One in 5 admissions to a healthcare institution for methicillin-resistant Staphylococcus aureus or vancomycin-resistant enterococci is based on data from a different healthcare system. A regional patient registry and electronic admission notifications can facilitate coordinated infectionprevention efforts.
Keywords: MRSA, VRE, infection control, health information exchange, clinical decision support
Abstract
Background. We developed and assessed the impact of a patient registry and electronic admission notification system relating to regional antimicrobial resistance (AMR) on regional AMR infection rates over time. We conducted an observational cohort study of all patients identified as infected or colonized with methicillin-resistant Staphylococcus aureus (MRSA) and/or vancomycin-resistant enterococci (VRE) on at least 1 occasion by any of 5 healthcare systems between 2003 and 2010. The 5 healthcare systems included 17 hospitals and associated clinics in the Indianapolis, Indiana, region.
Methods. We developed and standardized a registry of MRSA and VRE patients and created Web forms that infection preventionists (IPs) used to maintain the lists. We sent e-mail alerts to IPs whenever a patient previously infected or colonized with MRSA or VRE registered for admission to a study hospital from June 2007 through June 2010.
Results. Over a 3-year period, we delivered 12 748 e-mail alerts on 6270 unique patients to 24 IPs covering 17 hospitals. One in 5 (22%–23%) of all admission alerts was based on data from a healthcare system that was different from the admitting hospital; a few hospitals accounted for most of this crossover among facilities and systems.
Conclusions. Regional patient registries identify an important patient cohort with relevant prior antibiotic-resistant infection data from different healthcare institutions. Regional registries can identify trends and interinstitutional movement not otherwise apparent from single institution data. Importantly, electronic alerts can notify of the need to isolate early and to institute other measures to prevent transmission.
Methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) are 2 common antibiotic-resistant bacteria found in healthcare settings [1, 2]. The majority of studies on MRSA and VRE derive from single institutions. However, there is increasing recognition of the role of interfacility spread of infections, particularly in regions with multiple healthcare facilities [3–5].
To better monitor regional rates and track the spread of antibiotic-resistant bacterial infections, we built and embedded an antimicrobial-resistance registry and tracking system within a regional health information exchange (HIE) in order to register all known MRSA and VRE cases and identify when these patients were admitted to any healthcare facility within the region [6]. Since May 2007, we actively shared information on patient MRSA and VRE colonization or infection status among all major hospitals in Indianapolis, Indiana, and generated e-mail alerts when patients with a history of either were admitted to a hospital [7]. Here we describe initial findings from our citywide network.
METHODS
We conducted this study in Indianapolis and included all Indianapolis hospitals participating in the Indiana Network for Patient Care (INPC) at the time the study began [8]. The INPC is an operational HIE; it recently expanded beyond the original 5 hospital systems. This study involved the 5 major hospital systems (17 hospitals) in the Indianapolis (Marion County) area. The INPC has stored more than 1 billion data elements from the Indianapolis region, and more than 85% of the population in Marion County has some data in the system [9, 10].
We previously described the creation of a regional infection-control network tying together infection preventionists (IPs) among the 5 hospital systems in Indianapolis [6]. We created a common means for IPs to identify MRSA and VRE cases and to update information on cases as necessary.
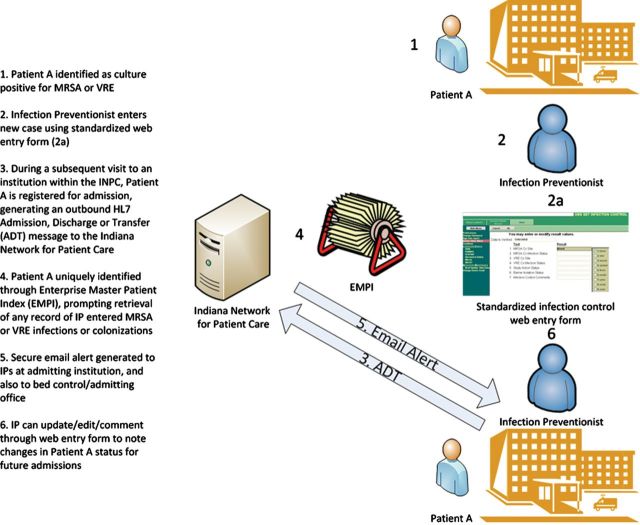
In mid-May 2007, we instituted e-mail alerts to notify infection-control personnel when a patient with a history of MRSA or VRE infection or colonization presented for admission at participating hospitals. If a patient had a history of both MRSA and VRE, a separate e-mail alert for each was sent at the time of admission. For analysis we included data from 1 June 2007 through 1 June 2010 to include only full months. A simplified flow diagram (Figure 1) outlines the process for generating an e-mail alert. Our system leveraged 2 key components of the INPC: a robust enterprise master patient index to uniquely link patients across institutions and the transmission of a standardized electronic message (an admission/discharge/transfer [ADT] message using the Health Level 7 [HL7] standard) whenever a patient was admitted to any participating institution [11]. The HL7 messaging standard is used in virtually all health systems, and ADT messages in particular are commonly generated at the time of patient registration within emergency departments [12].
Figure 1.
Flow diagram outlining process of generating regional e-mail alerts upon hospital admission for patients previously infected or colonized with methicillin-resistant Staphylococcus aureus or vancomycin-resistant enterococci. Abbreviations: ADT, admission/discharge/transfer; INPC, Indiana Network for Patient Care; IP, infection preventionist; MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant enterococci.
We “primed” the system with existing lists of patients with prior history of colonization or infection with MRSA or VRE from all participating institutions, as recorded by their infection-control teams. Our system recorded the initial laboratory result as entered by the IP, the source institution, and the culture site that prompted entry of the patient into the merged citywide list of MRSA and VRE cases. We similarly recorded the date/time and location of subsequent admissions of registry patients. Any infection-control provider could remove patients from the citywide list by documenting that the patient had been “cleared.” Each site used its institution's criteria to make this determination.
Although the standardized Web forms included a “pick list” (or “drop-down menu”) of the most common culture sites, IPs could use free text entry to describe the culture site in greater detail or to overwrite an option on the list. We therefore reclassified all free text entries as one of the common culture sites where possible (eg, blood, skin and soft tissue, urine, stool, sputum). For patients with more than 1 positive culture site, we included all cultures sites for analysis. We analyzed patients who had MRSA or VRE or both during the study period.
We created network diagrams to illustrate the connectivity among study hospitals using the open source GraphViz software [13]. Network diagrams can be used to visualize connections (edges) among entities (nodes) with applications in social network analysis or data flow diagrams. In this study, we used GraphViz software to visualize the flow of patients among institutions. We visualized nodes as circles, with area proportional to the number of unique patients identified with MRSA or VRE and admitted only within the same institution. We visualized edges or connections among the institutions with the width of the arrows proportional to the unique patients identified with MRSA or VRE at 1 institution but later admitted to a different institution (“crossover” patients).
We compared age, gender, and race for admitted crossover patients with patients who were admitted within the same institution. We used 2-sample t tests to compare mean ages among groups and χ² tests to compare gender and race. Missing values were negligible for age and gender. Missing race could not be imputed based on available data and was not included in tests of comparison.
Eighteen months after alerts went live, we surveyed IPs at all 5 participating hospital systems to determine overall burden of alerts, gauge perceived usefulness of the system, estimate time cost or savings in using the alerting system, and elicit suggestions for improvement.
From November 2006 to February 2008, 1 of the investigators (Bradley N. Doebbeling) led an Agency for Healthcare Research and Quality–funded project aimed at reducing MRSA infection and transmission in hospitals [14, 15]. We formed a regional collaborative to spread effective strategies for MRSA reduction; identify strategies for reducing healthcare-associated, community-onset (HACO) MRSA; and build a network of people and organizations devoted to MRSA prevention. They conducted a 2-phase project in order to identify and spread successful strategies for reducing MRSA infections in hospitals. The first phase involved 4 hospitals in Indianapolis over a 2-year period. The second phase, which began in mid-2009, was a multisite, multihospital quasi-experimental study of 7 hospital systems, including 4 systems in Indianapolis, over a 4-year period.
Doebbeling and colleagues also worked closely with hospital leaders and front-line staff in inpatient units to apply organizational change strategies and evidence-based infection-prevention precautions [14]. As part of this project, an intervention bundle was implemented in 4 of the 5 Indianapolis hospital systems. The intervention bundle consisted of active surveillance cultures (including nasal swabs) for all patients admitted to study units, preemptive barrier isolation of those identified as either infected or colonized with MRSA, and institution of strict hand hygiene before and after each patient contact.
RESULTS
From 2003 to 2010, the registry included 23 776 unique patients infected or colonized with MRSA, 3036 unique patients infected or colonized with VRE, and an additional 914 unique patients infected or colonized with both MRSA and VRE (Table 1). Data on race were missing for 19% of the study cohort.
Table 1.
Demographics of Patients in the Cohort (2003–2010) With Methicillin-Resistant Staphylococcus aureus or Vancomycin-Resistant Enterococci
| Demographic | MRSA |
VRE |
Both |
|||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Race | ||||||
| White | 13 212 | 55.6 | 2035 | 67 | 579 | 63.3 |
| Black | 4842 | 20.4 | 521 | 17.2 | 188 | 20.6 |
| Other | 846 | 3.6 | 89 | 2.9 | 11 | 1.2 |
| Missing | 4876 | 20.5 | 391 | 12.9 | 136 | 14.9 |
| Gender | ||||||
| Female | 11 663 | 49.1 | 1822 | 60 | 505 | 55.3 |
| Male | 12 096 | 50.9 | 1214 | 40 | 407 | 44.5 |
| Unknown | 17 | 0.1 | 0 | 0 | 2 | 0.2 |
| Age | ||||||
| <18 | 2595 | 10.9 | 23 | 0.8 | 17 | 1.9 |
| 18–35 | 4518 | 19 | 213 | 7 | 95 | 10.4 |
| 35–64 | 9341 | 39.3 | 1408 | 46.4 | 446 | 48.8 |
| ≧65 | 6967 | 29.3 | 1343 | 44.2 | 347 | 38 |
| Missing | 355 | 1.5 | 49 | 1.6 | 9 | 1 |
| Age | ||||||
| Mean | 46.4 | 60.8 | 51.6 | |||
| Median | 48 | 62 | 55.5 | |||
| SD | 24.5 | 17 | 24.5 | |||
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant enterococci.
From 1 June 2007 to 1 June 2010, 12 748 e-mail alerts on 6270 unique patients were delivered (Table 2). As with the larger cohort, race data were missing for a significant proportion (29%). Patients admitted with a history of MRSA colonization or infection were, on average, older than the overall cohort with a history of MRSA colonization or infection (57.0 years vs 46.4 years). The same was true among those who had a history of both MRSA and VRE (58.9 years for admitted patients vs 51.6 years for all patients).
Table 2.
Demographics of Patients Admitted (6/2007–6/2010) With a History of Methicillin-Resistant Staphylococcus aureus or Vancomycin-Resistant Enterococci Infection or Colonization
| Demographic | MRSA |
VRE |
Both |
|||
|---|---|---|---|---|---|---|
| N | % | N | % | N | % | |
| Race | ||||||
| White | 2414 | 49.7 | 435 | 64.1 | 424 | 57.8 |
| Black | 783 | 16.1 | 103 | 15.2 | 175 | 23.8 |
| Other | 91 | 1.9 | 19 | 2.8 | 6 | 0.8 |
| Missing | 1569 | 32.3 | 122 | 18 | 129 | 17.6 |
| Gender | ||||||
| Female | 2503 | 51.5 | 414 | 61 | 406 | 55.3 |
| Male | 2354 | 48.5 | 265 | 39 | 328 | 44.7 |
| Age | ||||||
| <18 | 110 | 2.3 | 0 | 0 | 1 | 0.1 |
| 18–34 | 610 | 12.6 | 45 | 6.6 | 64 | 8.7 |
| 35–64 | 2287 | 47.1 | 340 | 50.1 | 390 | 53.1 |
| ≧65 | 1849 | 38.1 | 294 | 43.3 | 279 | 38 |
| (1 age missing in MRSA group) | ||||||
| Age | ||||||
| Mean | 57 | 61.1 | 58.9 | |||
| Median | 58 | 61 | 60 | |||
| SD | 19.6 | 16.1 | 15.9 | |||
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; SD, standard deviation; VRE, vancomycin-resistant enterococci.
In 23% of admissions of patients with a previous history of MRSA, the MRSA had been identified at a hospital system different from the admitting hospital (range, 19%–30% of the admissions each year during the 3-year period). For VRE, this rate was 22% (range, 15%–35%). Patients in the MRSA group who were admitted to a hospital system that was different from where the MRSA information had been entered into the registry (ie, crossover patients) were younger (54.8 years vs 57.7 years, P < .001) and more often female (55% vs 50%, P = .003) than patients who stayed within the same system (Table 3). Patients with a history of VRE who were admitted to a hospital system different from where VRE had been entered into the registry were similar in age to patients who stayed within the same system and were more likely to be female (73% vs 59%, P = .004). Compared with those who stayed within the same hospital system, patients with a history of both MRSA and VRE admitted to a hospital system different from where MRSA and VRE had been entered into the registry were more likely to be black (38% vs 26%, P = .01), although race was missing for 17.6% of admitted patients.
Table 3.
Demographics of Crossover Patients vs Patients Staying Within Same Hospital System
| MRSA (n = 4857) |
VRE (n = 679) |
Both (n = 734) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Demographic | Crossover | Same | P Value | Crossover | Same | P Value | Crossover | Same | P Value |
| Age, in years | 54.8 | 57.7 | <.001 | 62.1 | 60.1 | .48 | 58.6 | 59.0 | .75 |
| % Female, n | 55.4 | 50.3 | .003 | 72.9 | 58.5 | .004 | 63.0 | 52.3 | .01 |
| % Black | 24.0 | 23.8 | .92 | 19.4 | 18.4 | .87 | 38.3 | 26.1 | .01 |
Abbreviations: MRSA, methicillin-resistant Staphylococcus aureus; VRE, vancomycin-resistant enterococci.
Evaluation of the cohort of new patients since the start of e-mail alerts (N = 4016) revealed that e-mail admission alerts occurred an average of 135 days after the patient was first identified as having MRSA or VRE in the registry (SD = 181) with a median of 57 days, and 60% of all alerts occurred within 365 days after the MRSA or VRE data were first entered into the registry. The maximum number of alerts across all participating hospitals in a single day was 29, with a maximum of 10 for a single hospital.
Of all patients who generated an alert, 57% had only a single alert during the 3-year period, 87% had 3 or fewer, and 99% had 9 or fewer. Sixty-eight patients generated 10 or more alerts, with 1 patient generating an alert at 47 distinct hospital admissions.
We created network diagrams that indicate the flow of patients from initial site of identification of colonization or infection with MRSA or VRE to sites of subsequent admissions (Figures 2 and 3). Every institution shared patients with every other institution, serving both as a source and as a receiver of patients. Different institutions accounted for the highest number of total admissions.
Figure 2.
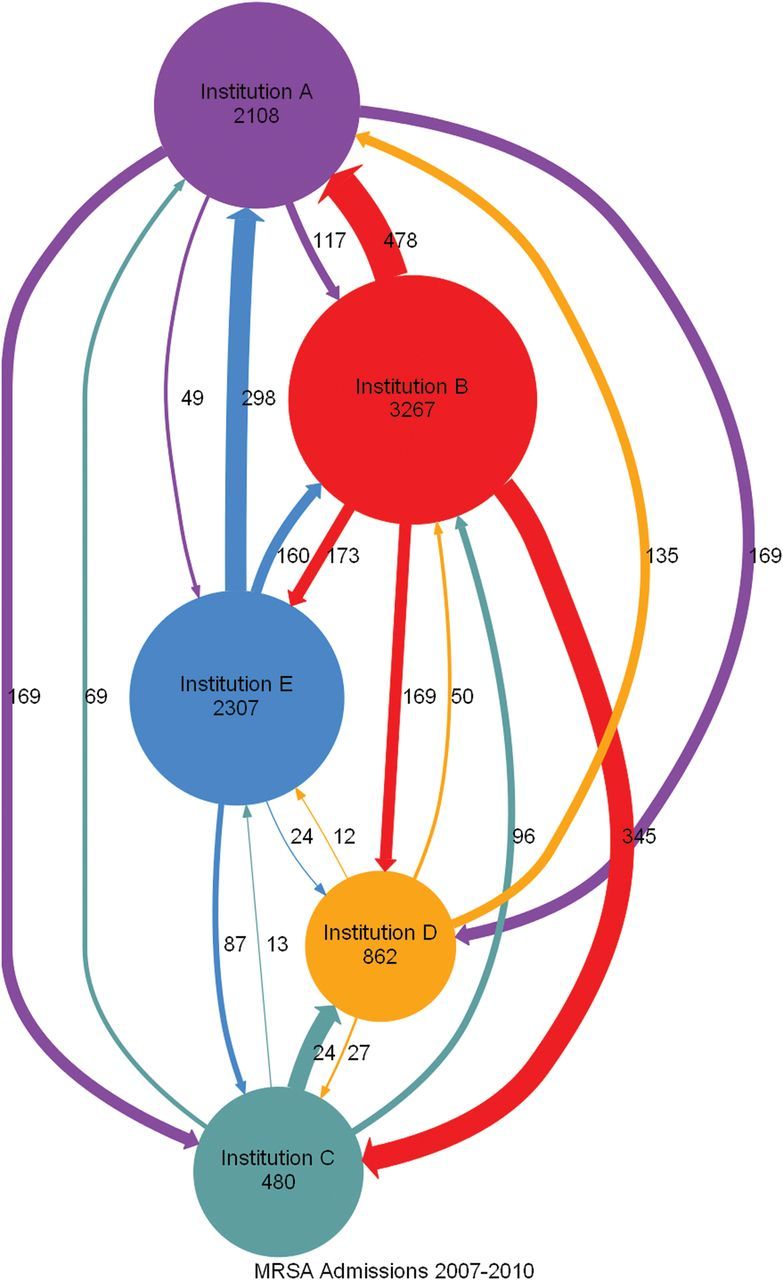
Directed graph of admissions for patients with a history of methicillin-resistant Staphylococcus aureus infection or colonization who stayed within a hospital system (circles or nodes) or who crossed over among hospital systems (arrows or edges). Abbreviation: MRSA, methicillin-resistant Staphylococcus aureus.
Figure 3.
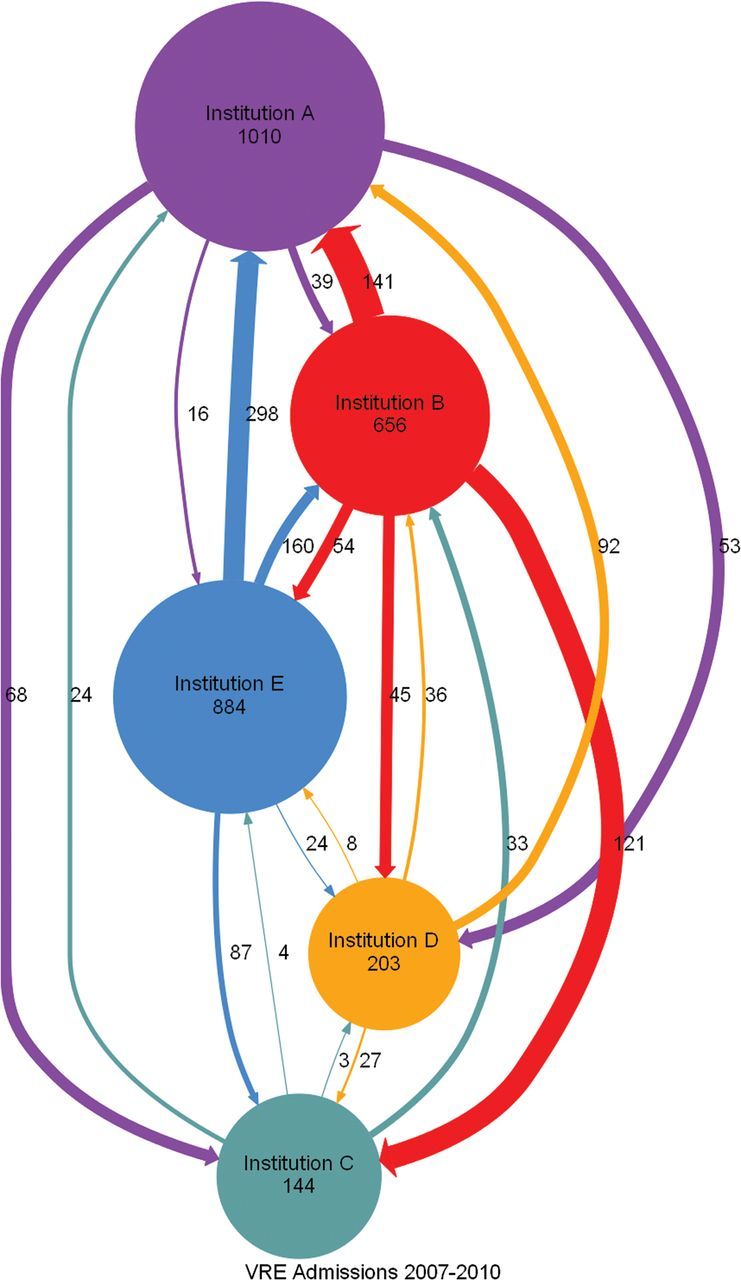
Directed graph of admissions for patients with a history of vancomycin-resistant enterococci infection or colonization who stayed within a hospital system (circles or nodes) or who crossed over among hospital systems (arrows or edges). Abbreviation: VRE, vancomycin-resistant enterococci.
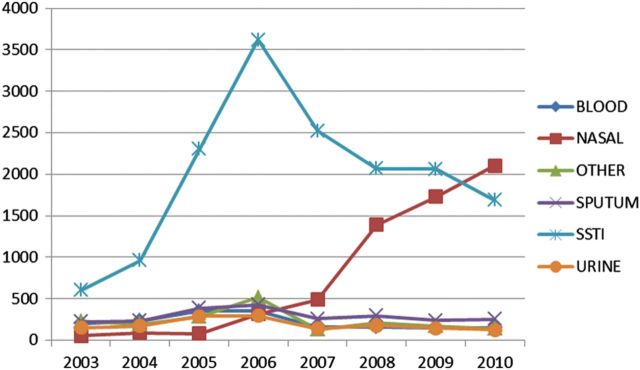
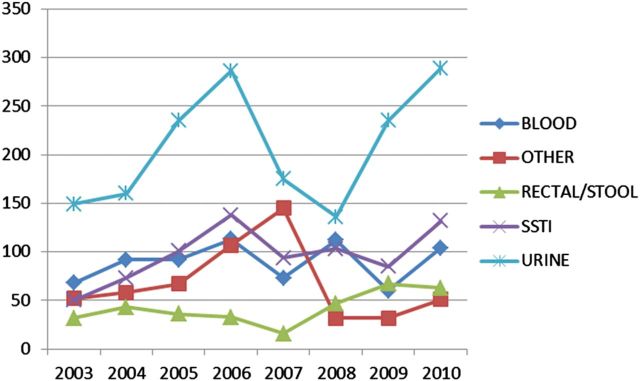
From 2003 to 2006, rates of positive cultures at sites associated with skin and soft tissue sites (SSTI) increased steadily as a proportion of total MRSA-positive cultures (Figure 4). From 2006 to 2010, rates of all MRSA-positive culture sites decreased, with sites associated with SSTIs decreasing most rapidly. Over the same period, rates of positive nasal cultures increased rapidly, coincident with regional implementation of the infection-control bundle. Over the same period, rates of SSTI, blood stream, and urinary tract culture sites positive for VRE steadily increased (Figure 5).
Figure 4.
Counts of positive methicillin-resistant Staphylococcus aureus culture sites by year. Abbreviation: SSTI, skin and soft tissue site.
Figure 5.
Counts of positive vancomycin-resistant enterococci culture sites by year. Abbreviation: SSTI, skin and soft tissue site.
Ten IPs representing all 5 institutions completed a subsequent survey at 18 months post going live. All responded “yes” to the question, “Do you find the e-mail alerts useful?” IPs estimated an average e-mail alert burden of 5 per day, of which just over half (55%) were already known to them from data at their own institution. The most common ways in which IPs used the alerts were to identify patients requiring intervention (eg, contact isolation) and to identify MRSA cases coming from outside institutions. In considering the time cost of the alerting system (e-mails and data entry), 6 IPs considered the system to be time neutral, 3 responded that use of the system added time, and 1 responded that the system was a time saver. The most common recommended improvement was automated capture of laboratory data into the system to reduce burden of manual entry of new cases.
DISCUSSION
Since May 2007, Indianapolis IPs have used a common system to collectively track more than 20 000 unique patients with a history of MRSA or VRE infection or colonization. Our network enabled IPs at participating institutions to benefit from the collective infection history of shared patients while continuing to maintain their own historical records. There have been a number of successful regional efforts to coordinate and implement regional infection control, although, to our knowledge, ours is the first to implement regional admission alerts [16–18].
Alerts based on regional data identified when a patient with a history of infection or colonization with a drug-resistant organism was readmitted to any network institution and may improve rates of compliance with contact precautions [19]. In this study, we demonstrated that approximately 1 in 5 patients with a relatively recent history of MRSA or VRE is readmitted at neighboring institutions, which corroborates and quantifies the estimates of other studies [3–5, 20]. Although we cannot directly relate our regional admission alerts to improved compliance with infection-control measures, our regional registry captured data that likely reflect compliance with increased surveillance cultures (nasal) as part of a coordinated effort to reduce MRSA infections. Recent models suggest that coordinated infection-control efforts in a region can help individual hospitals achieve better control than would be possible on their own [21].
Universal screening has been proposed as an effective means of controlling MRSA infections [22]. However, controversy over the optimal approach remains, given variation in how well infection-control measures are implemented and the significant investment in resources that is required [23–25]. In this example, data sharing on prior history of MRSA or VRE infection or colonization may have reduced the need for repeat culture and may have more quickly identified a patient who requires preemptive contact precautions. Our data demonstrate that local hospital interactions are asymmetric, with some hospital systems sharing a disproportionate burden of infected or colonized patients. Identification of higher-burden hospitals or hospital systems may help guide resources to match relative burden of disease in a community. Institutions also differed in their relative burden of MRSA and VRE patients. Further study may elucidate institutional factors associated with differing rates of drug resistance [26].
Creation of a common registry enabled regional tracking of new cases of MRSA or VRE. During the study period, the incidence of positive cultures involving SSTIs increased disproportionately, likely mirroring the increase in cases of community-acquired MRSA noted both locally and nationally [27, 28]. Overall rates of positive MRSA blood cultures decreased gradually over the same time period (a finding similar to that of other studies), although MRSA-positive blood culture rates increased slightly at individual institutions [29]. These changes may also have reflected an aggressive program of active surveillance and interventions to reduce hospital-based MRSA that were instituted during the study period. Notably, rates of positive VRE cultures did not change and, in fact, trended upward for urinary tract sites.
Regional surveillance of drug-resistant infections provides a broader and potentially more accurate view of infection burden than data from a single institution and can help coordinate the appropriate use of limited infection-prevention resources. Our system reflected national trends in MRSA incidence and documented evidence of dedicated active surveillance efforts and potential effects of these efforts on subsequent infection rates.
There are several limitations to our study. We designed our system to capture data entered and verified by IPs, rather than data taken directly from the laboratory information systems, based on preliminary work that showed that automating case capture could not be considered 100% reliable. As a result, entry dates for new cases sometimes lagged behind the actual time when the infection or colonization was recognized, depending on when the IP was able to manually enter the case information. The requirement for human review and entry into our standardized Web forms likely increased administrative burden on IPs. In fact, the IPs in 1 hospital system stopped entering data into the system in 2011. However, for the 2007–2010 study period, the system was in continuous use by IPs, which suggests that the benefits of the system may have outweighed (or may have been in approximate balance with) the additional burden of data entry, a fact supported by our mid-study survey. During the time of the study, molecular typing for MRSA strains was not routinely conducted and neither the hospitals nor the INPC reliably captured enough additional information electronically in 1 place to classify MRSA cases as healthcare associated, community associated, or HACO according to the Centers for Disease Control and Prevention clinical categories [1]. Instead, our system triggered alerts based on any prior history of MRSA or VRE, regardless of classification or when the original infection or colonization took place. IPs could remove a patient from the regional listing but may not have done so consistently. The majority of alerts (60%) were triggered based on historical data from within 1 year of the admission date, and limiting alerts to trigger based on no more than a 1-year window may reduce the risk of excess alerts or alert fatigue [30].
For this study we tracked only cases of MRSA and VRE. We recently expanded our focus to include infection or colonization with gram-negative organisms, recognizing that multidrug resistance in these organisms poses an impending threat [31–33]. We are developing a way to extract structured data on new infections directly from electronic messages generated by the laboratory information systems in order to limit manual entry only to unusual or uncertain cases and in turn reduce the burden on IPs of manual entry of all cases.
In this work we successfully implemented a system to track and coordinate infection-control efforts within an operational regional HIE. Although we benefited from a longstanding history of pioneering informatics work within Indianapolis, recent trends suggest that our work may be generalizable to other communities [8]. Government initiatives to stimulate adoption of electronic health records (EHRs) have yielded early success, with steady increase in EHR use nationally [34, 35]. Federal regulations that outline the “meaningful use” of EHRs may improve the quality and structure of data captured in EHRs for research and public health purposes [36, 37]. With increased adoption and improved use of EHRs, efforts to connect systems through local and regional HIEs are increasingly widespread, although significant barriers still remain [38]. Our work represents a specific-use case within a functioning HIE but one that leverages technology and standards commonly used in health systems and other HIEs (eg, an enterprise master patient index and HL7 ADT messages generated at admission). With the increasing implementation of EHRs and HIEs, other communities may be well positioned to develop similar electronically coordinated infection-control efforts.
Notes
Disclaimer. The opinions expressed in this document are those of the authors and do not reflect the official position of AHRQ or the US Department of Health and Human Services.
Financial support. This work was funded in part under contracts 290-04-0015 and HSA290200600013-3 Task Order 5 (A. N. K., B. N. D., M. B. R., D. C. S., and J. M. O.) from the Agency for Healthcare Research and Quality (AHRQ), US Department of Health and Human Services. J. M. O. was initially funded through the NLM T15 LM007117.
Potential conflicts of interest. All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Klevens RM, Morrison MA, Nadle J, et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–71. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 2.Cetinkaya Y, Falk P, Mayhall CG. Vancomycin-resistant enterococci. Clin Microbiol Rev. 2000;13:686–707. doi: 10.1128/cmr.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kho AN, Lemmon L, Commiskey M, Wilson SJ, McDonald CJ. Use of a regional health information exchange to detect crossover of patients with MRSA between urban hospitals. J Am Med Inform Assoc. 2008;15:212–6. doi: 10.1197/jamia.M2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang Susan SMD, Avery Taliser RBS, Song Y, et al. Quantifying interhospital patient sharing as a mechanism for infectious disease spread. Infect Control Hosp Epidemiol. 2010;31:1160–9. doi: 10.1086/656747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee BY, McGlone SM, Wong KF, et al. Modeling the spread of methicillin-resistant Staphylococcus aureus (MRSA) outbreaks throughout the hospitals in Orange County, California. Infect Control Hosp Epidemiol. 2011;32:562–72. doi: 10.1086/660014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kho AN, Dexter P, Lemmon L, et al. Connecting the dots: creation of an electronic regional infection control network. Stud Health Technol Inform. 2007;129(Pt 1):213–7. [PubMed] [Google Scholar]
- 7.Kho AN, Lemmon L, Dexter PR, Doebbeling B. An operational citywide electronic infection control network: results from the first year. AMIA Ann Symp Proc. 2008:1222. [PubMed] [Google Scholar]
- 8.McDonald CJ, Overhage JM, Barnes M, et al. The Indiana Network For Patient Care: a working local health information infrastructure. Health Affairs. 2005;24:1214–20. doi: 10.1377/hlthaff.24.5.1214. [DOI] [PubMed] [Google Scholar]
- 9.Overhage JM, Dexter PR, Perkins SM, et al. A randomized, controlled trial of clinical information shared from another institution. Ann Emerg Med. 2002;39:14–23. doi: 10.1067/mem.2002.120794. [DOI] [PubMed] [Google Scholar]
- 10.McGowan JJ, Overhage JM, Barnes M, McDonald CJ. Indianapolis I3: the third generation integrated advanced information management systems. J Med Libr Assoc. 2004;92:179–87. [PMC free article] [PubMed] [Google Scholar]
- 11.Grannis SJ, Overhage JM, Hui S, McDonald CJ. Analysis of a probabilistic record linkage technique without human review. AMIA Annu Symp Proc. 2003:259–63. [PMC free article] [PubMed] [Google Scholar]
- 12.Tsui F-C, Espino JU, Dato VM, Gesteland PH, Hutman J, Wagner MM. Technical description of RODS: a real-time public health surveillance system. J Am Med Inform Assoc. 2003;10:399–408. doi: 10.1197/jamia.M1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ellson J, Gansner E, Koutsofios L, North S, Woodhull G. Graph Drawing. Springer; 2002. Graphviz—open source graph drawing tools; pp. 594–7. [Google Scholar]
- 14.Welsh CA, Flanagan ME, Kiess C, Doebbeling BN. Implementing the MRSA bundle in ICUs: one citywide collaborative's key lessons learned. Infect Control Hosp Epidemiol. 2011;32:918–21. doi: 10.1086/661101. [DOI] [PubMed] [Google Scholar]
- 15.Hagg HW, Workman-Germann J, Flanagan M, et al. Implementation of systems redesign: approaches to spread and sustain adoption. Advances in patient safety: new directions and alternative approaches 2. Agency for Healthcare Research and Quality. 2008 Aug. [PubMed] [Google Scholar]
- 16.Ostrowsky BE, Trick WE, Sohn AH, et al. Control of vancomycin-resistant enterococcus in health care facilities in a region. N Engl J Med. 2001;344:1427–33. doi: 10.1056/NEJM200105103441903. [DOI] [PubMed] [Google Scholar]
- 17.Pronovost P, Needham D, Berenholtz S, et al. An intervention to decrease catheter-related bloodstream infections in the ICU. N Engl J Med. 2006;355:2725–32. doi: 10.1056/NEJMoa061115. [DOI] [PubMed] [Google Scholar]
- 18.Henriksen K, Battles JB, Marks ES, et al. Shared learning and the drive to improve patient safety: lessons learned from the Pittsburgh Regional Healthcare Initiative. 2005. [PubMed]
- 19.Kho AN, Dexter PR, Warvel JS, et al. An effective computerized reminder for contact isolation of patients colonized or infected with resistant organisms. Int J Med Inform. 2008;77:194–8. doi: 10.1016/j.ijmedinf.2007.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barnes SL, Harris AD, Golden BL, Wasil EA, Jon PF. Contribution of interfacility patient movement to overall methicillin-resistant Staphylococcus aureus prevalence levels. Infect Control Hosp Epidemiol. 2011;32:1073–8. doi: 10.1086/662375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee BY, Bartsch SM, Wong KF, et al. Simulation shows hospitals that cooperate on infection control obtain better results than hospitals acting alone. Health Affairs. 2012;31:2295–303. doi: 10.1377/hlthaff.2011.0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robicsek A, Beaumont JL, Paule SM, et al. Universal surveillance for methicillin-resistant Staphylococcus aureus in 3 affiliated hospitals. Ann Intern Med. 2008;148:409–18. doi: 10.7326/0003-4819-148-6-200803180-00003. [DOI] [PubMed] [Google Scholar]
- 23.Conterno LO, Shymanski J, Ramotar K, Toye B, Zvonar R, Roth V. Impact and cost of infection control measures to reduce nosocomial transmission of extended-spectrum β-lactamase-producing organisms in a non-outbreak setting. J Hosp Infect. 2007;65:354–60. doi: 10.1016/j.jhin.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Hubben G, Bootsma M, Luteijn M, et al. Modelling the costs and effects of selective and universal hospital admission screening for methicillin-resistant Staphylococcus aureus. Plos One. 2011;6:e14783. doi: 10.1371/journal.pone.0014783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richard P, Wenzel MDM, Gonzalo Bearman MM, Michael B, Edmond MMM. Screening for MRSA: a flawed hospital infection control intervention. Infect Control Hosp Epidemiol. 2008;29:1012–8. doi: 10.1086/593120. [DOI] [PubMed] [Google Scholar]
- 26.Ward MM, Diekema DJ, Yankey JW, et al. Implementation of strategies to prevent and control the emergence and spread of antimicrobial-resistant microorganisms in US hospitals. Infect Control Hosp Epidemiol. 2005;26:21–30. doi: 10.1086/502483. [DOI] [PubMed] [Google Scholar]
- 27.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clin Infect Dis. 2008;46:787–94. doi: 10.1086/528716. [DOI] [PubMed] [Google Scholar]
- 28.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev. 2010;23:616–87. doi: 10.1128/CMR.00081-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kallen AJ, Mu Y, Bulens S, et al. Health care-associated invasive MRSA infections, 2005–2008. JAMA. 2010;304:641–7. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- 30.van der Sijs H, Aarts J, Vulto A, Berg M. Overriding of drug safety alerts in computerized physician order entry. J Am Med Inform Assoc. 2006;13:138–47. doi: 10.1197/jamia.M1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kallen AJ, Srinivasan A. Current epidemiology of multidrug‐resistant gram‐negative bacilli in the United States. Infect Control Hosp Epidemiol. 2010;31(S1):S51–4. doi: 10.1086/655996. [DOI] [PubMed] [Google Scholar]
- 32.Kallen AJ, Hidron AI, Patel JP, Srinivasan A. Multidrug resistance among gram-negative pathogens that C\caused healthcare-associated infections reported to the National Healthcare Safety Network, 2006–2008. Infect Control Hosp Epidemiol. 2010;31:528–31. doi: 10.1086/652152. [DOI] [PubMed] [Google Scholar]
- 33.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–13. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hsiao CJ, Hing E, Socey TC, Cai B. Electronic medical record/electronic health record systems of office-based physicians: United States, 2009 and preliminary 2010 state estimates. National Center for Health Statistics; 2010. [Google Scholar]
- 35.Blumenthal D. Stimulating the adoption of health information technology. N Engl J Med. 2009;360:1477–9. doi: 10.1056/NEJMp0901592. [DOI] [PubMed] [Google Scholar]
- 36.Blumenthal D, Tavenner M. The “meaningful use” regulation for electronic health records. N Engl J Med. 2010;363:501–4. doi: 10.1056/NEJMp1006114. [DOI] [PubMed] [Google Scholar]
- 37.Kho AN, Pacheco JA, Peissig PL, et al. Electronic medical records for genetic research: results of the eMERGE Consortium. Sci Transl Med. 2011;3:79re1. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adler-Milstein J, Bates DW, Jha AK. U.S. regional health information organizations: progress and challenges. Health Aff. 2009;28:483–92. doi: 10.1377/hlthaff.28.2.483. [DOI] [PubMed] [Google Scholar]