expressed in Type II taste receptor cells, decreases the potency of bitter-stimulated T2R signaling in cultured cells, consistent with its in vitro GAP activity. However, the role of RGS21 in organismal responses to GPCR-mediated tastants was not established. Here, we characterized mice lacking the Rgs21 fifth exon. Eliminating Rgs21 expression had no effect on body mass accumulation (a measure of alimentation), fungiform papillae number and morphology, circumvallate papillae morphology, and taste bud number. Two-bottle preference tests, however, revealed that Rgs21-null mice have blunted aversion to quinine and denatonium, and blunted preference for monosodium glutamate, the sweeteners sucrose and SC45647, and (surprisingly) NaCl. Observed reductions in GPCR-mediated tastant responses upon Rgs21 loss are opposite to original expectations, given that loss of RGS21—a GPCR signaling negative regulator—should lead to increased responsiveness to tastant-mediated GPCR signaling (all else being equal). Yet, reduced organismal tastant responses are consistent with observations of reduced chorda tympani nerve recordings in Rgs21-null mice. Reduced tastant-mediated responses and behaviors exhibited by adult mice lacking Rgs21 expression since birth have thus revealed an underappreciated requirement for a GPCR GAP to establish the full character of tastant signaling.
Keywords: gustatory, mouse, regulator of G protein signaling-21 (RGS21), taste buds, taste perception
Introduction
Taste perception is initiated by clusters of cells in the oral cavity known as taste buds. Type II taste receptor cells within taste buds express the chemosensory transduction proteins responsive to stimuli for the mammalian tastes of sweet, bitter, and umami (Zhang et al. 2003; Clapp et al. 2004; DeFazio et al. 2006). Stimuli for sweet and umami tastes are detected by the T1R family of G protein-coupled receptors (GPCRs) (Nelson et al. 2001; Li et al. 2002; Nelson et al. 2002; Zhao et al. 2003), while stimuli for bitter tastes are detected by a family of approximately 30 GPCRs, the T2Rs, most of which are expressed in the same subset of taste bud cells (Adler et al. 2000; Chandrashekar et al. 2000; Bufe et al. 2002). Downstream signaling by both T1R and T2R receptors in Type II taste cells is typically mediated by activation of a heterotrimeric G protein complex composed of Gαgustducin (a.k.a. α-gustducin), Gβ3, and Gγ13 (McLaughlin et al. 1992; Wong et al. 1996; Ming et al. 1998; Huang et al. 1999). GPCR-mediated tastant signaling is primarily transduced by Gα nucleotide exchange and release of the Gβγ dimer, which in turn activates phospholipase C beta2 (PLCβ2) and, subsequently, the TRPM5 channel (Huang et al. 1999; Zhang et al. 2003). Opening of TRPM5 channels triggers membrane depolarization and extracellular release of ATP through the CALHM1 ion channel (Finger et al. 2005; Taruno et al. 2013), which in turn activates ATP-gated, ionotropic P2X2 and P2X3 receptors on gustatory afferent nerve fibers (Bo et al. 1999; Kinnamon and Finger 2013; Vandenbeuch et al. 2015b) and also metabotropic P2Y receptors on the Type II and Type III taste cells themselves (Kataoka et al. 2004; Bystrova et al. 2006; Huang et al. 2009).
Tastants, upon binding to T1R or T2R GPCRs, induce guanine nucleotide exchange on the underlying heterotrimeric G protein complex to release Gβγ from the Gα subunit (α-gustducin or a related Gα subfamily member) (Hoon et al. 1995; Wong et al. 1996; Kusakabe et al. 2000; Ruiz et al. 2003; Shindo et al. 2008; Tizzano et al. 2008). As in other conventional GPCR signaling cascades, signaling of taste receptor cells as initiated by freed Gβγ is terminated when the intrinsic GTPase activity of the Gα subunit hydrolyzes GTP to GDP, causing Gβγ re-association and thus restoration of the heterotrimeric G protein complex (reviewed in (Siderovski and Willard 2005; Palmer 2007). In this way, downstream taste signal transduction is critically regulated by the rate of GTP hydrolysis by the Gα subunit.
Alpha-gustducin is most closely related structurally to the transducin subunits Gαt(rod) and Gαt(cone) found within rod and cone photoreceptors, respectively (McLaughlin et al. 1992). All 3 Gα subunits have similar (and slow) intrinsic GTPase activity, which determines the lifetime of a Gα subunit in its GTP-bound (active) form and, thus, the duration of heterotrimeric G protein-based signal transduction. For α-transducin subunits, a large discrepancy was observed early on between their slow observed GTPase activity in vitro and the rapid inactivation of phototransduction in vivo (Angleson and Wensel 1993). This timing paradox was resolved upon discovery of the Regulator of G protein Signaling (RGS) proteins (Druey et al. 1996; Koelle and Horvitz 1996; Siderovski et al. 1996) (reviewed in Kimple et al. 2011), which accelerate the rate of GTP hydrolysis by Gα subunits like α-transducin in vitro (He et al. 1998) and in vivo (Chen et al. 2000). Thus, it is very likely that tastant signal transduction, which involves α-gustducin and other Gα subunits, is similarly regulated by RGS-mediated GTPase-accelerating protein (GAP) activity.
von Buchholtz and colleagues identified a putative Rgs gene transcript, Rgs21, from isolated rat foliate and fungiform papillae (von Buchholtz et al. 2004). In probing different rat tissues, these authors found that Rgs21 expression is restricted to taste tissue (von Buchholtz et al. 2004)—specifically in a subpopulation of taste cells within all types of taste papillae [foliate, fungiform, circumvallate (CV), and palate]. Furthermore, double-label in situ hybridization experiments revealed that virtually all cells expressing Rgs21 in rat lingual tissue also co-express Plcβ2, suggesting that Rgs21 is expressed exclusively in Type II taste receptor cells (von Buchholtz et al. 2004). Our subsequent biochemical analyses confirmed that purified RGS21 protein acts as a promiscuous GAP for multiple different Gα subunits of the Gαi/o and Gαq subfamilies, including α-gustducin and others involved in tastant signal transduction (Cohen et al. 2012; Kimple et al. 2014).
We demonstrated that Rgs21 is not only endogenously expressed in mouse taste buds, but is also expressed in primary airway epithelial cells known to express other components of the tastant signaling cascade (Cohen et al. 2012; Kimple et al. 2014). Measuring the separate effects of RGS21 over- and under-expression in the tastant-responsive, immortalized human epithelial cell line 16HBE confirmed that RGS21 acts, in cell culture, to oppose bitter signaling to calcium second messenger changes (Cohen et al. 2012), consistent with its demonstrable Gα-directed GAP activity in vitro. However, it remained unclear what role(s), if any, RGS21 plays in integrated tastant responses emanating from lingual tissue upon exposure to 1 of the 3 GPCR-mediated taste modalities of sweet, umami, and bitter. Here, we describe the characterization of a mouse strain constitutively deficient in Rgs21 expression, including 2-bottle choice tests for any altered preference/avoidance of umami (MSG), sweet, and bitter compounds; these behavioral findings were supported with complementary gustatory electrophysiology studies.
Materials and methods
Subjects and maintenance
All experiments involved Rgs21 wild-type and knockout mice—the latter mice lacked Rgs21 expression in a constitutive fashion, as described below. First, a conditional knockout of the Rgs21 gene was produced directly in the C57BL/6 background by genOway (Lyon, France). It involved insertion of loxP sites on either side of Rgs21 exon 5 (which encodes the final 67 amino-acids of the 152-amino acid RGS21 open-reading frame), as well as a neomycin-resistance gene (neo) flanked by “flippase recognition target” (FRT) sites, by homologous recombination in C57BL/6 embryonic stem cells. Drug-selected embryonic stem cells, with PCR and Southern blot evidence of homologous recombination, were subsequently injected into C57BL/6 blastocysts. Resulting chimeric male mice were crossed with females from the genOway proprietary ubiquitous Flp recombinase-expressing mice for in vivo removal of the FRT-flanked neo selection cassette. F1 heterozygous Rgs21fl/+ mice devoid of the neo selection cassette were delivered by genOway to the Siderovski lab at WVU.
This conditional “floxed” Rgs21 knockout strain was maintained at WVU by backcrossing to the C57BL/6J strain. For this study, to obtain constitutive Rgs21-null mice, the “floxed” Rgs21 mice were crossed with an ubiquitous Cre recombinase driver strain [B6.C-Tg(CMV-cre)1Cgn/J; JAX 006054] to generate Rgs21Δ5/+ mice. These Rgs21Δ5/+ mice were bred together to generate Rgs21Δ5/Δ5 (i.e., Rgs21-null) mice and Rgs21+/+ (i.e., wild-type) littermate control mice. Genotyping was performed via PCR using ear-snip tissue-derived genomic DNA and the following primer pairs: Shared Rgs21 allele forward primer 5′-CTGCCTTTGGGAAGCTTATG-3′ (nts. 144520871–144520890 of GenBank NC_000067.6 [C57BL/6J chromosome 1]) with either wild-type Rgs21 allele reverse primer 5′-TGGTATGGTGGTGGTGTTGT-3′ (nts. 144520654–144520673 of NC_000067.6) or with Rgs21Δ5 allele reverse primer 5′-CATTTCAGGGTTTGGAAAAGTT-3′ (nts. 144519608–144519629 of NC_000067.6).
Mice used in this study were maintained in a vivarium at 23°C on a 12 h/12 h light/dark cycle with lights off at 6 PM. They were housed in Allentown mouse cages (194 × 181 × 398 mm) with wire bar lids and corncob bedding, along with crinkled paper scattered on the floor for environmental enrichment. The mice had ad libitum access to Teklad Global 18% Protein Rodent Diet (Envigo, Inc.) and water (except during exposure tests, see below). Pups were weaned at 21 days and initially housed in groups of the same sex. Body mass was initially recorded at weaning and subsequently every 2 weeks until mice were 9 weeks of age. All mice were at least 9 weeks old prior to subsequent animal testing. Mice used for gustatory electrophysiology were shipped from WVU to Dr. Vandenbeuch’s laboratory at UC-Denver in Aurora, Colorado and allowed at least a week to recover before being tested. All procedures were approved by the Institutional Animal Care and Use Committee of the West Virginia University Health Sciences Center. Gustatory electrophysiological studies were also approved by the Animal Care and Use Committee of UC-Denver Anschutz Medical Campus.
RNA extraction
Tongues were excised from mice euthanized with Fatal-Plus® in accordance with guidelines from the National Institute of Health and with approval from the West Virginia University IACUC. After treatment with an enzyme cocktail consisting of Dispase (3 mg/mL; Gibco) and Elastase (2.5 mg/mL; Worthington) in Tyrode’s solution for 17 min, the epithelium was peeled from the underlying tissue. Gustatory tissue was isolated from the CV and non-gustatory tissue was isolated from equivalent-sized, non-taste epithelial tissue surrounding the CV prior to being flash frozen for subsequent RNA extraction. Tissue was homogenized in TRIzol reagent (Invitrogen™) using a bench top rotor stator. RNA was extracted according to manufacturer’s instructions using the Direct-zol™ RNA MiniPrep kit from Zymo Research. Reverse transcription and genomic DNA elimination was performed using the QuantiTect Reverse Transcription Kit from Qiagen.
Real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR)
qRT-PCR was performed to compare the expression levels of 18S rRNA and Rgs21 mRNA in Rgs21Δ5/Δ5 mice and Rgs21+/+ littermate controls. Additionally, transcript levels of taste cell-specific markers were also assessed: ecto-nucleoside triphosphate diphosphohydrolase 2 (Entpd2) for Type I cells; transient receptor potential cation channel subfamily M member 5 (Trpm5), phospholipase C β2 (Plb2), and calcium homeostasis modulator 1 (Calhm1) for Type II taste receptor cells; and synaptosomal-associated protein 25 (Snap25) for Type III cells. RNA extraction was performed as above. Two microliters of cDNA were used in each PCR reaction using the QuantiTect SYBR Green PCR Master Mix (Qiagen). RT2 qPCR Primer Assay for Mouse 18S rRNA was purchased from Qiagen. Primers (10 µM) were designed for Rgs21 mRNA (spanning exons 4–5) sequence elements (fwd primer: 5′-TCGTAGCTGATGCACCAAAA-3′; rev primer 5′-TACAGGAAAGGCAGCCATCT-3′) and purchased from ThermoFisher Scientific. Primer sequences for taste cell-specific markers were described previously (Huang et al. 2011; Taruno et al. 2013) and were purchased from ThermoFisher Scientific. PCR was performed (initial 15 min denaturation at 95°C followed by 40 cycles of 15 s denaturation at 95°C, 30 s annealing at 60°C, and 30 s extension at 72°C) in a Qiagen Rotor Gene-Q system. We utilized the SYBR green dye qPCR technique to detect double-stranded PCR amplicons as they accumulated during PCR cycling. Melting curves were obtained after each qRT-PCR experiment to assure specificity of resultant amplicons.
Tongue morphology
To assess general tongue morphology and number of fungiform papillae, tongues were dissected from Rgs21+/+ and Rgs21Δ5/Δ5 mice and briefly placed in 1% methylene blue (Ricca Chemical Co.). Tongues were then briefly rinsed in phosphate buffered saline. Methylene blue stains fungiform papillae more lightly than surrounding tissue, facilitating the quantification of taste papillae (Barnett et al. 2010). Images of the anterior dorsal surface of the tongue were taken using an Olympus SZX7 bright-field dissecting microscope with a Q-Fire CCD camera (Olympus America, Inc.). To ensure accuracy of scoring, 2 blinded investigators independently counted the number of fungiform papillae using the counter tool from ImageJ software to count and record all visible papillae on the anterior dorsal tongue. Subsequently, the independent counts were averaged. Images of the posterior tongue were also imported into ImageJ software to quantify the apical area of the CV papilla on each of these tongues. The freehand tool in ImageJ was used to trace around the interior wall of the papilla to quantify the total area of the papilla. Statistical analyses using unpaired Student’s t tests were applied to determine whether differences in fungiform number and apical CV area were significant between genotypes.
Immunohistochemistry
Tongues were excised from adult mice and placed in optimum cutting temperature (OCT) embedding compound (Sakura Finetech) prior to being flash frozen. Eight micrometers thick transverse sections of tongue containing the CV papilla were sliced using a cryostat and placed on Superfrost™ Microscope Slides (Fisherbrand™). Tissues sections were stained with hematoxylin and eosin (H&E) and examined under a bright-field microscope (EVOS FL Auto). To assess taste bud number, immunohistochemistry was performed on CV papillae sections using the Troma-1 antibody [Developmental Studies Hybridoma Bank (DSHB)]. The Troma-1 antibody has previously been characterized as specific using keratin-8 and -18 deficient mice (Tao et al. 2003). The Troma-1 antibody was used to label keratin-8, an intermediate filament protein in mature taste bud cells, to identify the boundary of taste buds (Toh et al. 1993; Zhang et al. 1995); Troma-1 has been shown to label taste bud cells with no labeling of stratified epithelium of the papilla wall (Liebl et al. 1999; Okubo et al. 2006; Seki et al. 2007). Labeling with Troma-1 allowed for counting of taste bud number. Tissue sections were fixed in 4% paraformaldehyde for 10 min at room temperature; membranes were then permeabilized in phosphate buffered saline (PBS) with 0.05% Triton-X100 for 10 min on ice. After blocking by incubation of slides for 60 min in PBS containing 5% goat serum at room temperature, sections were incubated at 4°C overnight with Troma-1 primary antibody (1:50). The following day, slides were incubated with Alexa Fluor-555-conjugated anti-mouse secondary antibody (1:500; Molecular Probes) for 60 min at room temperature. Slides were cover-slipped with Vectashield Hardset Mounting Media containing DAPI. Images were acquired using a Zeiss Violet Confocal microscope in the West Virginia University Microscope Imaging Facility, supported by the WVU Cancer Institute and NIH grants P30GM103488 and P20GM103434. Two blind, independent investigators used ImageJ software to quantify the taste bud number from these images. Taste bud number was counted on at least 4 sections per CV, with 80 µm between sections, from 4 separate mice using the ImageJ counter tool to avoid double counting buds. Statistical analysis using an unpaired Student’s t test was applied to determine whether the difference in taste bud number was significant between genotypes.
In situ hybridization
Tongue tissue sections from both genotypes of mice were provided to the Baylor College of Medicine’s RNA In Situ Hybridization Core which performed non-radioactive, RNA in situ hybridization (ISH) using Trpm5 sense and antisense probes, and a high-throughput protocol as previously described (Yaylaoglu et al. 2005). Images were acquired using a Zeiss Violet Confocal microscope, as stated above for IHC.
Two-bottle choice tests
Two-bottle choice tests were conducted in large Thoren mouse cages (30.80 × 40.60 × 15.88 cm). Individually housed mice were given access to 2 bottles containing autoclaved distilled water for 48 h prior to beginning all choice testing. Fluid was available through sipper spouts attached to 50-mL Corning™ conical-bottom centrifuge tubes, placed in separate bottle access slots on opposite sides of the food bin. Following the initial 48-h presentation of 2 bottles of water, mice were assessed over 48 h in tests with a choice between distilled water and ascending concentrations of a taste compound. The positions of the bottles were switched daily, and the fluid intakes were measured to the nearest 0.1 g by weighing the drinking bottles on an electronic balance. Preference ratio was calculated as volume of tastant solution consumed divided by volume of total solution consumed. The taste compounds were chosen as exemplars of the sweet, bitter, umami, salty, and sour taste qualities, and their concentrations spanned the range between indifference and marked acceptance or avoidance (Sucrose: 2.9, 14.6, 29.2, 58.5, and 292.4 mM; SC45647: 0.003, 0.01, 0.03, 0.1, 0.3, and 1 mM; Denatonium Benzoate: 0.01, 0.1, 0.3, 1, and 5 mM; Quinine Sulfate: 0.003, 0.01, 0.03, 0.1, 0.3, 0.6, and 1 mM; Monosodium Glutamate (MSG; containing 10 μM amiloride to block the taste of sodium) – 1, 10, 30, 100, 300, 600, and 1000 mM; NaCl: 18.75, 37.5, 75, 150, 300, and 600 mM; HCl: 0.1, 1, 10, and 30 mM). Mice naïve to the 2-bottle choice assay were used to assess NaCl taste preference. The mice were socially housed for 7 days between each test series. At least six mice per genotype were assessed at each concentration of taste solution. Results were analyzed using a 2-way ANOVA (genotype × concentration) with a Sidak multiple comparisons test (GraphPad Prism 7).
Nerve recording
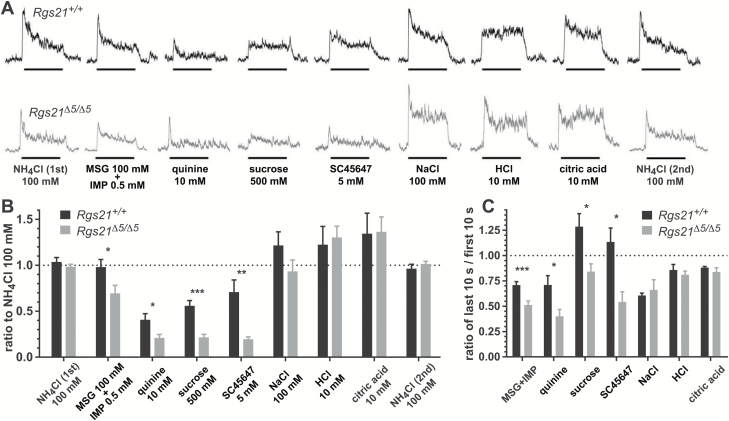
Chorda tympani nerve recordings were performed as previously described (Vandenbeuch et al. 2015a). Briefly, mice were anesthetized with urethane (2 g/kg), maintained in a head holder, and trachea cannulated to facilitate breathing. The chorda tympani nerve was exposed using a ventral approach, freed from surrounding tissue and cut near the tympanic bulla. The nerve was then placed on a platinum-iridium wire and a reference electrode was placed in a nearby tissue. The signal was fed to an amplifier (P511; Grass Instruments), integrated, and recorded using AcqKnowledge software (Biopac). For chorda tympani recordings, a total of 7–10 Rgs21Δ5/Δ5 mice and 7–9 wild-type littermate control mice were used. The fungiform papillae were stimulated with different tastants (applied for 30 s and then rinsed with water for 40 s) with a constant flow pump (Fisher Scientific). Each series of stimuli consisted of: 100 mM NH4Cl, 100 mM MSG + 0.5 mM inosine monophosphate (IMP), 10 mM quinine, 500 mM sucrose, 5 mM SC45657, 100 mM NaCl, 10 mM HCl, 10 mM citric acid (CA), and a second application of 100 mM NH4Cl. Stimuli were randomly applied between the 2 NH4Cl applications; each series was repeated 2–3 times on each animal. To analyze the data, the amplitude of each integrated response was first averaged over the 30-s application using AcqKnowledge software. Since no significant difference was observed between the separate NH4Cl responses at the beginning and at the end of each series (paired Student’s t test; P > 0.05), each tastant’s responses were normalized to the average amplitude of NH4Cl applications. Responses to each stimulus were averaged per animal and compared between Rgs21+/+ and Rgs21Δ5/Δ5 mice with an unpaired Student’s t test (GraphPad Prism 7). The grand mean was calculated and represented in Figure 6B. We also measured the amplitude of each response in the first 10 s and in the last 10 s; the ratio representing the last 10 s of response over the first 10 s of response was then calculated and compared between Rgs21+/+ and Rgs21Δ5/Δ5 mice with an unpaired Student’s t test.
Figure 6.
(A) Representative integrated chorda tympani nerve responses in Rgs21Δ5/Δ5 mice. Indicated taste stimuli were applied for 30 s with 40-s intervening washes. (B) Amplitude of the integrated response for each tastant. Responses were normalized to the responses to 100 mM NH4Cl, recorded both at the beginning and the end of each tastant series, to control for variability between mice. Responses show the mean ± SEM (n = 7–10 mice for each stimulus). No significant difference was observed between Rgs21+/+ and Rgs21Δ5/Δ5 mice for NH4Cl or the sour tastants HCl and citric acid (unpaired Student’s t tests: P > 0.7 for NH4Cl, P > 0.9 for sour tastants). Decreased responses of Rgs21Δ5/Δ5 mice to the umami agonists MSG + IMP, the bitterant quinine, and the sweeteners sucrose and SC45647 were statistically significant (unpaired Student’s t tests: P < 0.03 for MSG + IMP, P < 0.022 for quinine, P < 0.0002 for sucrose, P < 0.002 for SC45647). A trend toward a decreased response to 100 mM NaCl was observed in Rgs21Δ5/Δ5 mice, but the difference was not statistically significant (P < 0.16; Student’s t test). (C) Plotted are the ratios of the last 10 s to first 10 s of recorded CT nerve response (unpaired Student’s t tests: *P < 0.05; **P < 0.01; ***P < 0.001).
Results
Development of Rgs21-null mice
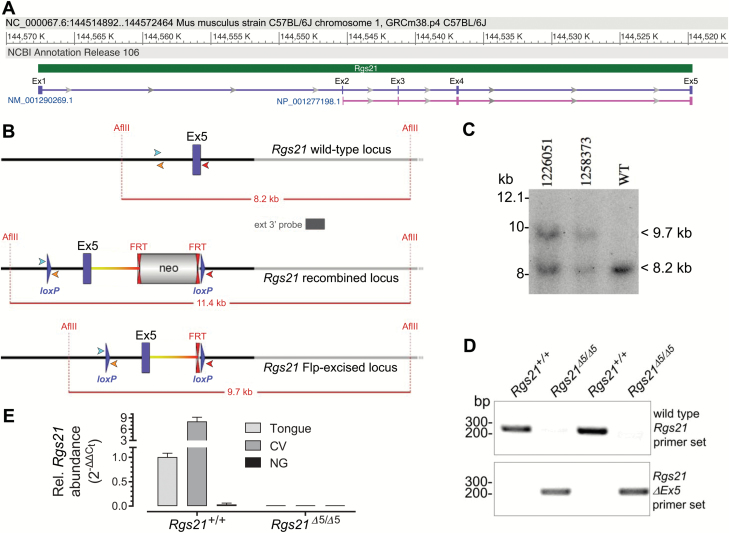
PCR amplification was used to determine the genotype of Rgs21+/+ and Rgs21Δ5/Δ5 mice. Specific primers, designed around the floxed exon 5 portion of Rgs21, were used to discriminate between mice with unexcised (Rgs21+/+) and excised (Rgs21Δ5/Δ5) genomic DNA (Figure 1). Real-time quantitative RT-PCR was used to confirm that Rgs21 mRNA expression is observable in tongue tissue and enriched in CV tissue, but absent in non-gustatory epithelial tissue (Figure 1E). Additionally, Rgs21Δ5/Δ5 mice lack the Rgs21 mRNA expression observable in Rgs21+l+ whole tongue and CV tissues (Figure 1E).
Figure 1.
Genotype and gene expression validation of successful excision of Rgs21 exon 5. (A) Architecture of Rgs21 locus on chromosome 1 of C57BL/6J mice, as obtained from GenBank record NC_000067.6 and spanning its 5 exons (Ex1–Ex5) as denoted by its RNA transcript (NM_001290269, purple) and its encoded RGS21 polypeptide (NP_001277198.1, pink). (B) Illustration of wild-type Rgs21 surrounding exon 5, the predicted insertion of loxP recombination sites and neomycin (“neo”) selection cassette by homologous recombination, and the predicted result of Flp recombinase excision. Forward (cyan) and reverse (orange, red) primers used in detecting Cre-mediated recombination of loxP sites are indicated as arrowheads. (C) Southern blot validation of successful Flp recombinase excision of neo cassette, based on AflII digestion of genomic DNA from indicated mice (or C57BL/6 control mouse, “WT”), subsequent blotting on nylon membrane, and hybridization with an external 3′ DNA probe (indicated in B). (D) Ear-punch DNA samples from 4 indicated progeny of an Rgs21Δ5/+ × Rgs21Δ5/+ mating were genotyped by PCR. Specific primers (denoted in B) were designed around the floxed exon 5 portion of Rgs21 to discriminate between unexcised (Rgs21+/+) and Cre recombinase-excised (Rgs21Δ5/Δ5) genomic DNA. (E) Data from qRT-PCR (SYBR Green detection) of the Rgs21 mRNA transcript, which is seen to be completely absent in Rgs21Δ5/Δ5 mice and absent in non-gustatory (“NG”) epithelial tissue from Rgs21+/+ mice, but detectable in tongue and enriched in circumvallate papillae (“CV”) tissue from Rgs21+/+ mice.
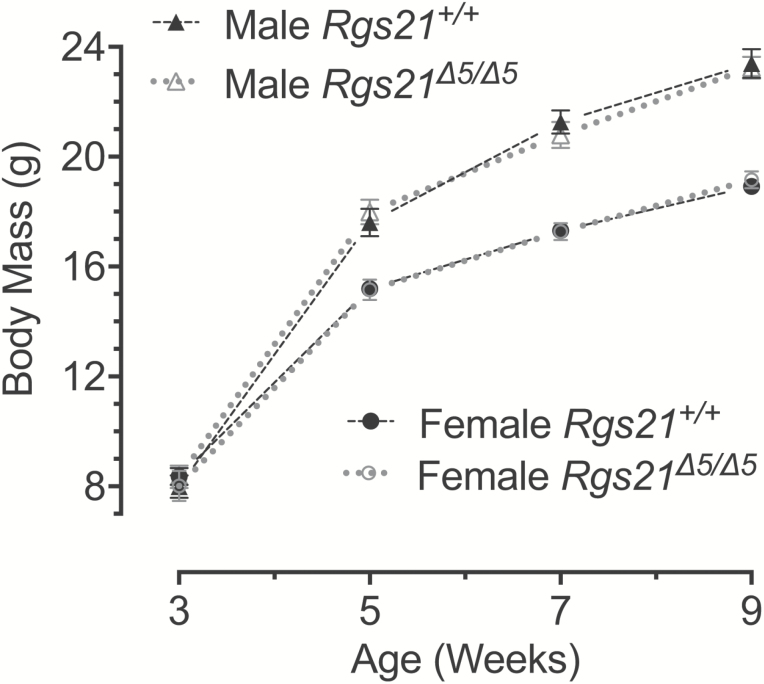
Rgs21 Δ5/Δ5 mice were observed to be grossly phenotypically normal and, when bred from Rgs21Δ5/+x Rgs21Δ5/+ crosses, were born in a normal Mendelian ratio of 1:2:1. In addition, Rgs21Δ5/Δ5 mice exhibited no differences in body weight gain compared with Rgs21+l+ littermates (Figure 2). These latter data suggest that no appetitive changes to regular nutritional sources (e.g., cage-delivered chow and water) occur upon the loss of Rgs21 expression.
Figure 2.
Mean ± SEM body mass of male (n = 10) and female (n = 10) Rgs21Δ5/Δ5 mice (and wild-type littermate controls; male n = 9, female n = 13) at 3, 5, 7, and 9 weeks of age. A 2-way ANOVA was used to compare the body mass of wild-type and Rgs21-null mice within each sex at the indicated time points. There was no difference found in body mass between Rgs21Δ5/Δ5 and Rgs21+/+ female (P = 0.8471) or male mice (P = 0.9384).
Normal lingual histology and detection of taste cell-specific markers in Rgs21-null mice
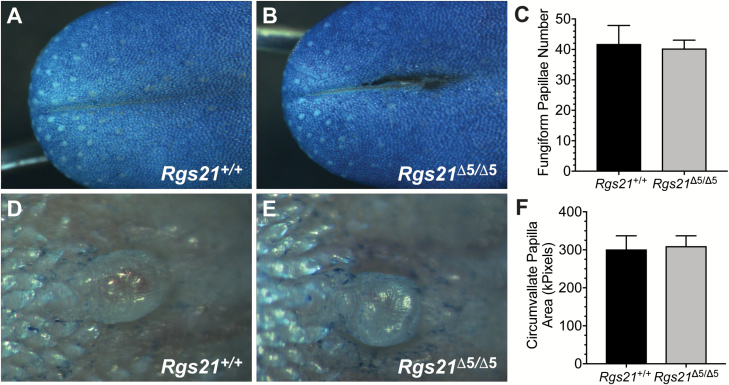
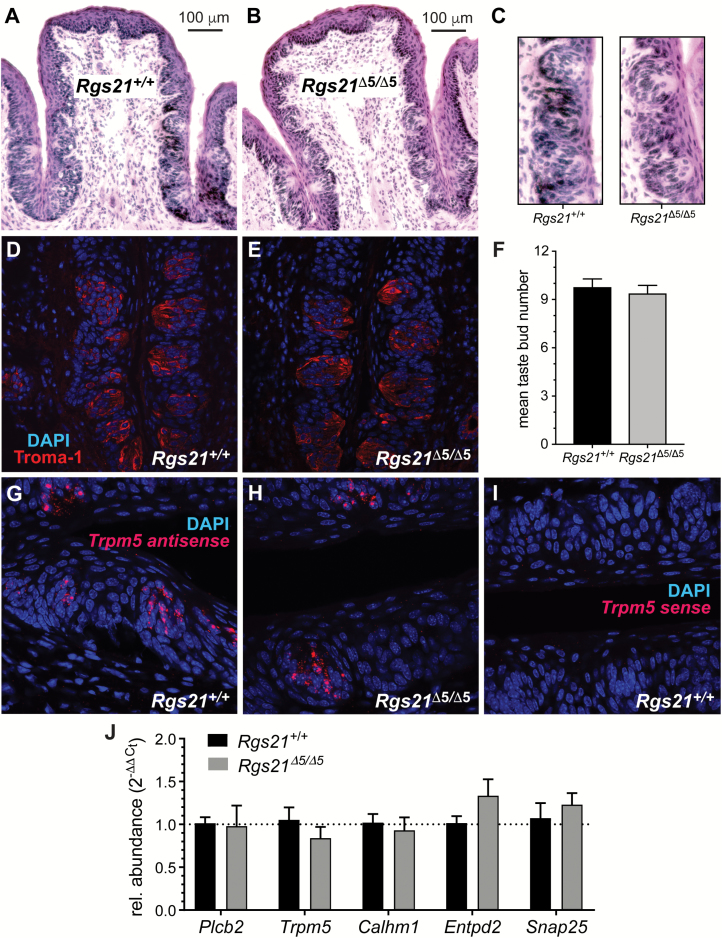
To assess any changes to tastant-responsive lingual tissue composition and/or morphology, we first assessed general taste papillae morphology of methylene blue stained Rgs21+/+ and Rgs21Δ5/Δ5 tongues. WT and Rgs21-null mice had an equivalent number of fungiform papillae on the anterior tongue (Figure 3A–C). Additionally, the apical size of the CV was similar between WT and Rgs21-null mice (Figure 3D–F).
Figure 3.
Normal tongue morphology in Rgs21-null mice. Tongues of wild-type (n = 6; e.g., A) and Rgs21Δ5/Δ5 mice (n = 6; e.g., B) were stained with methylene blue to visualize the fungiform papillae on the anterior portions. There was no difference found in mean fungiform papillae number or morphology between Rgs21-null and wild-type animals (C; unpaired Student’s t test; P = 0.8254). Circumvallate papillae of wild-type (n = 4) and Rgs21Δ5/Δ5 mice (n = 4) were observed by bright-field microscopy (D, E; areas plotted in F). There was no difference found in circumvallate papillae area (F; unpaired Student’s t test; P = 0.8578).
The CV was also stained with hemotoxylin and eosin to assess CV and taste bud morphology (Figure 4A–C). Taste bud and CV cross-sectional morphology appeared equivalent between Rgs21-null mice and wild-type littermate controls. Cross-sections of CV from Rgs21+/+ and Rgs21Δ5/Δ5 were also labeled with anti-keratin-8 (“Troma-1”; Figure 4D and E). The number of taste buds in each section were counted using Troma-1 staining; Rgs21Δ5/Δ5 taste buds did not differ from Rgs21+/+ taste buds in number (Figure 4F). Furthermore, ISH for the Type II taste receptor cell marker Trpm5 revealed no differences between genotypes (e.g., Figure 4G–I); quantitative RT-PCR detection of Trpm5 mRNA and transcripts of other taste cell markers (Plcb2, Calhm1 for Type II cells, Entpd2 for Type I cells, and Snap25 for Type III cells) within taste epithelium isolated from the CV also revealed no expression differences between genotypes (Figure 4J).
Figure 4.
Normal morphology and expression of taste cell markers within circumvallate papillae of Rgs21-null mice. Circumvallate papillae of wild-type (n = 4) and Rgs21Δ5/Δ5 mice (n = 4) were stained with haematoxylin and eosin (H&E) (A, B; magnification of taste buds in C), or visualized by dual-color indirect immunofluorescence with the taste cell marker Troma-1 (anti-keratin-8 in red) and DNA dye (DAPI in blue) (D, E). There was no difference found in mean taste bud number per section (F; unpaired Student’s t-test; P = 0.5879) between knockout and wild-type animals. Equivalent detection of the Type II taste cell marker Trpm5 within CV sections was observed in both genotypes via in situ hybridization and subsequent confocal microscopy (G, H with antisense probe; I with sense probe). Equivalent mRNA expression was also observed by qRT-PCR for additional markers of Type I (Entpd2), Type II (Plcb2, Trpm5, Calhm1), and Type III (Snap25) taste cells (J). Bar graph displays the mean ± SEM; no difference between genotypes was statistically significant by Student’s t test.
Reduced responsiveness in 2-bottle choice tests
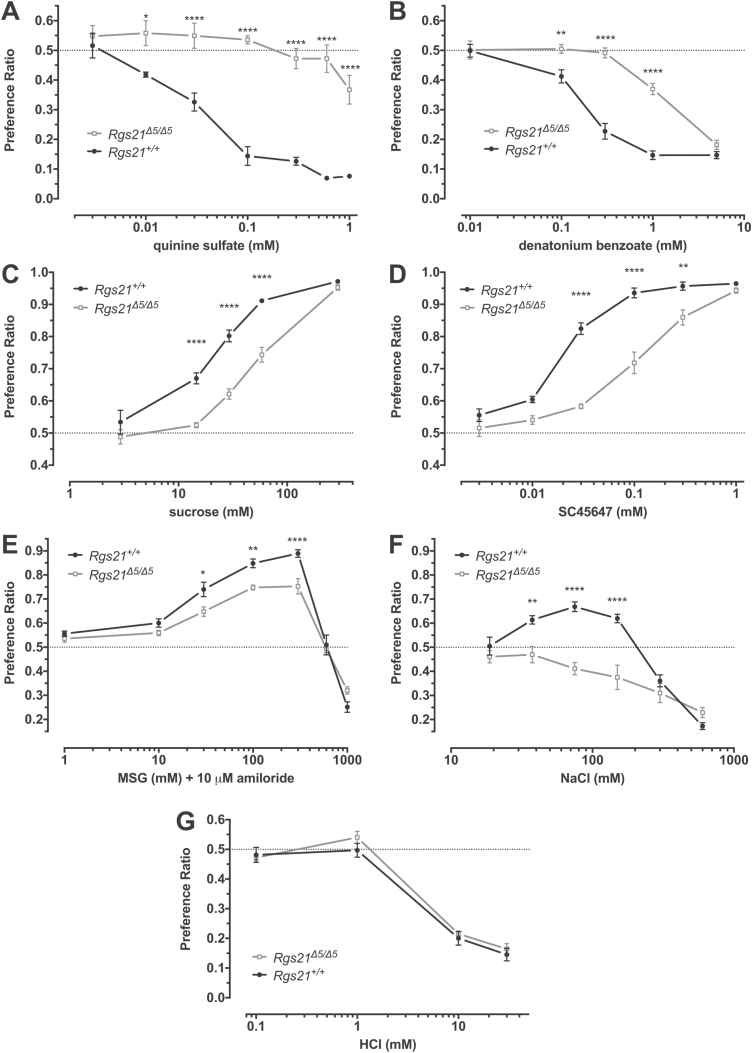
Rgs21 +/+ and Rgs21Δ5/Δ5 mice underwent 2-bottle choice testing to determine if Rgs21 contributes to the in vivo response to tastants sensed by GPCR taste receptors, that is, tastants described as bitter, sweet, or umami. Rgs21-null mice showed a lack of aversion to quinine sulfate (Figure 5A) and a reduced aversion to denatonium benzoate (Figure 5B), tastants representing the bitter taste quality, which are mediated by the T2R family of GPCRs. Additionally, Rgs21Δ5/Δ5 mice revealed reduced preference towards the sweetener sucrose (Figure 5C). To ensure that the post-ingestive influence of sucrose is not influencing the altered response observed with Rgs21-null mice, we also tested preference for the non-caloric sweetener SC45647. Rgs21Δ5/Δ5 mice also showed reduced preference for SC45647 (Figure 5D). To assess whether umami taste is altered in Rgs21Δ5/Δ5 mice, we measured 2-bottle choice using MSG. The preference for MSG was blunted in Rgs21-null mice, as they did not reach a peak preference ratio equivalent to that of wild-type mice (Figure 5E). All MSG solutions contained 10 μM amiloride to block the effects of the sodium ion. Surprisingly, Rgs21Δ5/Δ5 mice lacked the appetitive response to moderate concentrations of NaCl seen in wild-type mice (Figure 5F), but showed no change in aversion to the sour tastant hydrochloric acid (Figure 5G).
Figure 5.
Two-bottle choice preferences of Rgs21Δ5/Δ5 (n = 7) and wild-type mice (n = 7) for ascending concentration series of the bitterants quinine sulfate (A), denatonium benzoate (B), the sweeteners sucrose (C) and SC45647 (D), the L-amino acid umami tastant monosodium glutamate in the presence of 10 µM amiloride (E), the salt taste stimulus NaCl (F), and the sour tastant hydrochloric acid (G). Each test was assessed with a 2-way ANOVA. Differences between the groups in preference for specific concentrations of taste solution were determined using Sidak post hoc test to correct for multiple comparisons (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001).
Blunted responses in chorda tympani nerve recordings
To test whether Rgs21 loss affected the acute phase of taste responses post-lingual tissue engagement, we used Rgs21Δ5/Δ5 mice and Rgs21+l+ control littermates to perform chorda tympani whole-nerve recordings while stimulating the tongue with various taste stimuli. As shown in Figure 6, no significant difference was observed between the 2 mouse strains in the amplitude of responses to the sour tastants HCl and citric acid, as normalized to the average responses to NH4Cl recorded at the beginning and ending of the tastant series. In contrast, significantly depressed responses were seen in Rgs21Δ5/Δ5 mice for exposures to the bitterant quinine, the 2 sweeteners tested (sucrose and SC45647), and the joint application of 2 umami tastants (i.e., MSG and IMP; Figure 6B). Additionally, Rgs21Δ5/Δ5 mice are seen to have an altered ratio of the last 10 s to first 10 s of recorded response to sweet, bitter, and umami taste stimuli (Figure 6C).
In contrast to the behavioral response (Figure 5F), a significant difference was not found for Rgs21Δ5/Δ5 mice in response to 100 mM NaCl (Figure 6). However, the remainder of these data are consistent with the overall behavioral responses (i.e., reduction of GPCR-mediated events, but not sour signaling; Figure 5) and thereby suggest that Rgs21 functions at the level of peripheral taste input in these taste qualities. While Rgs21 expression is thought to be limited to lingual (and airway) gustatory tissue (von Buchholtz et al. 2004; Cohen et al. 2012; Kimple et al. 2014), blunted preference towards moderate concentrations of NaCl may instead be the result of altered post-ingestive factors, hedonics, and/or central integration caused by Rgs21 loss.
Discussion
The principal finding of this study is that Rgs21 expression is required for establishing the normal responsiveness of the mouse gustatory system to tastants that signal through GPCRs. Mouse and rat Rgs21 were each shown previously to be expressed exclusively in cells expressing markers found in Type II taste cells (von Buchholtz et al. 2004; Cohen et al. 2012). The mouse Rgs21 gene promoter was found to be active only in taste bud cells co-incident with α-gustducin expression (Cohen et al. 2012), whereas rat Rgs21 mRNA is seen to be present in all Plcb2-expressing taste bud cells (i.e., in addition to α-gustducin-expressing cells) (von Buchholtz et al. 2004). These observations may indicate a species difference, with mice expressing Rgs21 only in gustducin-positive Type II taste cells and rats expressing Rgs21 in all Type II taste cells. However, this difference may alternatively be the result of experimental limitations of the BAC transgenic mouse described by Cohen and colleagues (Cohen et al. 2012), which may lack all the necessary Rgs21 promoter/enhancer sequence elements within the 232-kb BAC clone for reflecting the totality of its native expression pattern.
Microscopy of tongues from Rgs21Δ5/Δ5 mice revealed apparently normal fungiform and CV papillae; taste bud number within the CV also did not differ between wild-type and Rgs21-null mice (Figures 3 and 4). The distribution of Trpm5+ cells and levels of other taste cell-specific transcripts, representing all 3 major classes of taste cells, are unchanged in lingual tissue from Rgs21Δ5/Δ5 mice, suggesting that Rgs21 knockout mice likely have a full complement of taste cells. These findings suggest that the reduced GPCR-mediated (and salt) taste responsiveness of Rgs21Δ5/Δ5 mice is unlikely to be explained by an abnormal development of lingual chemosensory tissue.
Observations of reduced responsiveness to bitter, sweet, and umami tastants upon Rgs21 loss run opposite to prior expectations built upon the general knowledge of RGS protein function as negative regulators of GPCR signaling (Siderovski and Willard 2005; Lambert et al. 2010; Woodard et al. 2015); moreover, these present observations from characterizing Rgs21Δ5/Δ5 mice run counter to our prior finding that acute Rgs21 knockdown in tastant-responsive epithelial cells leads to amplified intracellular second messenger responses upon exposure to bitter compounds (Cohen et al. 2012). Based centrally on the fact that RGS proteins like RGS21 accelerate inactivation of GPCR-activated Gα subunits (Berman et al. 1996; Watson et al. 1996; Snow et al. 1998; Cohen et al. 2012; Kimple et al. 2014), the absence of Rgs21 is expected to prolong, and therefore potentiate, the signaling of agonist-occupied GPCRs, such as the tastant-responsive T1Rs and T2Rs of taste receptor cells (Adler et al. 2000; Chandrashekar et al. 2000; Nelson et al. 2001; Bufe et al. 2002; Li et al. 2002; Nelson et al. 2002; Zhao et al. 2003). GPCR-targeting tastants should be more potent upon Rgs21 loss, and thus we originally expected to observe a leftward shift in the dose–response curves for sweet/umami preference and bitter avoidance in Rgs21-null mice compared with wild-type littermates. Instead, a rightward shift was seen for preference to sucrose, SC45647, and monosodium glutamate, and for aversion to the bitter compounds quinine and denatonium. Additionally, Rgs21Δ5/Δ5 mice were observed to have a reduced ratio of the last 10 s to the first 10 s of recorded CT nerve response upon exposure to these GPCR-mediated taste stimuli, suggesting that these mice have a less-prolonged signal output after initial tastant/receptor engagement.
As expected, Rgs21Δ5/Δ5 mice showed no change in aversion to hydrochloric acid (Figure 5G), a tastant representing the sour taste quality. Sour taste responses are not mediated by GPCR signaling; rather, they are mediated by a Zn2+-sensitive proton conductance in Type III taste cells, which blocks the inwardly rectifying K+ channel KIR2.1 (Chang et al. 2010; Ye et al. 2016; Tu et al. 2018).
An additional surprising observation was that Rgs21Δ5/Δ5 mice had a reduced appetitive response to moderate concentrations of NaCl (37.5–150 mM; Figure 5F). While recent work has suggested that bitter-responsive taste cells mediate a portion of the aversive response to high NaCl concentrations (Oka et al. 2013), appetitive responses to moderate NaCl concentrations are thought to be mediated by a separate population of amiloride-sensitive taste cells, independent of Types II and III taste cells (Vandenbeuch et al. 2008; Chandrashekar et al. 2010). Others have suggested that amiloride-sensitive taste cells may transduce salt taste information to the afferent nerve, at least in part, via GPCR-mediated communication with Type II cells (Tordoff et al. 2014). However, evidence of GPCR signaling in Type II taste cells mediating the appetitive response to moderate NaCl concentrations has been inconsistently observed (Wong et al. 1996; Zhang et al. 2003; Damak et al. 2006; Hisatsune et al. 2007). Additionally, Rgs21Δ5/Δ5 mice were not observed to have a statistically significant reduction of their CT nerve responses to 100 mM of NaCl, casting doubt on this hypothesized mechanism of amiloride-sensitive transmission. The reduced preference for moderate NaCl concentrations shown by Rgs21Δ5/Δ5 mice may therefore result from altered post-ingestive influences, including possible alterations to the renin–angiotensin system (Bachmanov et al. 2002; Sakamoto et al. 2016). This possibility is currently under investigation.
Does loss of Rgs21 lead to desensitization of the GPCR-mediated taste system?
Based upon the conventional function of RGS proteins, as well as our prior in vitro findings in tastant-responsive epithelial cells, loss of RGS21 in taste receptor cells is expected to result in prolonged signaling of taste GPCRs and, subsequently, a prolonged elevation of ATP release. Reduced (and less prolonged) responsiveness towards GPCR-mediated tastants in Rgs21Δ5/Δ5 mice may be explained by desensitization at the level of the taste receptor and/or downstream signaling components. Hyperactivity of GPCRs commonly leads to phosphorylation and subsequent desensitization and down-regulation of the receptor (Freedman and Lefkowitz 1996; Luttrell and Lefkowitz 2002; Rajagopal and Shenoy 2018). In the event that elevated signaling upon Rgs21 loss does not desensitize the taste receptor cell directly, prolonged elevation of ATP release could desensitize downstream purinergic receptors, including those on the gustatory nerve fiber (North 2002); there is precedence for desensitization and loss of taste responsiveness with elevated extracellular ATP levels caused by genetic ablation (e.g., of the ectonucleotidase NTPDase2; (Vandenbeuch et al. 2013).
Precedence of bidirectional effects on GPCR signaling upon RGS protein loss
In addition to the potential for Rgs21 loss to lead to desensitization, there is additional emerging evidence in the literature for bidirectional regulation of GPCR signaling by RGS proteins. For example, we used siRNA-mediated “knock-down” of each of the 17 different RGS proteins expressed in HEK293 cells to examine the effects of RGS protein deficiency on endogenous GPCR-mediated cellular signaling (Laroche et al. 2010). Consistent with the original model wherein RGS protein deficiency increases GPCR agonist-mediated signaling, we found that RGS11 knockdown increased maximal muscarinic receptor-induced calcium flux (Laroche et al. 2010). The same effect was observed for knockdown of RGS2 on PAR-1 receptor-induced calcium flux (Laroche et al. 2010). However, RGS9 knock-down resulted in a decrease in both the potency and efficacy of the agonist carbachol on endogenous muscarinic receptor responses. Similarly, RGS8 knock-down yielded a decrease in maximal PAR-1 receptor-induced calcium flux (Laroche et al. 2010). These 2 latter results are compatible with an alternative model wherein RGS protein deficiency can lead to a decrease of agonist potency and/or efficacy at some GPCRs, which may be the case in Rgs21-null mice with respect to proximal umami (T1R1/2), sweet (T1R2/3), and bitter (T2R) signaling and/or supportive, autocrine purinergic receptor (P2Y1 and P2Y2) signaling.
Parallel to Rgs-3 function in Caenorhabditis elegans?
There is precedence in another model organism for our present, mouse knockout strain-based observation of reduced taste responses upon the loss of an RGS protein. The selective distribution of Rgs21 in rodent and human taste cells (von Buchholtz et al. 2004; Cohen et al. 2012; Kimple et al. 2014) is reminiscent of the sensory-specific distribution in C. elegans of Rgs-3, which is found only in a subset of chemosensory neurons. Rgs-3-deficient C. elegans exhibit normal development and motor ability compared to wild-type nematodes, but they demonstrate an inability to respond to normal levels of chemoattractants such as isoamyl alcohol (Ferkey et al. 2007). RGS21 may play a similar role in dampening tastant signaling so that the mammalian gustatory system is not overwhelmed when tastants are too abundant on the lingual epithelium; in the absence of RGS protein GAP activity, normal exposure to GPCR-mediated tastants during lingual epithelium development may lead to compensatory desensitization or down-regulation of tastant response machinery. We are currently pursuing this particular hypothesis with temporally-controlled, conditional Rgs21 knockout approaches.
Funding
This work was supported, in part, by the West Virginia University E.J. Van Liere Medicine Professorship and William W. Fleming Pharmacology Trust endowments [to D.P.S. and A.S., respectively]. Trpm5 in situ hybridization data were generated by the Baylor College of Medicine’s RNA In Situ Hybridization Core supported by NIH grant S10 OD016167.
Acknowledgements
We thank Dr. Sue Kinnamon for provision of SC45647 and guidance throughout the conduct of this investigation.
References
- Adler E, Hoon MA, Mueller KL, Chandrashekar J, Ryba NJ, Zuker CS. 2000. A novel family of mammalian taste receptors. Cell. 100:693–702. [DOI] [PubMed] [Google Scholar]
- Angleson JK, Wensel TG. 1993. A GTPase-accelerating factor for transducin, distinct from its effector cGMP phosphodiesterase, in rod outer segment membranes. Neuron. 11:939–949. [DOI] [PubMed] [Google Scholar]
- Bachmanov AA, Beauchamp GK, Tordoff MG. 2002. Voluntary consumption of NaCl, KCl, CaCl2, and NH4Cl solutions by 28 mouse strains. Behav Genet. 32:445–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnett NP, Apodaca TR, Magill M, Colby SM, Gwaltney C, Rohsenow DJ, Monti PM. 2010. Moderators and mediators of two brief interventions for alcohol in the emergency department. Addiction. 105:452–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman DM, Wilkie TM, Gilman AG. 1996. GAIP and RGS4 are GTPase-activating proteins for the Gi subfamily of G protein alpha subunits. Cell. 86:445–452. [DOI] [PubMed] [Google Scholar]
- Bo X, Alavi A, Xiang Z, Oglesby I, Ford A, Burnstock G. 1999. Localization of ATP-gated P2X2 and P2X3 receptor immunoreactive nerves in rat taste buds. Neuroreport. 10:1107–1111. [DOI] [PubMed] [Google Scholar]
- Bufe B, Hofmann T, Krautwurst D, Raguse JD, Meyerhof W. 2002. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nat Genet. 32:397–401. [DOI] [PubMed] [Google Scholar]
- Bystrova MF, Yatzenko YE, Fedorov IV, Rogachevskaja OA, Kolesnikov SS. 2006. P2Y isoforms operative in mouse taste cells. Cell Tissue Res. 323:377–382. [DOI] [PubMed] [Google Scholar]
- Chandrashekar J, Kuhn C, Oka Y, Yarmolinsky DA, Hummler E, Ryba NJ, Zuker CS. 2010. The cells and peripheral representation of sodium taste in mice. Nature. 464:297–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J, Mueller KL, Hoon MA, Adler E, Feng L, Guo W, Zuker CS, Ryba NJ. 2000. T2Rs function as bitter taste receptors. Cell. 100:703–711. [DOI] [PubMed] [Google Scholar]
- Chang RB, Waters H, Liman ER. 2010. A proton current drives action potentials in genetically identified sour taste cells. Proc Natl Acad Sci USA. 107:22320–22325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CK, Burns ME, He W, Wensel TG, Baylor DA, Simon MI. 2000. Slowed recovery of rod photoresponse in mice lacking the GTPase accelerating protein RGS9-1. Nature. 403:557–560. [DOI] [PubMed] [Google Scholar]
- Clapp TR, Yang R, Stoick CL, Kinnamon SC, Kinnamon JC. 2004. Morphologic characterization of rat taste receptor cells that express components of the phospholipase C signaling pathway. J Comp Neurol. 468:311–321. [DOI] [PubMed] [Google Scholar]
- Cohen SP, Buckley BK, Kosloff M, Garland AL, Bosch DE, Cheng G Jr, Radhakrishna H, Brown MD, Willard FS, Arshavsky VY, et al. . 2012. Regulator of G-protein signaling-21 (RGS21) is an inhibitor of bitter gustatory signaling found in lingual and airway epithelia. J Biol Chem. 287:41706–41719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damak S, Rong M, Yasumatsu K, Kokrashvili Z, Pérez CA, Shigemura N, Yoshida R, Mosinger B Jr, Glendinning JI, Ninomiya Y, et al. . 2006. Trpm5 null mice respond to bitter, sweet, and umami compounds. Chem Senses. 31:253–264. [DOI] [PubMed] [Google Scholar]
- DeFazio RA, Dvoryanchikov G, Maruyama Y, Kim JW, Pereira E, Roper SD, Chaudhari N. 2006. Separate populations of receptor cells and presynaptic cells in mouse taste buds. J Neurosci. 26:3971–3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druey KM, Blumer KJ, Kang VH, Kehrl JH. 1996. Inhibition of G-protein-mediated MAP kinase activation by a new mammalian gene family. Nature. 379:742–746. [DOI] [PubMed] [Google Scholar]
- Ferkey DM, Hyde R, Haspel G, Dionne HM, Hess HA, Suzuki H, Schafer WR, Koelle MR, Hart AC. 2007. C. elegans G protein regulator RGS-3 controls sensitivity to sensory stimuli. Neuron. 53:39–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finger TE, Danilova V, Barrows J, Bartel DL, Vigers AJ, Stone L, Hellekant G, Kinnamon SC. 2005. ATP signaling is crucial for communication from taste buds to gustatory nerves. Science. 310:1495–1499. [DOI] [PubMed] [Google Scholar]
- Freedman NJ, Lefkowitz RJ. 1996. Desensitization of G protein-coupled receptors. Recent Prog Horm Res. 51:319–351; discussion 352. [PubMed] [Google Scholar]
- He W, Cowan CW, Wensel TG. 1998. RGS9, a GTPase accelerator for phototransduction. Neuron. 20:95–102. [DOI] [PubMed] [Google Scholar]
- Hisatsune C, Yasumatsu K, Takahashi-Iwanaga H, Ogawa N, Kuroda Y, Yoshida R, Ninomiya Y, Mikoshiba K. 2007. Abnormal taste perception in mice lacking the type 3 inositol 1,4,5-trisphosphate receptor. J Biol Chem. 282:37225–37231. [DOI] [PubMed] [Google Scholar]
- Hoon MA, Northup JK, Margolskee RF, Ryba NJ. 1995. Functional expression of the taste specific G-protein, alpha-gustducin. Biochem J. 309:629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Shanker YG, Dubauskaite J, Zheng JZ, Yan W, Rosenzweig S, Spielman AI, Max M, Margolskee RF. 1999. Ggamma13 colocalizes with gustducin in taste receptor cells and mediates IP3 responses to bitter denatonium. Nat Neurosci. 2:1055–1062. [DOI] [PubMed] [Google Scholar]
- Huang YA, Dando R, Roper SD. 2009. Autocrine and paracrine roles for ATP and serotonin in mouse taste buds. J Neurosci. 29:13909–13918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YA, Stone LM, Pereira E, Yang R, Kinnamon JC, Dvoryanchikov G, Chaudhari N, Finger TE, Kinnamon SC, Roper SD. 2011. Knocking out P2X receptors reduces transmitter secretion in taste buds. J Neurosci. 31:13654–13661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka S, Toyono T, Seta Y, Ogura T, Toyoshima K. 2004. Expression of P2Y1 receptors in rat taste buds. Histochem Cell Biol. 121:419–426. [DOI] [PubMed] [Google Scholar]
- Kimple AJ, Bosch DE, Giguère PM, Siderovski DP. 2011. Regulators of G-protein signaling and their Gα substrates: promises and challenges in their use as drug discovery targets. Pharmacol Rev. 63:728–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimple AJ, Garland AL, Cohen SP, Setola V, Willard FS, Zielinski T, Lowery RG, Tarran R, Siderovski DP. 2014. RGS21, a regulator of taste and mucociliary clearance?Laryngoscope. 124:E56–E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinnamon SC, Finger TE. 2013. A taste for ATP: neurotransmission in taste buds. Front Cell Neurosci. 7:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koelle MR, Horvitz HR. 1996. EGL-10 regulates G protein signaling in the C. elegans nervous system and shares a conserved domain with many mammalian proteins. Cell. 84:115–125. [DOI] [PubMed] [Google Scholar]
- Kusakabe Y, Yasuoka A, Asano-Miyoshi M, Iwabuchi K, Matsumoto I, Arai S, Emori Y, Abe K. 2000. Comprehensive study on G protein alpha-subunits in taste bud cells, with special reference to the occurrence of Galphai2 as a major Galpha species. Chem Senses. 25:525–531. [DOI] [PubMed] [Google Scholar]
- Lambert NA, Johnston CA, Cappell SD, Kuravi S, Kimple AJ, Willard FS, Siderovski DP. 2010. Regulators of G-protein signaling accelerate GPCR signaling kinetics and govern sensitivity solely by accelerating GTPase activity. Proc Natl Acad Sci USA. 107:7066–7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laroche G, Giguère PM, Roth BL, Trejo J, Siderovski DP. 2010. RNA interference screen for RGS protein specificity at muscarinic and protease-activated receptors reveals bidirectional modulation of signaling. Am J Physiol Cell Physiol. 299:C654–C664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Staszewski L, Xu H, Durick K, Zoller M, Adler E. 2002. Human receptors for sweet and umami taste. Proc Natl Acad Sci USA. 99:4692–4696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebl DJ, Mbiene JP, Parada LF. 1999. NT4/5 mutant mice have deficiency in gustatory papillae and taste bud formation. Dev Biol. 213:378–389. [DOI] [PubMed] [Google Scholar]
- Luttrell LM, Lefkowitz RJ. 2002. The role of beta-arrestins in the termination and transduction of G-protein-coupled receptor signals. J Cell Sci. 115:455–465. [DOI] [PubMed] [Google Scholar]
- McLaughlin SK, McKinnon PJ, Margolskee RF. 1992. Gustducin is a taste-cell-specific G protein closely related to the transducins. Nature. 357:563–569. [DOI] [PubMed] [Google Scholar]
- Ming D, Ruiz-Avila L, Margolskee RF. 1998. Characterization and solubilization of bitter-responsive receptors that couple to gustducin. Proc Natl Acad Sci USA. 95:8933–8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson G, Chandrashekar J, Hoon MA, Feng L, Zhao G, Ryba NJ, Zuker CS. 2002. An amino-acid taste receptor. Nature. 416:199–202. [DOI] [PubMed] [Google Scholar]
- Nelson G, Hoon MA, Chandrashekar J, Zhang Y, Ryba NJ, Zuker CS. 2001. Mammalian sweet taste receptors. Cell. 106:381–390. [DOI] [PubMed] [Google Scholar]
- North RA. 2002. Molecular physiology of P2X receptors. Physiol Rev. 82:1013–1067. [DOI] [PubMed] [Google Scholar]
- Oka Y, Butnaru M, von Buchholtz L, Ryba NJ, Zuker CS. 2013. High salt recruits aversive taste pathways. Nature. 494:472–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo T, Pevny LH, Hogan BL. 2006. Sox2 is required for development of taste bud sensory cells. Genes Dev. 20:2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer RK. 2007. The pharmacology and signaling of bitter, sweet, and umami taste sensing. Mol Interv. 7:87–98. [DOI] [PubMed] [Google Scholar]
- Rajagopal S, Shenoy SK. 2018. GPCR desensitization: acute and prolonged phases. Cell Signal. 41:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz CJ, Wray K, Delay E, Margolskee RF, Kinnamon SC. 2003. Behavioral evidence for a role of alpha-gustducin in glutamate taste. Chem Senses. 28:573–579. [DOI] [PubMed] [Google Scholar]
- Sakamoto T, Fujii A, Saito N, Kondo H, Ohuchi A. 2016. Alteration of amiloride-sensitive salt taste nerve responses in aldosterone/NaCl-induced hypertensive rats. Neurosci Res. 108:60–66. [DOI] [PubMed] [Google Scholar]
- Seki T, Namba T, Mochizuki H, Onodera M. 2007. Clustering, migration, and neurite formation of neural precursor cells in the adult rat hippocampus. J Comp Neurol. 502:275–290. [DOI] [PubMed] [Google Scholar]
- Shindo Y, Miura H, Carninci P, Kawai J, Hayashizaki Y, Ninomiya Y, Hino A, Kanda T, Kusakabe Y. 2008. G alpha14 is a candidate mediator of sweet/umami signal transduction in the posterior region of the mouse tongue. Biochem Biophys Res Commun. 376:504–508. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Hessel A, Chung S, Mak TW, Tyers M. 1996. A new family of regulators of G-protein-coupled receptors?Curr Biol. 6:211–212. [DOI] [PubMed] [Google Scholar]
- Siderovski DP, Willard FS. 2005. The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits. Int J Biol Sci. 1:51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Hall RA, Krumins AM, Brothers GM, Bouchard D, Brothers CA, Chung S, Mangion J, Gilman AG, Lefkowitz RJ, et al. . 1998. GTPase activating specificity of RGS12 and binding specificity of an alternatively spliced PDZ (PSD-95/Dlg/ZO-1) domain. J Biol Chem. 273:17749–17755. [DOI] [PubMed] [Google Scholar]
- Tao GZ, Toivola DM, Zhong B, Michie SA, Resurreccion EZ, Tamai Y, Taketo MM, Omary MB. 2003. Keratin-8 null mice have different gallbladder and liver susceptibility to lithogenic diet-induced injury. J Cell Sci. 116:4629–4638. [DOI] [PubMed] [Google Scholar]
- Taruno A, Vingtdeux V, Ohmoto M, Ma Z, Dvoryanchikov G, Li A, Adrien L, Zhao H, Leung S, Abernethy M, et al. . 2013. CALHM1 ion channel mediates purinergic neurotransmission of sweet, bitter and umami tastes. Nature. 495:223–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tizzano M, Dvoryanchikov G, Barrows JK, Kim S, Chaudhari N, Finger TE. 2008. Expression of Galpha14 in sweet-transducing taste cells of the posterior tongue. BMC Neurosci. 9:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh H, Rittman G, Mackenzie IC. 1993. Keratin expression in taste bud cells of the circumvallate and foliate papillae of adult mice. Epithelial Cell Biol. 2:126–133. [PubMed] [Google Scholar]
- Tordoff MG, Ellis HT, Aleman TR, Downing A, Marambaud P, Foskett JK, Dana RM, McCaughey SA. 2014. Salty taste deficits in CALHM1 knockout mice. Chem Senses. 39:515–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu YH, Cooper AJ, Teng B, Chang RB, Artiga DJ, Turner HN, Mulhall EM, Ye W, Smith AD, Liman ER. 2018. An evolutionarily conserved gene family encodes proton-selective ion channels. Science. 359:1047–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Anderson CB, Kinnamon SC. 2015a. Mice lacking pannexin 1 release ATP and respond normally to all taste qualities. Chem Senses. 40:461–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Anderson CB, Parnes J, Enjyoji K, Robson SC, Finger TE, Kinnamon SC. 2013. Role of the ectonucleotidase NTPDase2 in taste bud function. Proc Natl Acad Sci USA. 110:14789–14794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Clapp TR, Kinnamon SC. 2008. Amiloride-sensitive channels in type I fungiform taste cells in mouse. BMC Neurosci. 9:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbeuch A, Larson ED, Anderson CB, Smith SA, Ford AP, Finger TE, Kinnamon SC. 2015b. Postsynaptic P2X3-containing receptors in gustatory nerve fibres mediate responses to all taste qualities in mice. J Physiol. 593:1113–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Buchholtz L, Elischer A, Tareilus E, Gouka R, Kaiser C, Breer H, Conzelmann S. 2004. RGS21 is a novel regulator of G protein signalling selectively expressed in subpopulations of taste bud cells. Eur J Neurosci. 19:1535–1544. [DOI] [PubMed] [Google Scholar]
- Watson N, Linder ME, Druey KM, Kehrl JH, Blumer KJ. 1996. RGS family members: GTPase-activating proteins for heterotrimeric G-protein alpha-subunits. Nature. 383:172–175. [DOI] [PubMed] [Google Scholar]
- Wong GT, Gannon KS, Margolskee RF. 1996. Transduction of bitter and sweet taste by gustducin. Nature. 381:796–800. [DOI] [PubMed] [Google Scholar]
- Woodard GE, Jardín I, Berna-Erro A, Salido GM, Rosado JA. 2015. Regulators of G-protein-signaling proteins: negative modulators of G-protein-coupled receptor signaling. Int Rev Cell Mol Biol. 317:97–183. [DOI] [PubMed] [Google Scholar]
- Yaylaoglu MB, Titmus A, Visel A, Alvarez-Bolado G, Thaller C, Eichele G. 2005. Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev Dyn. 234:371–386. [DOI] [PubMed] [Google Scholar]
- Ye W, Chang RB, Bushman JD, Tu YH, Mulhall EM, Wilson CE, Cooper AJ, Chick WS, Hill-Eubanks DC, Nelson MT, et al. . 2016. The K+ channel KIR2.1 functions in tandem with proton influx to mediate sour taste transduction. Proc Natl Acad Sci USA. 113:E229–E238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Cotter M, Lawton A, Oakley B, Wong L, Zeng Q. 1995. Keratin 18 is associated with a subset of older taste cells in the rat. Differentiation. 59:155–162. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Hoon MA, Chandrashekar J, Mueller KL, Cook B, Wu D, Zuker CS, Ryba NJ. 2003. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell. 112: 293–301. [DOI] [PubMed] [Google Scholar]
- Zhao GQ, Zhang Y, Hoon MA, Chandrashekar J, Erlenbach I, Ryba NJ, Zuker CS. 2003. The receptors for mammalian sweet and umami taste. Cell. 115:255–266. [DOI] [PubMed] [Google Scholar]