High-depth targeted capture massively parallel sequencing analysis of plasma-derived circulating tumor DNA represents a potential tool to characterize the mutation repertoire of breast cancers and to monitor tumor burden and the somatic alterations in cancer cells during the course of targeted therapy.
Keywords: massively parallel sequencing, breast cancer, cell-free tumor DNA, intra-tumor heterogeneity, disease monitoring
Abstract
Background
Plasma-derived cell-free tumor DNA (ctDNA) constitutes a potential surrogate for tumor DNA obtained from tissue biopsies. We posit that massively parallel sequencing (MPS) analysis of ctDNA may help define the repertoire of mutations in breast cancer and monitor tumor somatic alterations during the course of targeted therapy.
Patient and methods
A 66-year-old patient presented with synchronous estrogen receptor-positive/HER2-negative, highly proliferative, grade 2, mixed invasive ductal–lobular carcinoma with bone and liver metastases at diagnosis. DNA extracted from archival tumor material, plasma and peripheral blood leukocytes was subjected to targeted MPS using a platform comprising 300 cancer genes known to harbor actionable mutations. Multiple plasma samples were collected during the fourth line of treatment with an AKT inhibitor.
Results
Average read depths of 287x were obtained from the archival primary tumor, 139x from the liver metastasis and between 200x and 900x from ctDNA samples. Sixteen somatic non-synonymous mutations were detected in the liver metastasis, of which 9 (CDKN2A, AKT1, TP53, JAK3, TSC1, NF1, CDH1, MML3 and CTNNB1) were also detected in >5% of the alleles found in the primary tumor sample. Not all mutations identified in the metastasis were reliably identified in the primary tumor (e.g. FLT4). Analysis of ctDNA, nevertheless, captured all mutations present in the primary tumor and/or liver metastasis. In the longitudinal monitoring of the patient, the mutant allele fractions identified in ctDNA samples varied over time and mirrored the pharmacodynamic response to the targeted therapy as assessed by positron emission tomography–computed tomography.
Conclusions
This proof-of-principle study is one of the first to demonstrate that high-depth targeted MPS of plasma-derived ctDNA constitutes a potential tool for de novo mutation identification and monitoring of somatic genetic alterations during the course of targeted therapy, and may be employed to overcome the challenges posed by intra-tumor genetic heterogeneity.
Registered clinical trial
introduction
Massively parallel sequencing (MPS) studies have revealed that cancers are characterized by remarkable genetic complexity, and that intra-tumor genetic heterogeneity is not an uncommon phenomenon [1–6]. The spatial and temporal intra-tumor genetic heterogeneity documented in breast cancers [4, 5, 7, 8] may have important implications for biomarker discovery programs and targeted cancer therapeutics [9, 10].
Given the spatial and temporal heterogeneity documented between primary cancers and metastatic lesions [2, 11], primary tumor biopsies may not constitute an ideal source for the genetic characterization of metastatic disease and extensive sampling of metastatic deposits is often unfeasible [2, 6, 9, 10]. Hence, approaches that provide a global assessment of the constellation of somatic genetic alterations in a cancer irrespective of its anatomical location would be required for the identification of potential therapeutic targets and mechanisms of resistance, in particular in the context of patients with metastatic disease.
Minimally invasive approaches that may help overcome the challenges posed by intra-tumor spatial and temporal heterogeneity and by the sampling bias stemming from the analysis of single-tumor biopsies have been developed [10, 12–15]. Plasma-derived cell-free tumor DNA (ctDNA) has been tested as a potential non-invasive surrogate for tumor tissue biopsies [10]. Given that ctDNA is believed to be shed into the circulation by cancer cells from both the primary tumor and/or its metastases, it may constitute a source of tumor material from all disease sites, offering a real time, easily obtainable and minimally invasive tool for the development of molecular biomarkers [9, 10].
Recent studies have shown that somatic genetic alterations can be identified by MPS-based analysis of ctDNA from plasma of breast cancer patients [13 - 18]. Thus, the genomic characterization of plasma ctDNA has introduced new means to investigate the metastatic process and mechanisms of therapeutic resistance, and to monitor actionable driver somatic genomic alterations during the course of therapy in breast cancer patients [9, 13–15, 17, 19, 20].
We hypothesize that MPS analysis of plasma-derived ctDNA of breast cancer patients would constitute a means to identify and monitor the presence of potentially actionable driver somatic genomic alterations during the course of therapy. In this proof-of-concept study, we demonstrate that MPS analysis of plasma-derived ctDNA resulted in the identification of the complete repertoire of mutations detected in the metastatic lesion, and that changes in mutant allele fractions (MAFs) in ctDNA mirrored the pharmacodynamic response to targeted monotherapy.
patient and methods
A 66-year-old, postmenopausal woman was diagnosed with an estrogen receptor (ER)-positive/HER2-negative, grade 2, mixed invasive ductal–lobular carcinoma of the breast and synchronous bone and liver metastases at Vall d'Hebron University Hospital (Barcelona, Spain) in July 2010. Imaging-guided biopsies of the primary tumor and liver metastasis were obtained before the initiation of systemic therapy. Following three lines of chemotherapy (i.e. paclitaxel-, anthracycline- and capecitabine-based therapies; Figure 1) and disease progression, the patient underwent a molecular pre-screening program in November 2011. The analysis of archival primary breast tumor material by Sequenom MassARRAY® revealed the presence of an AKT1 E17K mutation, with an MAF of 88% (supplementary Table S1, available at Annals of Oncology online). Based on these results, the patient was enrolled in the phase I study PAM4743g (Clinicaltrials.gov, NCT01090960) in January 2012 and treated with Ipatasertib (GDC-0068), a highly selective, orally available pan-AKT inhibitor [21] as the fourth line of therapy. A dose of 600 mg once daily (maximum tolerated dose in the expansion cohort was 400 mg) was administered on a 3-week-on/1-week-off treatment schedule, until documented disease progression in September 2012. Plasma samples were collected at baseline (i.e. before therapy), and during the treatment with Ipatasertib at 2 months, 6 months and at the time of disease progression. Response was assessed using the RECIST criteria (1.1) [22]. This study was approved by the IRB of Vall d'Hebron University Hospital.
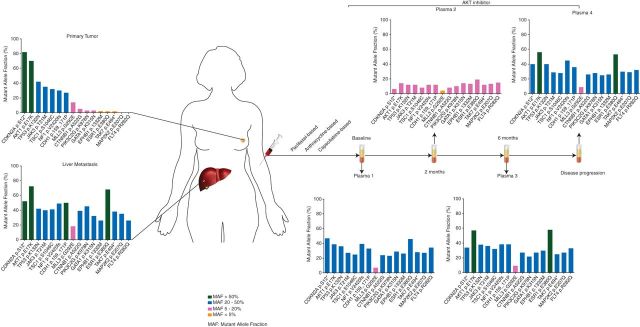
Figure 1.
Patient disease presentation, treatment timeline and mutant alleles in the primary breast tumor, liver metastasis and plasma-derived DNA. Biopsies of the primary breast cancer and its synchronous liver metastasis were obtained before initiation of therapy. Following three lines of chemotherapy, the patient was treated with the AKT inhibitor Ipatasertib, and multiple plasma samples were obtained during the course of treatment. DNA samples extracted from the primary tumor, metastasis and plasma samples were subjected to targeted high-depth massively parallel sequencing. Not all mutations identified in the metastasis were reliably identified in the primary breast tumor; however, all mutations present in the primary tumor and/or liver metastasis were found in ctDNA.
DNA extraction
The diagnosis of the primary breast tumor and synchronous liver metastasis was confirmed by histologic review (supplementary Figure S1, available at Annals of Oncology online). Five 10-µm thick formalin-fixed paraffin-embedded (FFPE) sections of the primary breast tumor and liver metastasis were microdissected with a needle under a stereomicroscope to ensure >80% of tumor cell content, as previously described 23. DNA from tumor samples was extracted using the RecoverAll™ Total Nucleic Acid Isolation Kit (Ambion, Austin, TX) for FFPE tissue, and germline DNA was extracted from peripheral blood leukocytes using the QIAamp DNA Blood Mini Kit (Qiagen, Valencia, CA), according to the manufacturers' instructions. Plasma-derived DNA was extracted with the QIAamp Circulating Nucleic Acid Kit (Qiagen), as per the manufacturer's protocol. DNA was quantified using the Qubit Fluorometer (Invitrogen, Life Technologies, Grand Island, NY).
targeted massively parallel sequencing
DNA samples from the primary breast cancer, liver metastasis, and from multiple plasma samples as well as germline DNA obtained from peripheral blood leukocytes were subjected to targeted capture MPS at the Integrated Genomics Operation (iGO), Memorial Sloan Kettering Cancer Center using the Integrated Mutation Profiling of Actionable Cancer Targets (IMPACT) platform [24], which comprises 300 cancer genes known to harbor actionable mutations as previously described (supplementary Table S2, available at Annals of Oncology online) [3]. In brief, barcoded sequence libraries were prepared (New England Biolabs, KapaBiosystems) using 22–250 ng of input DNA, pooled and captured using oligonucleotides for all protein-coding exons of the 300 genes (NimblegenSeqCap) [3]. Sequencing was carried out on an Illumina HiSeq2000 (San Diego, CA). Sequence alignment as well as calling of somatic single-nucleotide variants and small insertions and deletions were carried out as previously described [3, 24]. All candidate mutations were reviewed manually using the Integrative Genomics Viewer [25]. Mutations with an allele fraction of <1% and/or supported by ≤2 reads were disregarded.
Sequenom analysis
As a molecular pre-screening tool to select patients for clinical trials, 600 ng of DNA from the primary tumor was subjected to mutation profiling using a customized version of the OncoCarta Panel v1.0 (Sequenom MassARRAY®, San Diego, CA) (supplementary Table S1, available at Annals of Oncology online). Data analyses and mutation reports were generated using the Sequenom® software.
results
High-depth targeted MPS was carried out with DNA obtained from the ER-positive/HER2-negative primary breast cancer and synchronous liver metastasis sampled at the time of diagnosis, and multiple plasma samples collected during the fourth line of therapy with Ipatasertib. Ipatasertib monotherapy provided benefit in terms of long-lasting radiologic and biochemical responses as shown by CA15.3 levels (Figure 2), and stable disease as per RECIST 1.1 was the best response achieved by the patient and lasted for ∼8 months. We subsequently compared the patient's progression-free survival (PFS) on Ipatasertib (A) with the PFS for the most recent therapy on which the patient had experienced progression (i.e. capecitabine-based) (B) [26]. The ratio of the PFS of period B/PFS of period A was 3.1 (i.e. 7.8 months/2.5 months), superior to a ratio of >1.3, corroborating the potential clinical benefit of Ipatasertib monotherapy for this patient [26].
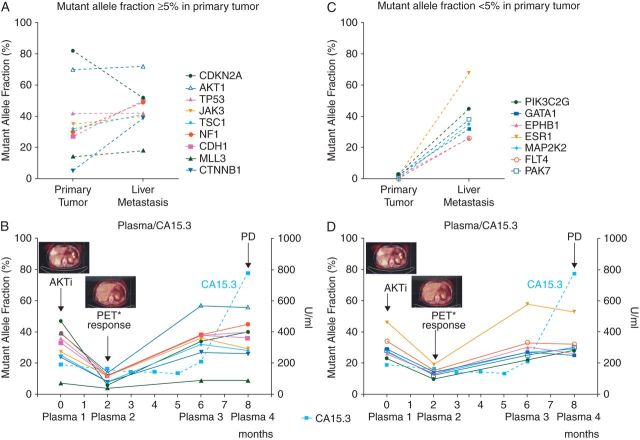
Figure 2.
Representation of the mutant alleles in the primary tumor and metastasis and longitudinal monitoring of CA15.3 levels in four plasma-derived ctDNA samples. Genes whose high confidence mutations were detected at a mutant allele fraction (MAF) of ≥5% in the primary tumor are depicted in (A) and (B), whereas genes whose high confidence mutations were detected in the plasma-derived ctDNA, but either absent or present at a MAF of <5% in the primary tumor, are illustrated in (C) and (D). In (B) and (D), representative PET–CT images obtained at baseline and 2 months after initiation of Ipatasertib monotherapy; CA15.3 levels assessed during the fourth line of systemic treatment with Ipatasertib monotherapy. Arrow, initiation of Ipatasertib (AKTi) monotherapy. PD, progressive disease; *PET–CT, pharmacodynamic response.
somatic genetic alterations are distinct between the primary tumor and its metastasis
Targeted capture MPS showed average read depths of 287x and 139x in the archival primary breast cancer and its liver metastasis, respectively, and 76x in the normal sample. Fifteen somatic non-synonymous mutations were detected in the primary tumor (CDKN2A, AKT1, TP53, JAK3, TSC1, NF1, CDH1, MLL3, CTNNB1, PIK3C2G, GATA1, EPHB1, ESR1 and PAK7), all of which were also detected in the liver metastasis (Table 1). Mutations affecting FLT4 and MAP2K2 were present in the liver metastasis (present in 12/47 reads and 40/113 reads, respectively), but could not be reliably detected in the primary tumor (found in 2/89 reads and 2/137 reads, respectively).
Table 1.
Allele fractions of somatic mutations identified in the primary breast tumor, liver metastasis and plasma samples
| Gene | Mutation (amino acid) | Primary tumor (287x) MAFs
(reads) |
Liver metastasis (139x) MAFs (reads) |
Plasma 1 (692x) MAFs (reads) |
Plasma 2 (728x) MAFs (reads) |
Plasma 3 (209x) MAFs (reads) |
Plasma 4 (918x) MAFs (reads) |
|---|---|---|---|---|---|---|---|
| CDKN2A | p.S12* | 82% (23/28) | 52% (11/21) | 47% (42/89) | 6% (7/117) | 34% (14/41) | 40% (55/137) |
| AKT1 | p.E17K | 70% (83/118) | 72% (79/110) | 39% (204/521) | 14% (83/593) | 57% (100/174) | 56% (373/663) |
| TP53 | p.K132N | 42% (101/241) | 42% (48/113) | 36% (228/625) | 12% (92/753) | 38% (78/204) | 40% (339/841) |
| JAK3 | p.T21M | 35% (60/172) | 40% (56/141) | 27% (253/939) | 12% (100/834) | 36% (122/340) | 29% (343/1181) |
| TSC1 | p.S1046C | 32% (31/98) | 41% (55/134) | 25% (132/521) | 8% (43/518) | 32% (59/182) | 28% (179/636) |
| NF1 | p.V2420fs | 30% (153/511) | 49% (61/124) | 39% (186/483) | 12% (92/761) | 38% (49/159) | 45% (328/726) |
| CDH1 | p.159_171 PPISCPENEKGPF>L | 27% (56/210) |
50% (46/92) | 33% (197/605) | 12% (93/758) | 38% (52/138) | 36% (265/731) |
| MLL3 | p.G292E | 14% (64/446) | 18% (30/168) | 7% (67/1002) | 4% (48/1183) | 9% (31/352) | 9% (73/831) |
| CTNNB1 | p.A522G | 5% (12/256) | 39% (60/155) | 24% (130/551) | 8% (47/618) | 27% (54/198) | 26% (164/641) |
| PIK3C2G | p.K978N | 3% (16/492) | 45% (113/250) | 23% (176/752) | 10% (80/803) | 22% (44/200) | 28% (268/960) |
| GATA1 | p.K315N | 3% (5/192) | 32% (35/111) | 29% (313/1071) | 14% (154/1067) | 27% (100/370) | 25% (419/1648) |
| EPHB1 | p.I332M | 2% (5/211) | 26% (25/96) | 26% (261/1015) | 13% (120/919) | 30% (102/343) | 26% (348/1322) |
| ESR1 | p.E380Q | 2% (7/287) | 68% (106/157) | 46% (339/737) | 19% (158/823) | 58% (160/275) | 53% (534/1009) |
| PAK7 | p.E494* | 2% (5/304) | 38% (56/148) | 28% (202/715) | 12% (83/701) | 25% (55/224) | 30% (273/897) |
| MAP2K2 | p.E207Q | NRD (2/137) | 35% (40/113) | 27% (221/815) | 13% (106/823) | 27% (72/270) | 29% (309/1076) |
| FLT4 | p.R282Q | NRD (2/89) | 26% (12/47) | 34% (225/667) | 15% (98/638) | 33% (89/270) | 32% (266/820) |
Color coding: dark gray cells, MAF > 50%; light gray cells, MAF 20%–50%; pale gray cells, MAF 5%–20% and white cells, MAF < 5% or no mutation identified (NRD, not reliably detected). All mutations detected in both the primary breast tumor and synchronous liver metastasis could be identified in the multiple plasma samples. Plasma 1, baseline; plasma 2, 2 months after initiation of AKT inhibitor Ipatasertib treatment; plasma 3, 6 months after initiation AKT inhibitor Ipatasertib treatment; plasma 4, at disease progression.
*, stop codon.
MAF, mutant allele fraction.
Analysis of the primary tumor and its liver metastasis revealed similar MAFs of the AKT1 (70% and 72% in the primary tumor and its metastasis, respectively) and TP53 (42% in the primary tumor and its metastasis) somatic mutations present in these samples (Table 1). On the basis of the allele fractions of the mutations affecting these two genes, it would be reasonable to hypothesize that these mutations were clonally distributed in the cancer cells of both the primary tumor and its liver metastasis. Evidence of intra-tumor genetic heterogeneity was, however, observed, given that the liver metastasis was enriched for mutations either only present at low allele fractions in the primary tumor (i.e. <5% MAF; PIK3C2G, GATA1, EPHB1, ESR1 and PAK7) or found at a MAF beyond the resolution obtained with the sequencing depth achieved for the primary tumor sample (i.e. FLT4 and MAP2K2 mutations; Table 1 and Figure 2). Targeted capture MPS also confirmed the presence of the AKT1 E17K mutation identified by Sequenom MassARRAY® during the molecular pre-screening program at Vall d'Hebron University Hospital (Table 1 and supplementary Table S1, available at Annals of Oncology online).
ctDNA analysis captures the heterogeneity of primary tumor and metastasis
Targeted capture MPS of the multiple plasma-derived DNA samples yielded average read depths ranging from 209x to 918x. Importantly, MAFs of up to 57% and 58% for AKT1 E17K and ESR1 E380Q were detected in the plasma DNA, respectively, providing evidence to suggest that most of the cell-free plasma DNA obtained from this patient was tumor-derived.
Interestingly, while not all mutations identified in the liver metastasis could accurately be detected in the primary tumor at the sequencing depth obtained, sequencing analysis of ctDNA from this patient captured the entire repertoire of mutations found in the primary tumor and/or metastatic deposit (Figures 1 and 2, and Table 1). For instance, a missense mutation in FLT4, present in the metastasis but found at a MAF beyond the resolution of targeted MPS of the primary tumor material, was captured in the plasma-derived ctDNA. All 16 mutations except MLL3 were found in the baseline plasma ctDNA sample with allelic fractions of >20% (Table 1). The presence of the AKT1 E17K mutation in the plasma samples 1 and 2 was validated independently by means of Sequenom MassARRAY® analysis (44% and 16% MAFs, respectively; supplementary Table S1, available at Annals of Oncology online). Further validation of the somatic genetic aberrations was not possible, given that no additional biological material was available for the remaining samples.
plasma-derived ctDNA for longitudinal disease monitoring
In the longitudinal monitoring of the patient during the course of Ipatasertib treatment, the MAFs identified in ctDNA samples varied following the administration of the targeted therapy. Two months after the initiation of the treatment (plasma 2), the fraction of all mutant alleles detected in the plasma-derived ctDNA decreased when compared with ctDNA analysis at baseline (plasma 1), mirroring the pharmacodynamic response as assessed by PET–CT (Figure 2). Assessment of ctDNA at 6 months of treatment (plasma 3) revealed an increase in the MAFs of all mutated genes similar to the levels observed at baseline before treatment (Figure 2 and Table 1). It should be noted, however, that the MAFs of AKT1 and ESR1 were increased in the ctDNA at 6 months when compared with baseline (AKT1 E17K 39% plasma 1 versus 57% plasma 3; ESR1 E380Q 46% plasma 1 versus 58% plasma 3; Figure 2 and Table 1). Furthermore, the increase in mutant alleles in plasma-derived ctDNA was observed before radiologic disease progression (data not shown), and before the increase in CA15.3 levels (Figure 2), providing evidence to suggest that increases in disease burden can be detected earlier by ctDNA analysis than by classical biochemical and radiologic assessments.
discussion
Here we demonstrate that high-depth targeted MPS of plasma-derived ctDNA contains representative tumor-derived genetic material that captured all mutations detected in the primary tumor and/or its synchronous liver metastasis, and provide a proof-of-principle that this approach can potentially be employed as a quantitative marker for disease monitoring of somatic genetic alterations during the course of targeted therapy.
At the time of diagnosis, at least a subset of breast cancers have been shown to constitute mosaics, being composed of heterogeneous populations of tumor cells that, in addition to the founder genetic events, harbor private mutations [4–8]. Consistent with this notion, here we demonstrate that mutations affecting ESR1, CTNNB1, PIK3C2G, GATA1, EPHB1, PAK7, MAP2K2 and FLT4, albeit present at allele fractions ≥26% in the metastatic lesion, were likely present in a minor clone of the primary tumor (i.e. MAFs ≤5%). It should be noted that activating ESR1 mutations have been identified in endocrine-resistant metastatic lesions while not detectable in the respective primary breast cancers [27]. In this study, the endocrine therapy resistance-associated ESR1 E380Q mutation [28] was present at a higher allele fraction in the ER-positive liver metastasis (MAF 68%) than in its synchronous ER-positive primary breast cancer (MAF 2%). Importantly, however, the biopsies of the synchronous primary and metastatic lesions were collected before any systemic therapy. It remains to be determined whether the ESR1 E380Q mutation provided a growth advantage at the metastatic site irrespective of treatment or merely co-segregated with other molecular alterations present in the clone that gave rise to the metastatic deposit.
Spatial and temporal intra-tumor genetic heterogeneity has been documented in cancers [2, 5–8, 11, 29], suggesting that genetic analysis of a single diagnostic biopsy of a primary tumor may not yield results that are representative of the somatic genetic aberrations present in the cancer cells of its metastases [2, 30]. Given that in the present case all mutations detectable by targeted MPS of the metastatic lesion were also detected in the plasma ctDNA samples, our findings lend credence to the notion that ctDNA may constitute an alternative to metastatic lesion sampling for MPS analysis.
Our study has several limitations. First, the analyses were carried out utilizing materials from a single patient with a high disease burden. The high MAFs observed in this study likely reflect this high tumor burden the patient presented, and therefore, an ideal setting for plasma DNA analysis. It is plausible that, in other settings and cancer types, this approach may not be sufficient, either due to lower tumor burden or due to the fact that cancer cells may not harbor mutations in any of the genes included in a given targeted capture panel [9]. In fact, in Bettegowda et al. [15], the vast majority of metastatic breast cancer patients had ctDNA detectable in plasma, whereas <50% of breast cancer patients with early disease had any detectable levels of ctDNA. Secondly, although ctDNA-targeted capture analysis was proven useful for disease monitoring, sequencing analysis of the plasma DNA sample at progression did not result in the identification of a genetic aberration causative of resistance to Ipatasertib monotherapy. Although resistance to AKT inhibition may be mediated by adaptive changes (e.g. activation of upstream receptor tyrosine kinases [31]), it is unknown whether this mechanism would induce resistance to the Ipatasertib monotherapy in patients harboring AKT1 mutations. Thirdly, given that all mutations found in the primary tumor were also detected in the liver metastasis, we cannot ascertain whether ctDNA not only capture the entire repertoire of mutations found in the metastatic lesion, but also that of the primary tumor. Regrettably, the amounts of plasma DNA obtained from this patient were insufficient for whole genome or whole exome sequencing analysis at the depth that would be required to determine whether a specific somatic genetic aberration was selected for during Ipatasertib monotherapy and to define whether there would be mutations affecting genes not included in the IMPACT assay that would be present in the primary tumor but not in the metastatic lesion.
In conclusion, we have demonstrated that plasma-derived DNA contains representative tumor genetic material that may be employed to uncover somatic genetic alterations present in cancer cells from patients with metastatic disease. Targeted capture MPS analysis of ctDNA may be a tool to combat intra-tumor genetic heterogeneity and to monitor tumor somatic alterations during the course of targeted therapy.
funding
LDM-A was supported in part by a Translational Research grant from the European Society for Medical Oncology (ESMO Translational Research Fellowship). The authors acknowledge Fundación Rafael del Pino for financial support (for LDMA and JT) and Asociación Española contra el Cáncer and the Steiner Foundation (for JS). No grant numbers apply.
disclosure
PP is an employee of Genentech/Roche. The other authors have no conflicts of interest.
Supplementary Material
acknowledgements
The authors thank A. Vivancos for the Sequenom data.
references
- 1.Swanton C. Intratumor heterogeneity: evolution through space and time. Cancer Res. 2012;72:4875–4882. doi: 10.1158/0008-5472.CAN-12-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Mattos-Arruda L, Bidard FC, Won HH, et al. Establishing the origin of metastatic deposits in the setting of multiple primary malignancies: the role of massively parallel sequencing. Mol Oncol. 2014;8:150–158. doi: 10.1016/j.molonc.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shah SP, Roth A, Goya R, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nik-Zainal S, Van Loo P, Wedge DC, et al. The life history of 21 breast cancers. Cell. 2012;149:994–1007. doi: 10.1016/j.cell.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martelotto LG, Ng CK, Piscuoglio S, et al. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014;16:210. doi: 10.1186/bcr3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geyer FC, Weigelt B, Natrajan R, et al. Molecular analysis reveals a genetic basis for the phenotypic diversity of metaplastic breast carcinomas. J Pathol. 2010;220:562–573. doi: 10.1002/path.2675. [DOI] [PubMed] [Google Scholar]
- 8.Navin N, Kendall J, Troge J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidard FC, Weigelt B, Reis-Filho JS. Going with the flow: from circulating tumor cells to DNA. Sci Transl Med. 2013;5:207ps214. doi: 10.1126/scitranslmed.3006305. [DOI] [PubMed] [Google Scholar]
- 10.De Mattos-Arruda L, Cortes J, Santarpia L, et al. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat Rev Clin Oncol. 2013;10:377–389. doi: 10.1038/nrclinonc.2013.80. [DOI] [PubMed] [Google Scholar]
- 11.Yap TA, Gerlinger M, Futreal PA, et al. Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med. 2012;4:127ps110. doi: 10.1126/scitranslmed.3003854. [DOI] [PubMed] [Google Scholar]
- 12.Powell AA, Talasaz AH, Zhang H, et al. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PLoS ONE. 2012;7:e33788. doi: 10.1371/journal.pone.0033788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forshew T, Murtaza M, Parkinson C, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra168. doi: 10.1126/scitranslmed.3003726. [DOI] [PubMed] [Google Scholar]
- 14.Dawson SJ, Rosenfeld N, Caldas C. Circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med. 2013;369:93–94. doi: 10.1056/NEJMc1306040. [DOI] [PubMed] [Google Scholar]
- 15.Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leary RJ, Sausen M, Kinde I, et al. Detection of chromosomal alterations in the circulation of cancer patients with whole-genome sequencing. Sci Transl Med. 2012;4:162ra154. doi: 10.1126/scitranslmed.3004742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murtaza M, Dawson SJ, Tsui DW, et al. Non-invasive analysis of acquired resistance to cancer therapy by sequencing of plasma DNA. Nature. 2013;497:108–112. doi: 10.1038/nature12065. [DOI] [PubMed] [Google Scholar]
- 18.Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem. 2013;59:211–224. doi: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 19.Misale S, Yaeger R, Hobor S, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–536. doi: 10.1038/nature11156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Ridgway LD, Wetzel MD, et al. The identification and characterization of breast cancer CTCs competent for brain metastasis. Sci Transl Med. 2013;5:180ra148. doi: 10.1126/scitranslmed.3005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin J, Sampath D, Nannini MA, et al. Targeting activated Akt with GDC-0068, a novel selective Akt inhibitor that is efficacious in multiple tumor models. Clin Cancer Res. 2013;19:1760–1772. doi: 10.1158/1078-0432.CCR-12-3072. [DOI] [PubMed] [Google Scholar]
- 22.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez L, Wilkerson PM, Lambros MB, et al. Genomic and mutational profiling of ductal carcinomas in situ and matched adjacent invasive breast cancers reveals intra-tumour genetic heterogeneity and clonal selection. J Pathol. 2012;227:42–52. doi: 10.1002/path.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Won HH, Scott SN, Brannon AR, et al. Detecting somatic genetic alterations in tumor specimens by exon capture and massively parallel sequencing. J Vis Exp. 2013;80:e50710. doi: 10.3791/50710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Von Hoff DD, Stephenson JJ, Jr, Rosen P, et al. Pilot study using molecular profiling of patients’ tumors to find potential targets and select treatments for their refractory cancers. J Clin Oncol. 2010;28:4877–4883. doi: 10.1200/JCO.2009.26.5983. [DOI] [PubMed] [Google Scholar]
- 27.Polyak K. Tumor heterogeneity confounds and illuminates: a case for Darwinian tumor evolution. Nat Med. 2014;20:344–346. doi: 10.1038/nm.3518. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Shen D, Shao J, et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013;4:1116–1130. doi: 10.1016/j.celrep.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gerlinger M, Horswell S, Larkin J, et al. Genomic architecture and evolution of clear cell renal cell carcinomas defined by multiregion sequencing. Nat Genet. 2014;46:225–233. doi: 10.1038/ng.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sottoriva A, Spiteri I, Piccirillo SG, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci USA. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandarlapaty S, Sawai A, Scaltriti M, et al. AKT inhibition relieves feedback suppression of receptor tyrosine kinase expression and activity. Cancer Cell. 2011;19:58–71. doi: 10.1016/j.ccr.2010.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.