Abstract
Summary
Data-parallel programming techniques can dramatically decrease the time needed to analyze large datasets. While these methods have provided significant improvements for sequencing-based analyses, other areas of biological informatics have not yet adopted them. Here, we introduce Biospark, a new framework for performing data-parallel analysis on large numerical datasets. Biospark builds upon the open source Hadoop and Spark projects, bringing domain-specific features for biology.
Availability and Implementation
Source code is licensed under the Apache 2.0 open source license and is available at the project website: https://www.assembla.com/spaces/roberts-lab-public/wiki/Biospark
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Data-intensive statistical analysis, colloquially known as ‘Big Data,’ has seen increasing use in genomics and bioinformatics since the arrival of high-throughput sequencing (O’Driscoll et al., 2013). New technologies for accelerating data-intensive computation have been developed to speed up the analysis of large sequencing datasets (McKenna et al., 2010; Nordberg et al., 2013). Other areas of biology generating large numerical datasets from simulations or experiments have not adopted these new technologies as quickly (Ekanayake et al., 2008; Nothaft et al., 2015).
The primary advance associated with data-intensive computing is to reverse the conventional high-performance computing model, where data are distributed to the nodes of a cluster from high-performance dedicated storage. Instead, data are partitioned among the individual nodes and calculations are scheduled so as to be performed on the node where the requisite data are stored. Such a data-parallel approach can provide high performance using commodity hardware. Hadoop is an open source implementation of a distributed filesystem (HDFS) and the MapReduce data-parallel programming model (Dean and Ghemawat, 2004).
MapReduce, though powerful, has limitations. Most notably, since intermediate results are not cached, MapReduce is not efficient at executing algorithms that require multiple iterations through the data. Recently, more general-purpose data-parallel programming models have been introduced, including Spark (Zaharia et al., 2012). Spark introduced the concept of a resilient distributed dataset (RDD), a collection of data that can be operated on in parallel. RDDs can be cached to store intermediate results in distributed memory for iterative processing.
We have developed a new framework, called Biospark, for storing and analyzing large binary numerical datasets using Hadoop and Spark. Biospark is comprised of a set of Java, C ++ and Python libraries that support writing scripts for analyzing large numerical datasets. The project also includes reference implementations of a number of analyses that are commonly used by computational biologists.
2 Design and implementation
2.1 SFile file format
A key component of the Biospark framework is the SFile file format for storing binary numerical data in HDFS. Compared to a traditional filesystem, files stored in HDFS are more restricted in terms of random access. Files are split into blocks, typically 64–256 MB in size, and the blocks are stored across all of the nodes in the Hadoop cluster. In order to achieve maximum performance, data residing in any block should be accessible without the need to read an index or other data structure.
Traditionally, Hadoop was designed to process text files where records are separated by a special character, such as a newline. This approach is problematic for binary numerical data, since any given character may appear in the data stream. Hadoop file formats for storing binary data have been defined, but are often specific to one application domain, e.g. (Niemenmaa et al., 2012). Additionally, it is desirable to store metadata alongside the numerical data. To address these issues we have defined the SFile file format. An SFile is composed of a series of sequential records, where each record starts with a 16-byte record separator followed by the record name, the record type and the binary data, as shown in Supplementary Figure S1. A comparison of the SFile format with other Hadoop-compatible binary file formats is given in the SI.
2.2 Multidimensional array storage
The unit of storage in scientific computing is often a multidimensional array containing numerical data. To make storing and retrieving such data as easy as possible within SFiles, we have defined serialization formats for a multidimensional array, called an NDArray, using the Protocol Buffers and Avro languages (see Supplementary Figs. S3 and S4). The serialized message stores all needed metadata, including the array shape and data type, and also specifies whether the array is stored in one of a variety of compressed formats. Compression of numerical data can be especially useful for sparse arrays.
2.3 Parallel analysis algorithms
To showcase the capabilities of the Biospark framework, here we present parallel analyses of three different computational biology datasets: kinetic Monte Carlo simulation data, molecular dynamics (MD) trajectories and cellular time-lapse microscopy images. Each of these application domains can generate large datasets that are often challenging to store and analyze. We tested six different analyses on these datasets. Four algorithms were MapReduce-like and two were iterative, details are given in the SI.
[map-reduce] Read the data without performing any calculations.
[map-reduce] Estimate a probability density function (PDF) from kinetic Monte Carlo simulations by counting the frequency of states.
[map-reduce] Calculate the root-mean-square deviation (RMSD) of a protein with respect to the crystal structure across an MD trajectory.
[iterative] Divide an MD trajectory into windows and calculate the RMSD between the average structure of every pair of windows.
[map-reduce] Align a set of time-lapse microscopy images to a reference image based on a fiducial marker, e.g. a bead.
[iterative] Propagate cell segmentation boundaries forward in time for a set of time-lapse microscopy images.
3 Performance
To test the performance of Hadoop, Spark and Biospark on the analyses described above, we performed scaling tests using the MARCC Bluecrab cluster. Each node had 24 CPU cores, 128 GB RAM and 1 hard disk drive (HDD). Unlike statistical inference methods, not all computational biology analyses are limited by data I/O. Therefore, we first tested CPU/HDD ratios to determine the optimal value for each of the analyses, see SI.
Figure 1 shows the scaling results. All of the MapReduce-like algorithms scaled well, with a slight drop at 128 nodes. The read, PDF and RMSD operations became essentially I/O-bound at 2, 3 and 8 cores per HDD, respectively. Image alignment was compute-bound when using all 24 cores. The sustained throughput for the I/O-bound applications was ∼6 to 8 GB/s when using 128 HDDs, analyzing 1 TB of data in ∼2 min.
Fig. 1.
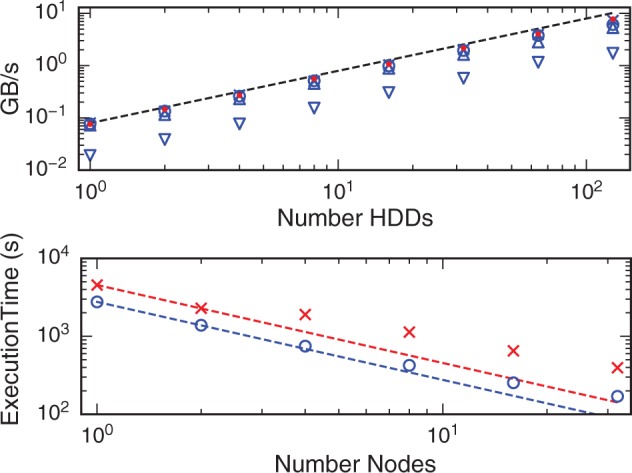
(Top) Performance of map-reduce algorithms: (crosses) read throughput [2 cores per HDD], (circle) PDF calculation [3 cores per HDD], (triangle) RMSD calculation [8 cores per HDD], (inverted triangle) image alignment [24 cores per HDD]. Also, (dot) performance using Lustre [1 core per node]. (bottom) Performance of iterative algorithms: (crosses) pairwise RMSD, (circle) cell segmentation. The lines show perfect scaling
The iterative algorithms can take advantage of the total memory of the cluster for storing intermediate results to process large datasets in a reasonable amount of time. Here, we calculated a 12 000 × 12 000 pairwise RMSD matrix and segmented 640 cell boundaries. The iterative algorithms scaled with a parallel efficiency of ∼0.5 at 32 nodes, see SI.
Biospark analyses can also be run using Spark backed by a traditional distributed filesystem. As can be seen in Figure 1, Lustre and HDFS provide similar performance under the test conditions (see SI for details).
4 Conclusion
Deployment of a Hadoop cluster can be a cost-effective and high-performance way to store and analyze biological data. The Biospark framework will help enable data-parallel analysis in computational biology research. The framework provides abstractions for parallel analysis of standard data types, including multidimensional arrays and images. It also provides APIs and file conversion tools to assist in parallel analysis of some common datasets, including Monte Carlo and molecular dynamics simulations and time-lapse microscopy. To assist users in learning how to take advantage of these new technologies in custom scripts, we have written and made available on the project website a series of detailed tutorials and protocols. We hope Biospark will expand by developing into a community focused on sharing custom parallel analysis code.
Conflict of Interest: none declared.
Supplementary Material
References
- Dean J., Ghemawat S. (2004) MapReduce: Simplified data processing on large clusters. In: OSDI ’04: 6th Symposium on Operating Systems Design and Implementation.
- Ekanayake J. et al. (2008) MapReduce for data intensive scientific analyses. In: IEEE Fourth International Conference on eScience.
- McKenna A. et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemenmaa M. et al. (2012) Hadoop-BAM: directly manipulating next generation sequencing data in the cloud. Bioinformatics, 28, 876–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordberg H. et al. (2013) BioPig: a Hadoop-based analytic toolkit for large-scale sequence data. Bioinformatics, 29, 3014–3019. [DOI] [PubMed] [Google Scholar]
- Nothaft F.A. et al. (2015). Rethinking data-intensive science using scalable analytics systems. In: SIGMOD ’15.
- O’Driscoll A. et al. (2013) ‘ Big data’, Hadoop and cloud computing in genomics. J. Biomed. Inf., 46, 774–781. [DOI] [PubMed] [Google Scholar]
- Zaharia M. et al. (2012). Resilient distributed datasets: a fault-tolerant abstraction for in-memory cluster computing. In: Proceedings of the 9th USENIX Conference on Networked Systems Design and Implementation.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


