Abstract
Immune checkpoint inhibitors, including those targeting programmed cell death protein 1 (PD-1), are reshaping cancer therapeutic strategies. Evidence suggests, however, that tumor response and patient survival are determined by tumor programmed death-ligand 1 (PD-L1) expression. We hypothesized that preconditioning of the tumor immune microenvironment using targeted, virus-mediated interferon (IFN) stimulation, would upregulate tumor PD-L1 protein expression and increase cytotoxic T cell infiltration, improving the efficacy of subsequent checkpoint blockade. Oncolytic viruses (OVs) represent a promising form of cancer immunotherapy. For brain tumors, almost all studies to date have used direct intralesional injection of OV, because of the largely untested belief that intravenous (i.v.) administration will not deliver virus to this site. Here we show, in a window-of-opportunity clinical study, that i.v. infusion of oncolytic human Orthoreovirus (referred to herein as reovirus), leads to infection of tumor cells subsequently resected as part of standard clinical care, both in high-grade glioma (HGG) and in brain metastases, and increases cytotoxic T cell tumor infiltration relative to patients not treated with virus. We further show that reovirus upregulates IFN-regulated gene expression, as well as the PD-1/PD-L1 axis in tumors, via an IFN-mediated mechanism. Finally, we show that addition of PD-1 blockade to reovirus enhances systemic therapy in a preclinical glioma model. These results support the development of combined systemic immunovirotherapy strategies for the treatment of both primary and secondary tumors in the brain.
Introduction
Therapies targeting T cell inhibitory checkpoint signalling pathways, including PD-1 monoclonal antibodies, have produced unprecedented results in recent years in solid malignancies (1–4). Unfortunately, only a minority of patients benefit, with mounting evidence that tumor response and patient survival are associated with tumor PD-L1 expression (5) and pre-existing tumor-infiltrating cytotoxic T cells (CD8+) (6). OV immunotherapy uses wild-type or genetically-modified viruses selectively to kill tumor cells and promote tumor-directed innate and adaptive immune responses (7, 8). The first OV to receive US Food and Drug Administration (FDA) approval was talimogene laherparepvec (T-VEC), after a phase III trial demonstrating superior outcomes in patients with advanced melanoma treated with intratumoral T-VEC compared to subcutaneous granulocyte-macrophage colony-stimulating factor (GM-CSF) (9). Major challenges remain, including the optimization of combination therapies and routes of virus delivery. In particular, the combination of OV with immune checkpoint blockade deserves attention, because a number of OVs stimulate the secretion of IFNs (10, 11), intermediary cytokines in PD-1/PD-L1 expression. Furthermore, OV delivery to tumor can enhance T cell infiltration (11), hence priming the tumor immune microenvironment for immune-mediated therapy when combined with PD-1/PD-L1 axis blockade.
For patients with brain tumors, concerns that the blood-brain barrier (BBB) may inhibit OV delivery have, thus far, limited studies using i.v. administration, notwithstanding the infiltrative and/or multifocal nature of such tumors. A number of OVs, including HSV-1716 (12–14), HSV-G207 (15), adenovirus-dl1520 (ONYX-015)(16), and reovirus (17, 18), have been trialled in glioma patients by surgical intratumoral or intracavity injection. These techniques require careful patient selection and technically challenging neurosurgery, limiting repeat administration. Yet, the need for effective therapies in this group of patients cannot be overemphasized; median survival for grade IV gliomas (glioblastoma multiforme - GBM) after tumor-directed surgery and chemoradiotherapy is 14.6 months (19), and those with a single brain metastasis and controlled extracranial disease survive only nine to 10 months, despite optimal treatment (20). The clinical trial described herein tested whether i.v. reovirus could infect recurrent HGGs and metastatic brain tumors in patients and examined the ensuing immunological sequelae, with particular focus upon the tumor microenvironment.
Results
i.v. injected reovirus accesses brain tumors in mice
Preclinical experiments confirmed that i.v. reovirus selectively accesses intracranially implanted malignant melanoma in immunocompetent mice, albeit to varying degrees (fig. S1). Reovirus σ3 capsid protein and reovirus RNA were strongly detected after i.v. infusion in mice 1 and 2, suggesting viral genome replication and translation, but were only detectable in extremely low amounts in mouse 3. Lower magnification pictures revealed high reovirus protein expression clustered in small areas of tumor, with lower expression in a larger number of tumor cells (fig. S1, middle row). Reovirus was not detected in normal peri-tumor murine brain tissue or PBS control.
i.v. reovirus associates with multiple peripheral white blood cell subsets in patients
On the basis of the murine experiment results, we recruited nine patients to a phase Ib window of opportunity trial (table S1), where each patient was treated with a single, one hour i.v. infusion of 1x1010 TCID50 (50 % tissue culture infectious dose) reovirus ahead of planned surgical resection of his or her brain tumor. Treatment was well tolerated in all cases, and surgery was undertaken three to 17 days after reovirus infusion. The most commonly observed adverse events were lymphopenia (grade 1-2 in all nine patients, grade 3-4 in six patients) and flu-like symptoms. Median overall survival from the day of reovirus infusion to death was 469 days (range 118 to 1079 days), which is consistent with the expected survival for this group of patients that have variable cancer diagnoses.
Extending upon findings from our previous study (21), where we demonstrated i.v. reovirus carriage and protection from neutralizing antibody by peripheral blood mononuclear cells (PBMCs), granulocytes, and platelets, we examined white blood cell subsets taken mid-reovirus-infusion for reovirus RNA by RT-PCR (fig. S2A). In addition to granulocytes, we confirmed the association of reovirus RNA with CD14+ (monocytes, which are pivotal for reovirus cell carriage in mice (22)), CD19+ (B cells), and CD56+ (NK/NKT cells) fractions, but viral RNA could not be detected on CD3+ (T cells) in this subset of trial patients, for whom samples were available. Time-course analysis of IFN-α concentrations in patient sera taken before and after reovirus, revealed significantly increased IFN-α (P=0.0153) two days after infusion, in comparison to baseline (fig. S2B). This indicates reovirus engagement of pathogen recognition receptors, potentially during carriage by peripheral white blood cells, resulting in systemic IFN release. Plasma concentrations of other inflammatory cytokines were also increased two days after reovirus infusion, relative to pre-infusion concentrations (table S2).
Reovirus is detected in resected brain tumors from trial patients
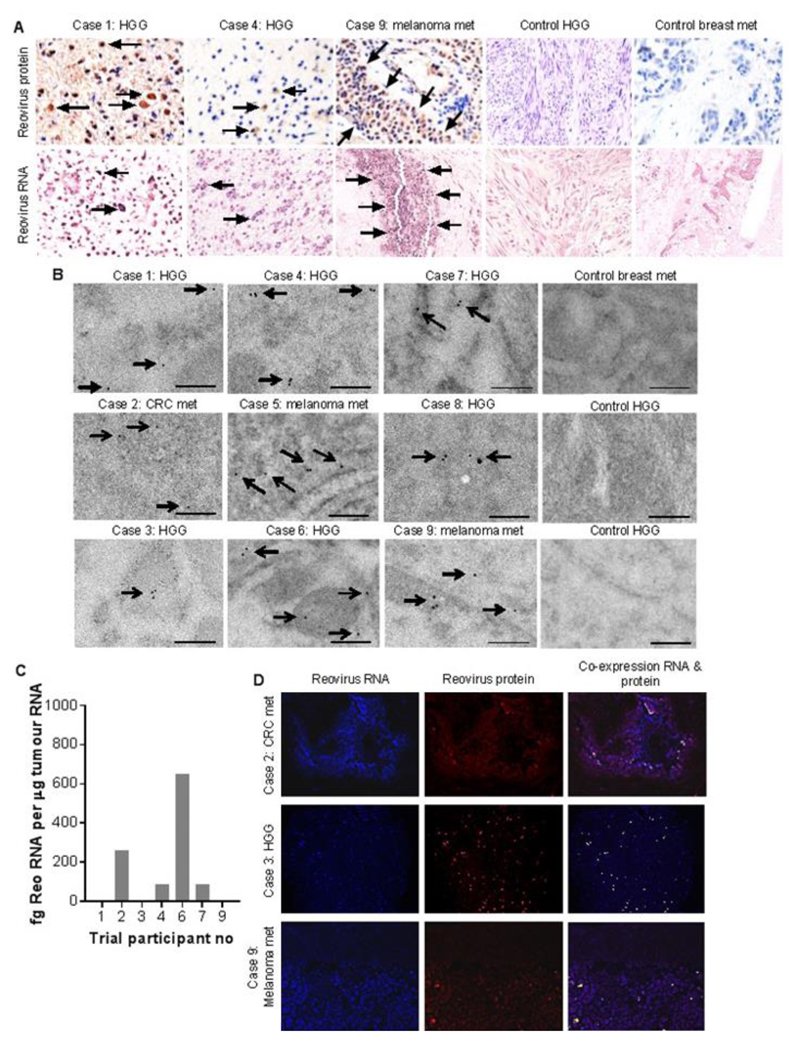
Examination of resected brain tumors by immunohistochemistry (IHC) revealed the presence of reovirus σ3 capsid protein in low amounts in six out of nine tumours (Fig. 1A upper row and table S3) and nine out of nine tumors by immunogold transmission electron microscopy (TEM) (Fig. 1B). Resected brain tumor specimens from patients outside the trial served as controls. Secondary antibody-only controls for background immuno-gold staining in trial patient tumors are shown in fig. S3. The vast majority of reovirus protein was localized to tumor cells, with only 0-6 % localizing to endothelial cells (table S3 and fig. S4). Examination of the specimens by in-situ hybridization (ISH) revealed eight out of nine tumors to be positive for reovirus RNA (Fig. 1A lower row), with reovirus RNA being detected in a higher percentage of cells than reovirus σ3 protein in all cases (table S3), consistent with the findings in mice (fig. S1). In comparison, control brain tumors showed no reovirus RNA staining (Fig. 1A lower panel). The presence of reovirus RNA in tumors was further examined by qRT-PCR amplification of the S4 genome segment (encoding σ3), confirming four of the seven available tumor samples to be positive (Fig. 1C). Despite some variation in detection limits for different techniques, together these data convincingly support delivery of systemically administered reovirus to patient brain tumors. The distribution of reovirus RNA and protein within tumors was further examined using immunofluorescence (IF) (Fig. 1D for trial tumors and fig. S5A for control). Reovirus RNA was detected in a large proportion of cells, whereas reovirus protein and protein-RNA co-localization were only detected in discrete areas of tumor, suggesting that reovirus protein translation and/or productive infection occurred only in small areas of tumor, at least by the snapshot timepoint of surgical resection.
Fig. 1. Intravenous delivery of reovirus to primary and secondary brain tumors.
A) Representative IHC and ISH trial and control patient tumor sections stained for reovirus σ3 protein (brown - top row) and reovirus RNA (blue - bottom row). The ‘control breast met’ is a metastasis to the brain from a breast cancer primary. Arrows point to examples of positive cells / positive areas of tissue. Scale bars = 20 μm. B) Trial and control patient tumor immunogold-TEM images for reovirus σ3 protein (arrows). Scale bar = 200 nm. C) qRT-PCR for reovirus σ3 gene, using whole tumor RNA. Data indicate fg reovirus RNA per µg of whole tumor RNA. Histogram shows the mean of triplicate samples, and error bars indicate standard deviation. D) Representative IF of trial patient tumor sections showing staining for reovirus RNA (blue), reovirus σ3 protein (red), and their co-expression (yellow). Scale bars = 80 μm.
The presence of reovirus RNA and protein in tumors correlates with Ki67
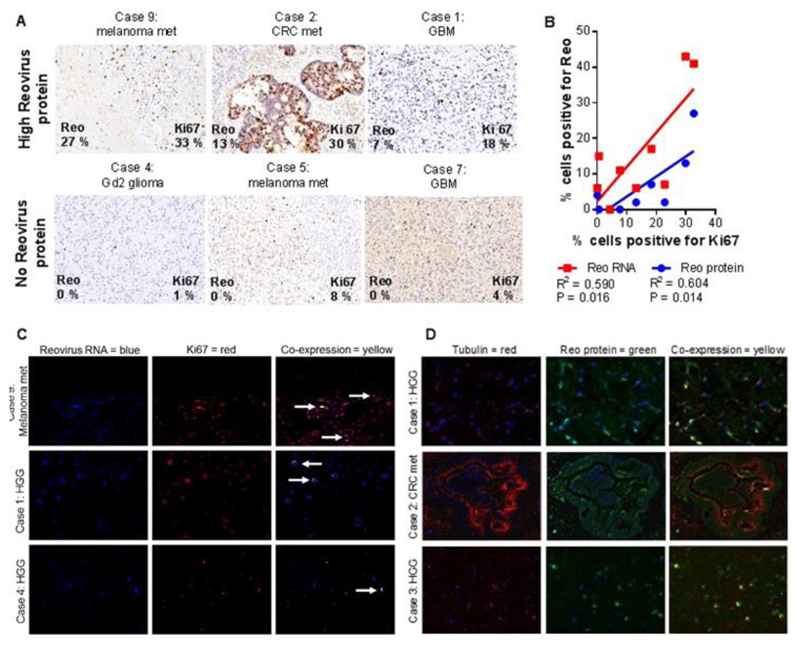
The overall proportion of reovirus σ3 protein- and RNA-positive cells within individual tumors varied widely between the nine trial patients (table S3). Because actively dividing cells preferentially support reovirus replication in comparison to quiescent cells (23), (24), we analyzed resected trial patient and control tumors for expression of the proliferation marker Ki67 relative to reovirus protein/RNA (Fig. 2A, Fig. 2B, and table S4). The amounts of both reovirus σ3 protein and RNA correlated with tumors containing a high proportion of Ki67-positive cells (P=0.014 for σ3 protein and P=0.016 for reovirus RNA). However, immunofluorescence analysis of tumors revealed little co-expression of reovirus RNA and Ki67 (Fig. 2C for trial tumors and fig. S5B for control), potentially because Ki67 staining is restricted to particular phases of the cell cycle (25). Further IF examination of tumors confirmed the presence of low amounts of reovirus σ3 protein, as was detected by IHC and TEM, and showed reovirus protein to frequently co-localize with tubulin, a key component of reovirus replication factories (Fig. 2D for trial tumors and fig. S5C for control) (26). Altogether, these results indicate that tumors with a higher proliferation index are more susceptible to reovirus infection, but that reovirus protein translation and/or productive infection overall occur at relatively low rates only. In keeping with these observations, and in contrast to our previous trial in resected colorectal liver metastases (21), replication-competent reovirus could not be retrieved from any of the nine trial tumors. Analysis of the number of days between reovirus administration and surgery revealed no significant change in reovirus RNA and protein over time from reovirus infusion (fig. S6).
Fig. 2. Correlation of reovirus RNA / protein with proliferating tumor cells.
A) Trial patient IHC tumor sections stained for Ki67 (brown) with indicated percentages of cells positive for Ki67 and reovirus σ3 protein (from table S3), showing examples of tumors with high reovirus σ3 staining (top row) and no reovirus σ3 protein staining (bottom row). Scale bars = 40 μm. B) Scatter plot and line of best fit, correlating the percentages of tumor cells positive by IHC for reovirus RNA or σ3 protein and for Ki67. C) Representative tumor sections derived from trial patient nine (high Ki67, top row), trial patient one (intermediate Ki67, middle row), and trial patient four (low Ki67, bottom row), showing IF staining for reovirus RNA (blue), Ki67 (red), or their co-expression (yellow, arrows). Scale bars = 40 μm. D) Representative trial patient tumor IF staining for tubulin (fluorescent red), reovirus σ3 protein (fluorescent green) and their co-expression (yellow). Nuclear counterstaining is blue. Top and bottom row scale bars = 40 μm. Middle row scale bar = 80 μm.
Reovirus treatment increases tumor leukocyte infiltration
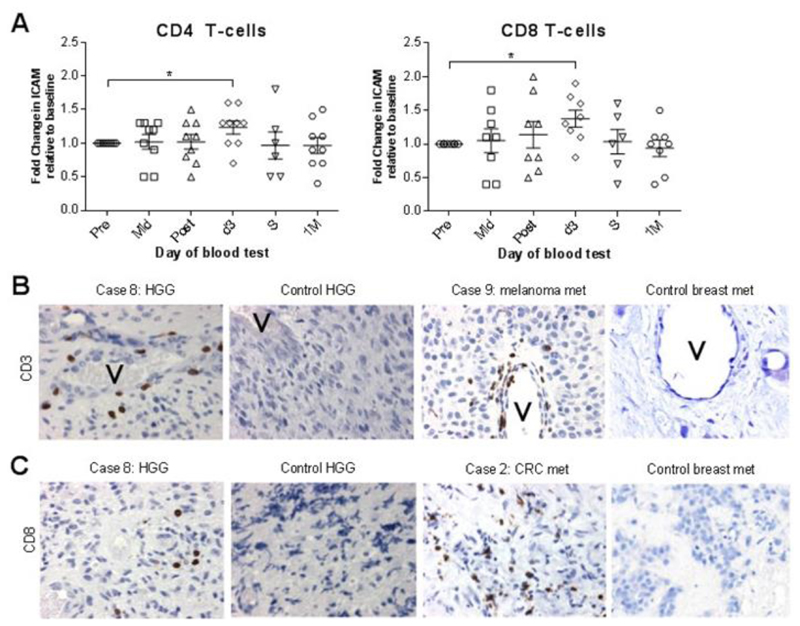
We used RNAseq to compare expression of coding and non-coding transcripts in whole tumor RNA from three GBM trial patient samples (cases one, six, and seven), to that of three control GBM tumors. Given that sample numbers were small, criteria for statistically significant differential gene expression between treatment and control tumors were stringently set as described in the methods section (q<0.1). Of the 2366 sequenced transcripts, 102 genes were differentially expressed between reovirus-treated and untreated GBM groups (table S5). Two of these transcripts were CCL3 (mean control group expression = 4.2, mean treatment group expression = 34.3, q=0.0188) and CCL4 (mean control group expression = 2.5, mean treatment group expression = 19.2, q=0.0188), which both function to recruit CD8+ T cells and other leukocytes to sites of immunization (27). CCL4 protein and a number of other chemokines were higher in trial patient plasma two days after reovirus infusion, relative to pre-infusion concentrations (table S2). Furthermore, peripheral blood assessment of CD4+ and CD8+ T cell populations revealed increased cell surface expression of intercellular adhesion molecule (ICAM) two days after i.v. reovirus infusion, in comparison to baseline expression (Fig. 3A). ICAM expression is upregulated by inflammatory cytokines, enhancing leukocyte interaction with vascular endothelial cells to enable migration to sites of inflammation (28). In keeping with these observations, IHC analysis of trial patient and control tumors revealed CD3+ T cells in and around blood vessel walls, in virus-treated but not untreated controls, consistent with reovirus-induced chemotaxis of T cells into infected brain tumors (Fig. 3B). Further IHC assessment for tumor-infiltrating cytotoxic T cells (CD8+), which are critical for PD-1/PD-L1 directed immunotherapy (29), revealed their presence in eight of the nine trial patient tumors, four of which showed more staining (2+ or 3+), in comparison to the control cases, where CD8+ T cell infiltration was detected only in three of the six tumors, all in low amounts (Fig. 3C and table S4).
Fig. 3. Tumor immune cell infiltration.
A) Fold change in cell-surface ICAM expression on CD4 and CD8 T cells from trial patients’ peripheral blood. B) Trial and control patient IHC tumor sections stained for CD3 (brown). Scale bars = 20 μm. ‘V’ indicates blood vessel. C) Trial and control patient IHC tumor sections stained for CD8 (brown). Scale bars = 20 μm.
We also examined tumours for the presence of CD68+ microglia/infiltrating macrophages, and found these to be present in higher numbers in tumors from reovirus-treated patients in comparison to controls (fig. S7 and table S4). Very few tumor-infiltrating CD56+ natural killer cells and CD19+ B cells were found in any tumor. The daily dose of dexamethasone taken by patients within and outside the trial did not appear to correlate with tumour immune cell infiltration in the examined surgical specimens (table S4).
Genes associated with programmed cell death are more highly expressed in GBM tumors from reovirus-treated patients than in matched controls
Functional analysis of the differentially expressed genes found by RNAseq indicated significant enrichment in members of several biological processes, including those governing programmed cell death (P=0.0003), regulation of viral transcription (P=0.0000502), and cytokine activity (P=0.0129)(table S6). Consistent with these RNA expression data and preclinical models (30), IHC analysis of trial HGG samples revealed a higher proportion of tumor cells to be positive for cleaved caspase 3, albeit in a small number of patients, than in controls, suggesting the specific induction of apoptosis within tumors after i.v. reovirus infusion (Fig. 4A and table S4). A similar pattern was observed for the three trial brain metastases in comparison to controls (fig. S8A, table S4).
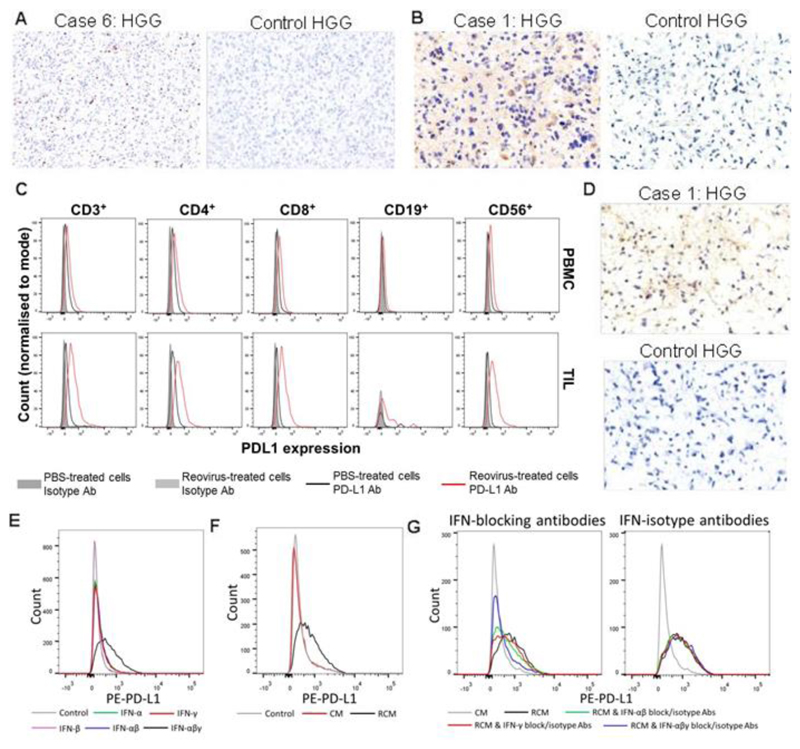
Fig. 4. Expression of cleaved caspase 3, PD-L1, and PD-1 in high grade gliomas after reovirus treatment.
A) Representative trial and control patient HGG sections stained for cleaved caspase 3 (brown) by IHC. Scale bars = 60 μm. B) Representative trial and control patient HGG sections stained by IHC for PD-L1 (brown). Scale bars = 30 μm. C) Representative (one of three samples tested) flow cytometry for PD-L1 on GBM TILs (bottom row) or PBMCs (top row) derived from the same patient, after stimulation for 48 hours using 1 PFU/cell reovirus. D) Representative trial and control patient HGG sections stained by IHC for PD-1 (brown). Scale bars = 30 μm. E) Representative flow cytometry for PD-L1 on GBM1 cells after stimulation with combinations of purified IFN-α/-β/-γ for 24 hr, each at 100 pg/ml. F) Representative flow cytometry for PD-L1 on GBM1 cells after stimulation with ex vivo HGG-derived CM or RCM for 24 hours (at a concentration of 1:4 of conditioned medium to native medium). G) Representative flow cytometry for PD-L1 on GBM1 cells after stimulation using PBMC-derived CM or RCM for 24 hours (at a concentration of 1:4 of conditioned medium to native medium) with blockade of IFN-α+β/γ/ α+β+γ or equivalent isotype controls.
PD-L1 expression is higher in tumors resected from reovirus-treated patients than in controls
We next sought to determine whether reovirus treatment results in the upregulation of IFN-regulated genes (IRGs). Of the 23,366 genes expressed in our samples, 5031 are IRGs (31), in contrast to 48 of the 102 genes that were differentially expressed between trial and control samples (chi-squared test, p=1.28x10-8).
Interferon transcripts and PD-1 mRNA were not detected in our analysis, perhaps due to a transient rise and fall in expression before the timepoint of surgical resection. The expression of PD-L1 was, on average, about twice as high in reovirus-treated patient GBM samples than controls, but the difference was not statistically significant (1.310 in trial samples vs. 0.668 in controls). However, protein analysis by IHC revealed consistent PD-L1 expression in trial HGGs, but not in controls (Fig. 4B and table S4), and a similar pattern was observed for brain metastases (fig. S8B, table S4). Of the two melanoma metastases in the trial, case nine, which stained most intensely for Ki67 and reovirus RNA, also displayed the strongest PD-L1 expression. Case five, in contrast, displayed relatively low expression of Ki67, reovirus RNA, and PD-L1 (fig. S8C). We sought to confirm these clinical findings in vitro; direct reovirus treatment of the established glioma cell line U87, primary human GBM cells (GBM1 and GBM4 (32)), and cell lines derived from metastatic breast cancer, colon cancer, and melanoma (MCF-7, SW620, and Mel624) significantly increased PD-L1 expression in U87 (P=0.0021), GBM4 (P=0.0275), MCF-7 (P=0.0002), and SW620 cells (P=0.0062), with no significant differences in GBM1 and Mel624 cells (fig. S9A).
We reasoned that reovirus treatment would also promote checkpoint protein expression within tumor-infiltrating immune cell populations. In vitro reovirus treatment of patient-derived mixed HGG cell cultures from control patients outside the trial induced PD-L1 upregulation within tumor-infiltrating lymphocytes (TILs), including helper/cytotoxic T cells, B cells, and NK cells (Fig. 4C, lower panel). This was also observed within PBMCs derived from the same patients (Fig. 4C upper panel), and in healthy donor PBMCs (fig. S9B). With regards to PD-1, higher protein expression was seen in both reovirus-treated HGGs and metastatic tumors in comparison to controls (Fig. 4D and table S4), as well as in healthy donor PBMC subsets after in vitro reovirus treatment (fig. S9C).
Reovirus induces PD-L1 expression via an IFN-based mechanism
We used GBM1 cells as an in vitro model of HGG tumor cells, to confirm an IFN-dependent mechanism for reovirus-induced stimulation of PD-L1 expression. GBM1 cells were treated using type I IFNs (IFN-α, IFN-β) and type II IFN (IFN-γ) in isolation or in combination. Whereas type I or type II IFN treatments each induced 50-70 % upregulation of PD-L1, the combination of type I and type II IFNs induced a 250 % increase in cell surface PD-L1 expression. In contrast, combining IFN-α and -β, which bind the same type I IFN receptor, induced no further increase in PD-L1 expression over IFN-α alone (Fig. 4E and fig. S10A). Reovirus treatment of fresh patient-derived HGG single-cell suspensions (including all cell types contributing to the tumor immune microenvironment) generated reovirus-conditioned medium (RCM), which contained high concentrations of type I and II IFNs (fig. S10B). RCM was filtered to remove reovirus, and conditioned medium (CM) controls were also filtered, to maintain experimental consistency. Soluble factors within HGG-RCM significantly upregulated GBM1 PD-L1 expression in comparison to CM (P=0.0481) (Fig. 4F and fig. S10C). To establish the relative contributions of type I and type II reovirus-induced interferons in the upregulation of PD-L1, we used PBMC-RCM (containing IFN-α, -β, and –γ) to treat GBM1 cells with concurrent blockade of interferon receptors and soluble interferons. Blockade of type I or II IFNs partially reduced PBMC-RCM-induced PD-L1 expression on GBM1 cells, whereas blockade of both type I and II IFNs greatly diminished PD-L1 expression, confirming that type I and II IFNs co-operate to induce PD-L1 in patient-derived glioma cells (Fig. 4G and fig. S10D).
Sequential treatment using i.v. reovirus followed by PD-1/PD-L1 axis blockade improves survival in mice with brain tumors
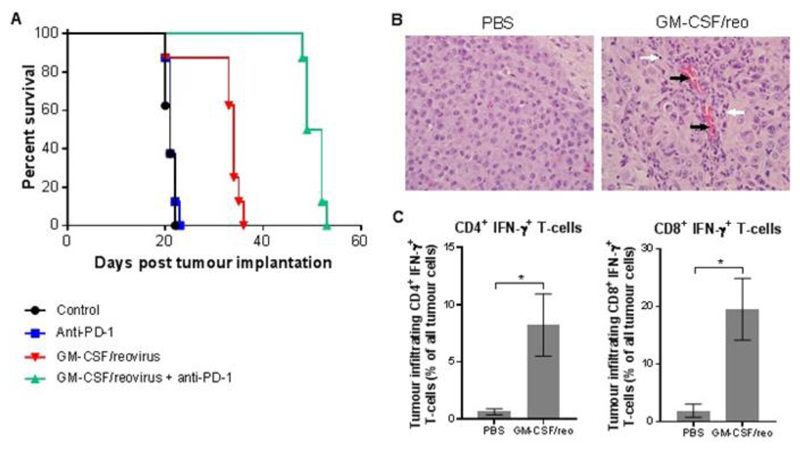
Based on the above data showing immune cell infiltration and upregulation of the PD-1/PD-L1 axis by systemic reovirus in brain tumor patients, we sought to test survival using sequential OV-checkpoint inhibitor blockade in an immunocompetent orthotopic animal model of glioma. Consistent with our trial data, C57/BL6 mice implanted intracranially with GL261 glioma cells exhibited improved survival using i.v. GM-CSF/reovirus (our optimal systemic reovirus regime (22)) over a two-week period, followed by a one-week period of PD-1 antibody treatment, compared to treatment with either virotherapy or checkpoint blockade alone (Fig. 5A). Comparison of hematoxylin and eosin-stained sections from brain tumors taken post-mortem from GM-CSF/reovirus-treated mice revealed prominent perivascular and intratumoral inflammatory infiltrate, containing lymphocytes (Fig. 5B). Further flow cytometry analysis of brain tumor single-cell suspensions revealed significantly higher active (IFN-γ+) helper (CD3+CD4+) and cytotoxic (CD3+CD8+) T cells within GM-CSF/reovirus-treated tumors in comparison to PBS-treated tumors (P=0.0089 and P=0.0125, respectively, for helper and cytotoxic T cells, Fig. 5C).
Fig. 5. Combination i.v. reovirus and checkpoint inhibition in an orthotopic syngeneic brain tumor model.
C57/BL6 reovirus-vaccinated mice (22) were injected with GL261 cells intracranially on day one and treated using combinations of GM-CSF plus i.v. reovirus and/or anti-PD-1 antibody. A) Kaplan-Meier survival plot, with Mantel-Cox comparison of survival curves: control vs. Anti-PD-1 P=0.4617, control vs. GM-CSF/reovirus P=0.0012, control vs. GM-CSF/reovirus + anti-PD-1 P<0.0001, GM-CSF/reovirus vs. GM-CSF/reovirus + anti-PD-1 P<0.0001, anti-PD-1 vs. GM-CSF/reovirus + anti-PD-1 P<0.0001. B) Representative brain tumor hematoxylin and eosin stained sections from PBS- and GM-CSF/reovirus treated mice. Black arrows mark vascular endothelial cells; white arrows mark lymphocytes. Scale bars = 30 μm. C) Flow cytometry quantification of CD3+ CD4+ IFN-γ+ or CD3+ CD8+ IFN-γ+ tumor-infiltrating lymphocytes from PBS or GM-CSF/reovirus-treated mice. Graph shows the mean ± SD of four samples.
Discussion
Our data provide evidence of an OV, reovirus, gaining access to brain tumors after i.v. administration to patients. Reovirus RNA was widely detected in tumor cells of differing histological types. The i.v. route, therefore, holds promise as an efficient means of delivering OV to brain tumors, enabling regular scheduled treatments to be administered, while avoiding the need for neurosurgical methods of access.
The tight window of opportunity between clinical presentation and planned brain surgery limited the number of patients able to participate in this study. Nonetheless, tumors from all nine treated patients, across a range of histological tumor types, showed evidence of reovirus infection. An ongoing clinical trial is assessing the safety and efficacy of i.v. reovirus in combination with post-operative chemoradiotherapy for patients with GBM (ReoGlio, ISRCTN70044565). Other OVs, including parvovirus H-1, are also being tested by i.v. infusion in patients with brain tumors (33).
Previous studies have shown that apoptosis in malignant cells is induced after reovirus receptor binding and disassembly to form intermediate sub-virus particles, but that viral genome transcription and translation are not required (34). Hence, the lack of evidence for major reovirus productive infection in our study does not necessarily mean the absence of direct viral cytotoxicity. Indeed our RNAseq analysis and IHC for cleaved caspase 3 do indicate the induction of apoptosis. Moreover, from an immunotherapy perspective, intracellular reovirus RNA is sufficient to engage pathogen recognition receptors, inducing interferon expression. Interferons are critical mediators of immune-mediated anti-cancer effects, for example by activating NK, T cell, and dendritic cell populations and enhancing antigen presentation (35). Future studies should, therefore, look to optimize reovirus treatment schedules to further increase the delivery of reovirus to tumor cells, given the absence of major reovirus productive infection.
The BBB is known to be disrupted in brain tumors, as indicated by the presence of vasogenic edema (36). Nonetheless, many systemic anti-cancer agents, including monoclonal antibodies, are thought to be excluded by the BBB due to their higher molecular mass (37, 38). The mechanism(s) by which reovirus enters brain tumors in comparison to tumors outside of the brain and its relation to the integrity of the BBB remain unclear, although the association of reovirus with multiple peripheral white blood cell subsets supports the idea that these immune cells may play a role in the delivery of virus to tumor. This is additionally supported by our observations of enhanced immune cell tumor infiltration after reovirus, and also by the clinically observed lymphopenia in all nine treated patients, a phenomenon that could be attributed at least in part to the accumulation of lymphocytes at the site of infection in tumor, consistent with previous reports of lymphopenia in acute viral infections (39). We found upregulation of CCL3 and CCL4 mRNA in brain tumors from reovirus-treated patients. These chemokines are upregulated in other acute viral encephalitis infections, including Semliki Forest Virus and West Nile Virus (40), whereas blockade of CCR5, whose ligands include CCL3 and CCL4 (41), decreases leukocyte migration into murine brains after viral infection (40). Pre-existing CD3+ and CD8+ TILs have a positive effect on survival in solid tumors, and infiltrating cytotoxic T cells are critical for PD-1/PD-L1 directed immunotherapy (29, 42). In our trial, a proportion of brain tumors contained high numbers of cytotoxic T cells after reovirus treatment, a finding consistent with our murine data showing increased T cell infiltration into tumors after systemic reovirus treatment. Our data indicate lower baseline CD68 staining than in previous reports (43), potentially due to differences in staining methods. The functional relevance of increased CD68 cells in brain tumors after i.v. reovirus infusion and their origin remain unclear, warranting further investigation.
Tumors from reovirus-treated patients exhibited more intense staining for both PD-1 and PD-L1, immune checkpoint proteins that are induced by IFNs. We further found evidence for both peripheral and tumor induction of IFNs, key cytokines in reovirus-mediated activation of immune cell populations (44). It has previously been shown that effector T cells upregulate PD-1 during the acute phase of viral infections, whilst maintaining activity, and that this protective response does not correspond to adverse outcomes (45, 46). One of the determinants of efficacy in PD-1 checkpoint blockade is tumor expression of PD-L1 (47). In GBM, PD-L1 expression is relatively weak in the majority of tumors (48). We found PD-L1 to be strongly upregulated by type I IFNs in combination with IFN-γ, cytokines that were secreted after ex vivo reovirus treatment of HGG cells. Hence, reovirus therapy may be used to improve clinical outcomes in patients with brain tumors by activating white blood cells, enhancing T cell infiltration into tumors, and upregulating PD-L1 there, in preparation for subsequent anti-PD-1 therapy. In support of our findings, a recent article by Ribas et al. has reported findings from the phase Ib portion of the Masterkey-265 study in extracranial advanced melanoma, where patients were treated using intratumoral injections of T-VEC (HSV type I encoding GM-CSF), with concomitant anti-PD1 therapy beginning six weeks after the start of OV therapy (49). Patients who responded to treatment in this study had increased CD8+ T cells and elevated PD-L1 protein expression on several cell subsets in tumors after OV therapy.
The combination of i.v. OV with anti-PD-1 therapy for the treatment of brain tumors is the focus of investigations in our laboratory. Rational drug combinations that include anti-PD-1 therapy for the treatment of brain tumors are all the more pertinent in light of the failure of single-agent nivolumab to deliver a survival advantage in patients with GBM (NCT02017717) (50). The challenges of local delivery of OVs into brain tumors are substantial, and direct intratumoral injection is inevitably suboptimal for multifocal/infiltrative disease. Systemic virus-based immunotherapy provides a pragmatic alternative and appears capable of altering the immune microenvironment within brain tumors, which, in turn, could potentially improve cancer therapy when combined with immune checkpoint blockade.
Materials and Methods
Study design
Clinical trial number (EudraCT) 2011-005635-10. This was an open-label, non-randomized, single center study, which recruited nine adult patients between July 2013 and November 2014 at The Leeds Teaching Hospitals NHS Trust (LTHT), Leeds, UK. Patients were planned for debulking neurosurgery either for recurrent HGG or for metastatic tumor to the brain, as part of routine clinical care. A single one hour i.v. infusion of 1x1010 TCID50 reovirus was administered to patients ahead of surgery. The primary endpoint of the study was the presence of reovirus in the resected tumor sample. Tumors were initially analyzed for the presence of reovirus in batches of three. The trial achieved its primary endpoint and was closed after the recruitment of six patients with recurrent HGG and three with metastatic tumors to the brain, because reovirus was detected in all nine tumors. Inclusion criteria included adequate hematological and organ function and an Eastern Cooperative Oncology Group (ECOG) performance status ≤1. All patients gave written informed consent according to good clinical practice guidelines. Protocol, patient information sheet, and consent forms were approved by the United Kingdom Medicines and Healthcare products Regulatory Authority (MHRA), regional ethics review committee, as well as institutional review board at St James’s University Hospital. The trial management committee met on a monthly basis to discuss study progress, including patient safety and adverse events. Clinical patient safety assessments were performed within one week of start of treatment, on the day of reovirus infusion (day one), day three, the day of surgery and one month after surgery. Imaging was performed as for standard clinical care only. Control brain tumors were obtained from patients undergoing routine planned surgery at LTHT. Written informed consent was obtained in accordance with local institutional ethics review and approval. After surgery, all brain tumors were transported in L-glutamine-containing RPMI-1640 medium (Sigma) supplemented with 10 % FCS (Biosera) and 1 % (v/v) antibiotic antimycotic solution (Sigma).
Reovirus
Clinical-grade reovirus Dearing type 3 (Reolysin) was provided by Oncolytics Biotech Inc.
Animal experiments
In vivo animal models were approved by the University of Leeds Local Ethics Review Committee or the Mayo Foundation Institutional Animal Care and Use Committee. For fig. S1, six- to 10-week old C57/BL6 mice (Jackson Laboratories) were injected intracranially with 1x105 B16 melanoma cells and eight days later treated with a single injection of 1x108 PFU i.v. reovirus or PBS. Mice were sacrificed three days after treatment. For Figure 5, eight mice in each group were used; six- to eight-week old C57/BL6 reovirus-vaccinated mice (22) were injected intracranially with GL261 cells on day one. On day five, mice were treated using daily i.v. injections of 300 ng GM-CSF (Peprotech) and i.v. reovirus at 5x107 PFU or PBS as a control for five days. Treatments were repeated for a further five consecutive days starting on day 12. On days 19, 21, and 23, mice were treated with anti-PD-1 antibody (clone RMP1-14; Bio X Cell) or IgG isotype control (clone MPC11; Bio X Cell) by i.p. injections. Mice were regularly monitored for any signs of deterioration or weight loss, upon which animals were sacrificed and the duration of survival recorded.
IHC, ISH, and IF
These techniques were performed on formalin-fixed paraffin-embedded tissue sampled randomly from one to two areas of resected patient or animal tumor. Tissue for IHC was processed using an automated Bond Max system (Leica Biosystems) as described (51). Reovirus σ3 and cleaved caspase 3 antibodies were diluted 1:1000. IHC detection of PD-1, PD-L1, CD3, CD8, CD68, and Ki67 used an automated Bond Max system (Leica Biosystems) according to manufacturer’s protocol. Primary antibodies (all Abcam) were diluted either 1:500 (PD1 and PD-L1) or 1:200 (CD3, CD31, CD8, CD68, Ki67). The percentage of positive cells was determined using the InForm system (Perkin Elmer). ISH for reovirus RNA and IF for reovirus σ3 protein, reovirus RNA, and tubulin were performed as previously described (51), (21). IF nuclear counterstaining was with DAPI diluted 1:10,000 (ThermoFisher Scientific). One to two slides per tumor were examined for quantification of reovirus σ3 protein and reovirus RNA, whereas two to three slides per tumor were examined for quantification of Ki67, PD-1, PD-L1, and CD markers.
Peripheral reovirus carriage RT-PCR
Peripheral blood samples taken from study patients mid-reovirus-infusion were collected, fractionated for PBMCs and granulocytes, then RNA extracted as previously described (21). PBMC subsets were isolated by MACS Microbead selection (Miltenyi Biotec) according to manufacturer’s instructions. PCR conditions and primers were also previously described (52). Samples were run on 2 % agarose gels alongside a 100 bp DNA ladder (New England Biolabs).
Tumor reovirus qRT-PCR
Tumor tissue was disaggregated using a Cell Dissociation Sieve & Tissue Grinder Kit (Sigma). 1 µg of Trizol (Sigma) -extracted RNA was reverse-transcribed (Bioline SensiFast cDNA Synthesis kit) according to manufacturer’s instructions in 20 µl reactions. 2 µl of the resultant reaction containing cDNA was subjected to standard qPCR using SYBR Green PCR Master Mix (Life Technologies) and primers specific to the reovirus σ3 gene, to amplify a 61 bp product (forward 5’ GATGCGCCAATGTCTAATCA 3’, reverse 5’ CTCCTCGCAATACAACTCGT 3’; both Sigma). QuantiTect primers (Qiagen) specific to 18S rRNA served as cellular RNA controls.
RNAseq
Three control GBM tumors were compared to three GBM tumors from reovirus-treated trial patients. RNA sequencing libraries were prepared using the Truseq Stranded Total RNA Library Prep Kit (Illumina), and 150 bp paired end reads were sequenced on a lane of the Illumina HiSeq3000. Reads were quality-processed and aligned as described previously (53). Expression analysis was performed using the Cufflinks suite of programs (54). Briefly, expression was quantified using cuffquant with multi-read correction and abundant non-informative transcripts (ribosomal and mitochondrial RNAs) masked. Differential expression analysis between the control (patients without reovirus) and treated (patients receiving reovirus) groups was then performed using cuffdiff with a false discovery rate (FDR) of 0.1. Gene set analysis of the resulting significantly differentially expressed genes was performed using hypergeometric testing via WebGestalt (55), with the set of genes expressed in any sample as the reference set.
Processing of HGG single-cell suspensions and PBMCs
Written informed consent was obtained in accordance with local institutional ethics review and approval. HGG tumor samples were processed using a Brain Tumor Dissociation Kit (Miltenyi Biotec), according to the manufacturer’s instructions. Briefly, tumor tissue was homogenized and digested in gentleMACS C tubes using a dissociator and buffers and enzymes supplied by the manufacturer, then incubated at 37°C. Single-cell suspensions were passed through a 70 µm filter. Where appropriate, samples were demyelinated using Myelin Removal Beads II (Miltenyi Biotec). Paired PBMC samples were derived from the same patients by step density centrifugation over Lymphoprep (Axis-Shield).
Culture of GBM1, GBM4, MCF-7, U87, SW620, and Mel624 cell lines
All cells were cultured at 37 °C with 5 % CO2. The human breast adenocarcinoma cell line, MCF-7, human colorectal cancer cell line, SW620, human glioblastoma cell line, U87, and the human melanoma cell line, Mel624, were maintained in L-glutamine-containing Dulbecco’s Modified Eagle’s Medium (DMEM; Sigma) supplemented with 10 % (v/v) fetal calf serum (FCS). GBM1 and GBM4 cells were adherently propagated on plasticware coated with poly-L-ornithine (Sigma; 5 μg/ml) and laminin (Invitrogen; 5 mg/ml) and cultured in Neurobasal medium supplemented with 0.5X B27, 0.5X N2 (all from Thermofisher), recombinant human basic fibroblast growth factor (bFGF; 40 ng/mL; Gibco), and epidermal growth factor (EGF; 40 ng/ml; R&D Systems).
Generation of CM and RCM from fresh patient HGG samples and PBMCs
HGG single-cell suspensions or PBMCs were cultured for 24 hrs at 2x106/ml alone (CM), with 50 PFU/cell reovirus (HGG-RCM), or with 1 PFU/cell reovirus (PBMC-RCM) in L-glutamine-containing RPMI supplemented with 10 % (v/v) FCS. Cell-free supernatants were collected, and reovirus was removed by filtration through Millipore OptiScale 25 filters. CM and RCM were stored at -80 °C until required.
Flow cytometry
Flow cytometry was performed on an LSRII flow cytometer, and data were analysed using FACSDiva (both Beckton Dickinson) or FlowJo (Treestar). Relative fluorescence shift (RFS) was calculated using the formula where MFI is median fluorescence intensity. To create overlay plots in FlowJo, the y-axis was normalized to mode.
Direct reovirus stimulation of patient HGG and PBMC
HGG single-cell suspensions and PBMCs were plated separately at a density of 2x106/ml and cultured with or without reovirus at 1 PFU/cell for 48 hrs in RPMI supplemented with 10 % FCS. Samples were harvested and washed with isotonic buffer (PBS/0.05 % BSA) before staining with the following antibodies: CD45-FITC (H130), CD8-BV421 (RPA-T8), CD20-BUV395 (2H7), NKp46-APC (9E2/Nkp46), and PD1-PE (MIH4) from BD Biosciences; CD3PerCP (BW264/56) and CD4-PEvio770 (M-T466) from Miltenyi Biotec; CD69-BV421 FN50 and PDL1-PE (29E.2A3) from Biolegend; mouse CD3-FITC, CD4-Percp, and CD8-PE (eBioscience); mouse intracellular IFN-γ-PE-Cy7 (Biolegend).
Direct reovirus stimulation of primary human cells and cell lines
GBM1, GBM4, MCF-7, U87, SW620, and Mel624 cells were cultured overnight at 1x105 cells/well. Reovirus (0, 1, or 10 PFU/cell) was then added for 24 hrs before cells were harvested and stained with anti-human PD-L1-PE (clone MIH1; eBioscience) as above.
Treatment of GBM1 using exogenous, purified IFNs
GBM1 cells were cultured overnight at 1x105 cells/well. Cells were then treated with 100 pg of recombinant human IFN-α (R&D Systems), IFN-β (PBL Interferon Source), or IFN-γ (Peprotech) alone or in combination. After 24 hrs, cells were harvested and stained with anti-PD-L1-PE as above.
Treatment of GBM1 using HGG-derived CM / RCM
GBM1 cells were cultured as above, then treated with 500 µl of a 1:16 dilution of CM or RCM for 24 hr before being stained for PD-L1 expression as above.
Treatment of GBM1 cells using PBMC-derived CM/RCM and IFN blockade
GBM1 cells were cultured at 5x104 cells/well. After 24 hrs, thawed CM and RCM were incubated for 1 hr at 4 °C with 0.75 % anti-human-IFNα, -IFNβ, or -IFNγ blocking antibody (alone or in combination; all Pestka Biomedical Laboratories, PBL) or isotype control (rabbit serum (IFNγ) or sheep serum (IFNα and IFNβ) alone or in combination; both Sigma). Simultaneously, when GBM1 cells were to be treated with anti-human-IFNα/IFNβ blocking antibodies, 1.25 % anti-human IFNα/β receptor chain 2 antibody (PBL), or IgG2a isotype control (R&D Systems) were added to cells for 1 hr at 37 °C. Prepared CM/RCM (including block/isotype) was then added for 4 hrs at 37 °C before medium was removed and replaced with full growth medium. After 24 hrs, cells were harvested and stained for PD-L1 expression as described above.
Determination of cytokine and chemokine concentrations
BioRad Bio-Plex Pro Cytokine and Chemokine Assays (21-plex, human group I and 27-plex, human group II) were used to determine concentrations of soluble mediators in plasma, as per manufacturer’s instructions.
Cytokine secretion from fresh ex-vivo HGG single-cell suspensions
IFN-α (Mabtech) and IFN-γ (BD Biosciences) were detected by ELISA using matched antibody pairs, whereas IFN-β was detected using a Verikine Human IFN Beta ELISA Kit (PBL) as previously described (30).
Cytokine secretion from patient serum
IFN-α was detected using a Verikine Human IFN-α Multi Sub-type Serum ELISA Kit, as per manufacturer’s instructions.
Transmission electron microscopy
This method was performed as previously described (21).
Statistical analysis
All statistical analysis was conducted using GraphPad Prism software. Asterisks in Fig. 5C, fig. S2B, fig. S8, and fig. S9 represent P < 0.05 using a t-test. The statistical correlation of reovirus RNA / protein with Ki67 (Fig. 2B) and with time between infusion and surgery (fig. S6) was determined by linear regression.
Supplementary Material
RNAseq expression data (provided as an Excel file)
One Sentence Summary.
Intravenous infusion of oncolytic reovirus in patients leads to infection of brain tumors, infiltration by cytotoxic T cells, and up-regulation of PD-L1.
Acknowledgments
We are grateful to all the patients that participated in this trial.
Funding: AS was the recipient of a Cancer Research UK Clinical Research Fellowship. Clinical and translational costs were kindly donated by Brain Tumour Research and Support Across Yorkshire. We are also grateful for support from The Brain Tumour Charity, Yorkshire Cancer Research, the Cancer Research UK Leeds Centre, the Leeds Experimental Cancer Medicine Centre, and the NIHR Leeds Clinical Research Facility. KJH acknowledges support from the Institute of Cancer Research/Royal Marsden NIHR Biomedical Research Facility.
Footnotes
Author Contributions: Study design: AS, AM, SCS, MC. Enrolment and management of Patients: AS, ST, RC, RM, JC, PVH, GS, DB, FC, CR, AA, CT. Laboratory work: AS, KS, DT, EW, EJW, GN, MF, TK, JT, EI, EP, ST, EA, GM, AR, MC, AF, SQ, HW, FEM, LS. Data analysis and study write up: AS, AM, KS, DT, LS, EW, GN, KH, HP, PS, SG, SCS, RV.
Competing interests: MC is an employee of Oncolytics Biotech Inc., Calgary, Canada, from which SG, SCS, AM, KH and HP have received research grants. MC and colleagues are inventors on several patents (including patents 6110461, 6136307, 6261555, 6344195, 6455038) held by Oncolytics Biotech Inc. that cover reovirus treatment of neoplasia. All other authors declare that they have no competing interests.
Materials and data availability: Correspondence and requests for materials should be addressed to AS/AAM/SCS. Oncolytic reovirus (REOLYSIN) was supplied by Oncolytics Biotech Inc. under a material transfer agreement with the University of Leeds, UK.
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior written permission of AAAS.
References and Notes
- 1.Robert C, Long GV, Brady B, Dutriaux C, Maio M, Mortier L, Hassel JC, Rutkowski P, McNeil C, Kalinka-Warzocha E, Savage KJ, et al. Nivolumab in Previously Untreated Melanoma without BRAF Mutation. N Engl J Med. 2014 doi: 10.1056/NEJMoa1412082. 141116004513004. [DOI] [PubMed] [Google Scholar]
- 2.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, Gonzalez R, Robert C, Schadendorf D, Hassel JC, Akerley W, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. 2010 doi: 10.1056/NEJMoa1003466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brahmer J, Reckamp KL, Baas P, Crinò L, Eberhardt WEE, Poddubskaya E, Antonia S, Pluzanski A, Vokes EE, Holgado E, Waterhouse D, et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N Engl J Med. 2015 doi: 10.1056/NEJMoa1504627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, Tykodi SS, Sosman JA, Procopio G, Plimack ER, Castellano D, et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N Engl J Med. 2015 150925150201006. [Google Scholar]
- 5.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, Chow LQ, Vokes EE, Felip E, Holgado E, Barlesi F, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non–Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627–1639. doi: 10.1056/NEJMoa1507643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJM, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature. 2014;515:568–571. doi: 10.1038/nature13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melcher A, Parato K, Rooney CM, Bell JC. Thunder and lightning: immunotherapy and oncolytic viruses collide. Mol Ther. 2011;19:1008–16. doi: 10.1038/mt.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaufman HL, Kohlhapp FJ, Zloza A. Oncolytic viruses: a new class of immunotherapy drugs. Nat Rev Drug Discov. 2015;14:642–662. doi: 10.1038/nrd4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andtbacka RHI, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS, Milhem M, et al. Talimogene Laherparepvec Improves Durable Response Rate in Patients With Advanced Melanoma. J Clin Oncol. 2015 doi: 10.1200/JCO.2014.58.3377. JCO.2014.58.3377- [DOI] [PubMed] [Google Scholar]
- 10.Steele L, Errington F, Prestwich R, Ilett E, Harrington K, Pandha H, Coffey M, Selby P, Vile R, Melcher A. Pro-inflammatory cytokine/chemokine production by reovirus treated melanoma cells is PKR/NF-κB mediated and supports innate and adaptive anti-tumour immune priming. Mol Cancer. 2011;10:20. doi: 10.1186/1476-4598-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benencia F, Courrèges MC, Conejo-García JR, Mohamed-Hadley A, Zhang L, Buckanovich RJ, Carroll R, Fraser N, Coukos G. HSV oncolytic therapy upregulates interferon-inducible chemokines and recruits immune effector cells in ovarian cancer. Mol Ther. 2005;12:789–802. doi: 10.1016/j.ymthe.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 12.Rampling R, Cruickshank G, Papanastassiou V, Nicoll J, Hadley D, Brennan D, Petty R, MacLean A, Harland J, McKie E, Mabbs R, et al. Toxicity evaluation of replication-competent herpes simplex virus (ICP 34.5 null mutant 1716) in patients with recurrent malignant glioma. Gene Ther. 2000;7:859–66. doi: 10.1038/sj.gt.3301184. [DOI] [PubMed] [Google Scholar]
- 13.Papanastassiou V, Rampling R, Fraser M, Petty R, Hadley D, Nicoll J, Harland J, Mabbs R, Brown M. The potential for efficacy of the modified (ICP 34.5(-)) herpes simplex virus HSV1716 following intratumoural injection into human malignant glioma: a proof of principle study. Gene Ther. 2002;9:398–406. doi: 10.1038/sj.gt.3301664. [DOI] [PubMed] [Google Scholar]
- 14.Harrow S, Papanastassiou V, Harland J, Mabbs R, Petty R, Fraser M, Hadley D, Patterson J, Brown SM, Rampling R. HSV1716 injection into the brain adjacent to tumour following surgical resection of high-grade glioma: safety data and long-term survival. Gene Ther. 2004;11:1648–58. doi: 10.1038/sj.gt.3302289. [DOI] [PubMed] [Google Scholar]
- 15.Markert JM, Medlock MD, Rabkin SD, Gillespie GY, Todo T, Hunter WD, Palmer CA, Feigenbaum F, Tornatore C, Tufaro F, Martuza RL. Conditionally replicating herpes simplex virus mutant, G207 for the treatment of malignant glioma: results of a phase I trial. Gene Ther. 2000;7:867–74. doi: 10.1038/sj.gt.3301205. [DOI] [PubMed] [Google Scholar]
- 16.Chiocca EA, Abbed KM, Tatter S, Louis DN, Hochberg FH, Barker F, Kracher J, Grossman SA, Fisher JD, Carson K, Rosenblum M, et al. A phase I open-label, dose-escalation, multi-institutional trial of injection with an E1B-Attenuated adenovirus, ONYX-015, into the peritumoral region of recurrent malignant gliomas, in the adjuvant setting. Mol Ther. 2004;10:958–66. doi: 10.1016/j.ymthe.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 17.Forsyth P, Roldán G, George D, Wallace C, Palmer CA, Morris D, Cairncross G, Matthews MV, Markert J, Gillespie Y, Coffey M, et al. A phase I trial of intratumoral administration of reovirus in patients with histologically confirmed recurrent malignant gliomas. Mol Ther. 2008;16:627–32. doi: 10.1038/sj.mt.6300403. [DOI] [PubMed] [Google Scholar]
- 18.Kicielinski KP, Chiocca EA, Yu JS, Gill GM, Coffey M, Markert JM. Phase 1 clinical trial of intratumoral reovirus infusion for the treatment of recurrent malignant gliomas in adults. Mol Ther. 2014;22:1056–62. doi: 10.1038/mt.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stupp R, Hegi ME, Mason WP, van den Bent MJ, Taphoorn MJB, Janzer RC, Ludwin SK, Allgeier A, Fisher B, Belanger K, Hau P, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10:459–66. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 20.Mintz A, Perry J, Spithoff K, Chambers A, Laperriere N. Management of single brain metastasis: a practice guideline. Curr Oncol. 2007;14:131–43. doi: 10.3747/co.2007.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Adair RA, Roulstone V, Scott KJ, Morgan R, Nuovo GJ, Fuller M, Beirne D, West EJ, Jennings VA, Rose A, Kyula J, et al. Cell carriage, delivery, and selective replication of an oncolytic virus in tumor in patients. Sci Transl Med. 2012;4:138ra77. doi: 10.1126/scitranslmed.3003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ilett E, Kottke T, Donnelly O, Thompson J, Willmon C, Diaz R, Zaidi S, Coffey M, Selby P, Harrington K, Pandha H, et al. Cytokine conditioning enhances systemic delivery and therapy of an oncolytic virus. Mol Ther. 2014;22:1851–63. doi: 10.1038/mt.2014.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taterka J, Sutcliffe M, Rubin DH. Selective reovirus infection of murine hepatocarcinoma cells during cell division. A model of viral liver infection. J Clin Invest. 1994;94:353–60. doi: 10.1172/JCI117329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parato KA, Breitbach CJ, Le Boeuf F, Wang J, Storbeck C, Ilkow C, Diallo J-S, Falls T, Burns J, Garcia V, Kanji F, et al. ki67. Mol Ther. 2012;20:749–58. doi: 10.1038/mt.2011.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun N, Papadopoulos T, Müller-Hermelink HK. Cell cycle dependent distribution of the proliferation-associated Ki-67 antigen in human embryonic lung cells. Virchows Arch B Cell Pathol Incl Mol Pathol. 1988;56:25–33. doi: 10.1007/BF02889998. [DOI] [PubMed] [Google Scholar]
- 26.Parker JSL, Broering TJ, Kim J, Higgins DE, Nibert ML. Reovirus core protein mu2 determines the filamentous morphology of viral inclusion bodies by interacting with and stabilizing microtubules. J Virol. 2002;76:4483–96. doi: 10.1128/JVI.76.9.4483-4496.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castellino F, Huang AY, Altan-Bonnet G, Stoll S, Scheinecker C, Germain RN. Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell–dendritic cell interaction. Nature. 2006;440:890–895. doi: 10.1038/nature04651. [DOI] [PubMed] [Google Scholar]
- 28.Long EO. ICAM-1: getting a grip on leukocyte adhesion. J Immunol. 2011;186:5021–3. doi: 10.4049/jimmunol.1100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rajani K, Parrish C, Kottke T, Thompson J, Zaidi S, Ilett L, Shim KG, Diaz R-M, Pandha H, Harrington K, Coffey M, et al. Combination Therapy With Reovirus and Anti-PD-1 Blockade Controls Tumor Growth Through Innate and Adaptive Immune Responses. Mol Ther. 2016;24:166–74. doi: 10.1038/mt.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Errington F. Inflammatory tumour cell killing by oncolytic reovirus for the treatment of melanoma. Gene Ther. 2008;15:1257–1270. doi: 10.1038/gt.2008.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rusinova I, Forster S, Yu S, Kannan A, Masse M, Cumming H, Chapman R, Hertzog P. INTERFEROME v2. 0: an updated database of annotated interferon-regulated genes. Nucleic Acids Res. 41:D1040–D1046. doi: 10.1093/nar/gks1215. (databa, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wurdak H, Zhu S, Romero A, Lorger M, Watson J, Chiang C-Y, Zhang J, Natu VS, Lairson LL, Walker JR, Trussell CM, et al. An RNAi screen identifies TRRAP as a regulator of brain tumor-initiating cell differentiation. Cell Stem Cell. 2010;6:37–47. doi: 10.1016/j.stem.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 33.Geletneky K, Huesing J, Rommelaere J, Schlehofer JR, Leuchs B, Dahm M, Krebs O, von Knebel Doeberitz M, Huber B, Hajda J. Phase I/IIa study of intratumoral/intracerebral or intravenous/intracerebral administration of Parvovirus H-1 (ParvOryx) in patients with progressive primary or recurrent glioblastoma multiforme: ParvOryx01 protocol. BMC Cancer. 2012;12:99. doi: 10.1186/1471-2407-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Connolly JL, Dermody TS. Virion disassembly is required for apoptosis induced by reovirus. J Virol. 2002;76:1632–41. doi: 10.1128/JVI.76.4.1632-1641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker BS, Rautela J, Hertzog PJ. Antitumour actions of interferons: implications for cancer therapy. Nat Rev Cancer. 2016;16:131–144. doi: 10.1038/nrc.2016.14. [DOI] [PubMed] [Google Scholar]
- 36.Papadopoulos MC, Saadoun S, Binder DK, Manley GT, Krishna S, Verkman AS. Molecular mechanisms of brain tumor edema. Neuroscience. 2004;129:1011–20. doi: 10.1016/j.neuroscience.2004.05.044. [DOI] [PubMed] [Google Scholar]
- 37.Yau T, Swanton C, Chua S, Sue A, Walsh G, Rostom A, Johnston SR, O’Brien MER, Smith IE. Incidence, pattern and timing of brain metastases among patients with advanced breast cancer treated with trastuzumab. Acta Oncol. 2006;45:196–201. doi: 10.1080/02841860500486630. [DOI] [PubMed] [Google Scholar]
- 38.Lampson LA. Monoclonal antibodies in neuro-oncology: Getting past the blood-brain barrier. MAbs. 3:153–60. doi: 10.4161/mabs.3.2.14239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dolin R, Reichman RC, Fauci AS. Lymphocyte Populations in Acute Viral Gastroenteritis. Infect Immun. 1976;14:422–428. doi: 10.1128/iai.14.2.422-428.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michlmayr D, McKimmie CS, Pingen M, Haxton B, Mansfield K, Johnson N, Fooks AR, Graham GJ. Defining the Chemokine Basis for Leukocyte Recruitment during Viral Encephalitis. J Virol. 2014;88:9553–9567. doi: 10.1128/JVI.03421-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu L, LaRosa G, Kassam N, Gordon CJ, Heath H, Ruffing N, Chen H, Humblias J, Samson M, Parmentier M, Moore JP, et al. Interaction of chemokine receptor CCR5 with its ligands: multiple domains for HIV-1 gp120 binding and a single domain for chemokine binding. J Exp Med. 1997;186:1373–81. doi: 10.1084/jem.186.8.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gooden MJM, de Bock GH, Leffers N, Daemen T, Nijman HW. The prognostic influence of tumour-infiltrating lymphocytes in cancer: a systematic review with meta-analysis. Br J Cancer. 2011;105:93–103. doi: 10.1038/bjc.2011.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lapa C, Linsenmann T, Lückerath K, Samnick S, Herrmann K, Stoffer C, Ernestus R-I, Buck AK, Löhr M, Monoranu C-M. Tumor-associated macrophages in glioblastoma multiforme-a suitable target for somatostatin receptor-based imaging and therapy? PLoS One. 2015;10:e0122269. doi: 10.1371/journal.pone.0122269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adair RA, Scott KJ, Fraser S, Errington-Mais F, Pandha H, Coffey M, Selby P, Cook GP, Vile R, Harrington KJ, Toogood G, et al. Cytotoxic and immune-mediated killing of human colorectal cancer by reovirus-loaded blood and liver mononuclear cells. Int J Cancer. 2013;132:2327–38. doi: 10.1002/ijc.27918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kasprowicz V, Schulze Zur Wiesch J, Kuntzen T, Nolan BE, Longworth S, Berical A, Blum J, McMahon C, Reyor LL, Elias N, Kwok WW, et al. High level of PD-1 expression on hepatitis C virus (HCV)-specific CD8+ and CD4+ T cells during acute HCV infection, irrespective of clinical outcome. J Virol. 2008;82:3154–60. doi: 10.1128/JVI.02474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brown KE, Freeman GJ, Wherry EJ, Sharpe AH. Role of PD-1 in regulating acute infections. Curr Opin Immunol. 2010;22:397–401. doi: 10.1016/j.coi.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diggs LP, Hsueh EC. Utility of PD-L1 immunohistochemistry assays for predicting PD-1/PD-L1 inhibitor response. Biomark Res. 2017;5:12. doi: 10.1186/s40364-017-0093-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nduom EK, Wei J, Yaghi NK, Huang N, Kong L-Y, Gabrusiewicz K, Ling X, Zhou S, Ivan C, Chen JQ, Burks JK, et al. PD-L1 expression and prognostic impact in glioblastoma. Neuro Oncol. 2016;18:195–205. doi: 10.1093/neuonc/nov172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ribas A. Oncolytic Virotherapy Promotes Intratumoral T Cell Infiltration and Improves Anti-PD-1 Immunotherapy. Cell. 2017 doi: 10.1016/j.cell.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bristol Myers Squibb. Bristol-Myers Squibb Announces Results from CheckMate -143, a Phase 3 Study of Opdivo (nivolumab) in Patients with Glioblastoma Multiforme. 2017 https://news.bms.com/press-release/bmy/bristol-myers-squibb-announces-results-checkmate-143-phase-3-study-opdivo-nivoluma.
- 51.Nuovo GJ, Garofalo M, Valeri N, Roulstone V, Volinia S, Cohn DE, Phelps M, Harrington KJ, Vile R, Melcher A, Galanis E, et al. Reovirus-associated reduction of microRNA-let-7d is related to the increased apoptotic death of cancer cells in clinical samples. Mod Pathol. 2012;25:1333–44. doi: 10.1038/modpathol.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vidal L, Pandha HS, Yap TA, White CL, Twigger K, Vile RG, Melcher A, Coffey M, Harrington KJ, DeBono JS. A phase I study of intravenous oncolytic reovirus type 3 Dearing in patients with advanced cancer. Clin Cancer Res. 2008;14:7127–37. doi: 10.1158/1078-0432.CCR-08-0524. [DOI] [PubMed] [Google Scholar]
- 53.Conway C, Graham JL, Chengot P, Daly C, Chalkley R, Ross L, Droop A, Rabbitts P, Stead LF. Elucidating drivers of oral epithelial dysplasia formation and malignant transformation to cancer using RNAseq. Oncotarget. 2015;6:40186–201. doi: 10.18632/oncotarget.5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012;7:562–78. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Zhang B. WEB-based GEne SeT AnaLysis Toolkit. 2016 doi: 10.1093/nar/gkt439. (available at http://bioinfo.vanderbilt.edu/webgestalt/option.php%3E) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RNAseq expression data (provided as an Excel file)