Abstract
Enteroendocrine cells (EECs) control a wide range of physiological processes linked to metabolism1. We show that EECs hormones are differentially expressed between crypts (e.g. Glp1) and villi (e.g. Secretin). As demonstrated by single cell mRNA sequencing, BMP4 signals can reprogram the expressed repertoire of hormones of individual EECs in murine Lgr5+ intestinal stem cell-derived organoids. Accordingly, BMP4 induces hormone switching of EECs migrating up the crypt-villus axis in vivo. This implies the existence of fewer EEC lineages in the small intestine than proposed previously. We also describe a protocol to generate human EECs in organoids and demonstrate that BMP signaling regulates hormone expression in a similar fashion. These observations suggest strategies to pharmacologically manipulate numbers of EECs expressing therapeutically relevant hormones, such as Glp1.
The EECs of the intestine together constitute the largest hormone-producing organ in mammals. EECs are classified according to their hormone products1. Enterochromaffin cells (ECs) produce Serotonin, a regulator of intestinal motility, and Tachykinin 1 (Tac1), also known as Substance P, a peptide which is believed to play a role in muscle contraction and inflammation2. L-cells produce Glp1, an inducer of insulin release encoded by the Glucagon (Gcg) gene, and can co-express Pyy1. Other EEC subtypes include Gastric inhibitory protein (Gip)-producing K-cells, Somatostatin (Sst)-producing D-cells, Cholestocystokinin (Cck)-producing I-cells, Neurotensin (Nts)-producing N-cells and Secretin (Sct)-producing S-cells1. Although this classification suggests well-defined, distinct subtypes of EECs, these hormones are often co-expressed suggesting considerable overlap between lineages3,4. Like all other cell types of the intestinal epithelium, the short-lived EECs are constitutively produced by Lgr5+ crypt stem cells5. Lgr5 stem cells can be cultured to generate epithelial organoids that faithfully recapitulate gut epithelial biology6. Single cell sequencing has shown that a complete set of EECs subtypes is produced in these mini-guts, including some that had previously gone unnoticed in intact gut7,8. However, extrinsic factors that control EEC subtype identity have remained largely unknown.
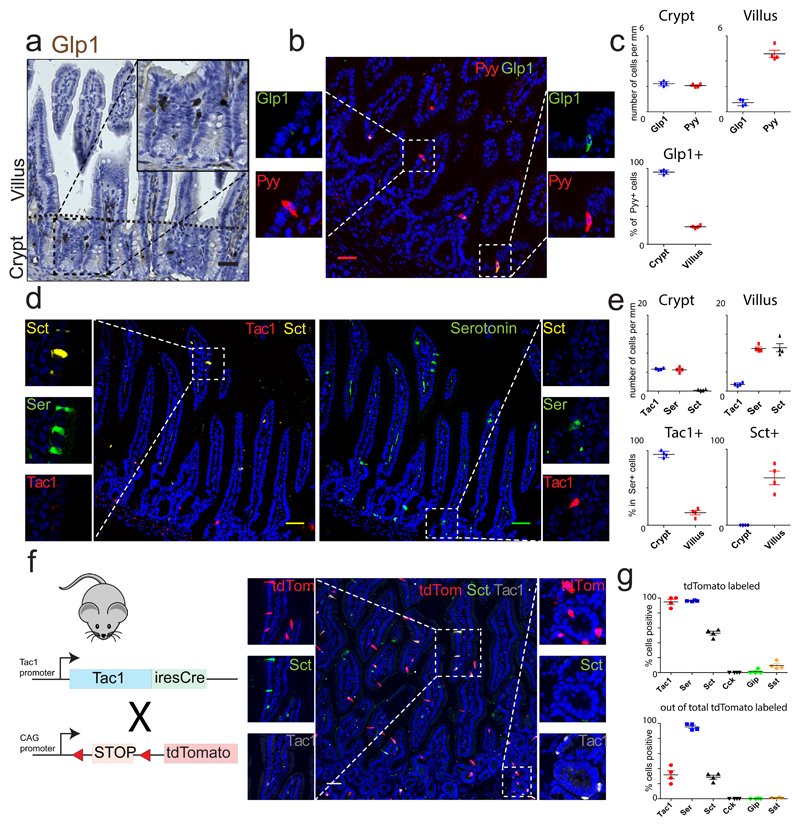
Previous work has suggested that EECs expressing Tac1 and Glp1 are restricted to crypts, while Sct-, Pyy- and Nts-producing EECs are enriched in villi9,10. In agreement with these studies, we found that ileal L-cells co-express Glp1 and Pyy in the crypt, but mostly lack Glp1 in the villus (Fig. 1a-c, Fig. S1a). Serotonin-producing ECs occur along the length of the crypt-villus axis, but selectively co-express Tac1 in the crypt and Sct in the villus (Fig. 1d-e, Fig. S1a). To address if hormone switching occurs during migration of EECs along the crypt-villus axis, we analyzed intestines from Tac1iresCre /RosatdTomato mice, an allele which faithfully labels all Tac1+/Serotonin+ cells in the crypt (Fig. 1f)11. In adult intestines, almost all Serotonin+ cells were marked by tdTomato (Fig.1g). Importantly, >55% of Sct+ cells on villi were also traced, while being negative for Tac1 (Fig. 1f-g, S1b). The rarity of tdTomato+ cells that were negative for Serotonin suggests that ECs do not lose Serotonin during their lifetime to become single Sct+ as previously suggested9,12. Sct+ cells that are not traced by Tac1 and are Serotonin-negative must thus be part of another EEC lineage. EECs producing other hormones, including Cck, Gip and Sst, were only very rarely derived from Tac1+ progenitors (Fig. 1g, S1b). These tracing data implied lineage relationships between crypt and villus EECs, i.e that Tac1-/Sct+ ECs on villi derive from Tac1+ crypt ECs. This in turn suggested that local niche signals could induce shifts in hormone expression.
Fig. 1. Enteroendocrine cells switch hormone expression while migrating from crypt to villus.
a and b, Immunohistochemical analysis reveals Glp1+ cells are enriched in the ileal crypt, where they co-express Pyy. Experiment in (a) was repeated two times independently with similar results. c, Quantification of b. The percentage of Pyy+ cells that co-express Glp1 are represented in the lower chart. d, ECs express Tac1 in the crypt and Sct in the villus, while Serotonin is produced in both locations. e, Quantification of d. The percentage of Serotonin+ cells that co-express Tac1 or Sct are represented in the lower charts. f, Intestine of Tac1iresCre /RosaAi14 mice reveals that ECs lose Tac1 and gain Sct expression from crypt to villus. g, Quantification of (f) and Fig S1. The percentages of each hormone that is tdTomato+ (upper chart) and of tdTomato+ cells that are hormone positive (lower chart) are shown. The mean values are depicted in graphs c, e and g, and error bars present SD for n=4 mice for each experiment. Scale bar is 50 µm.
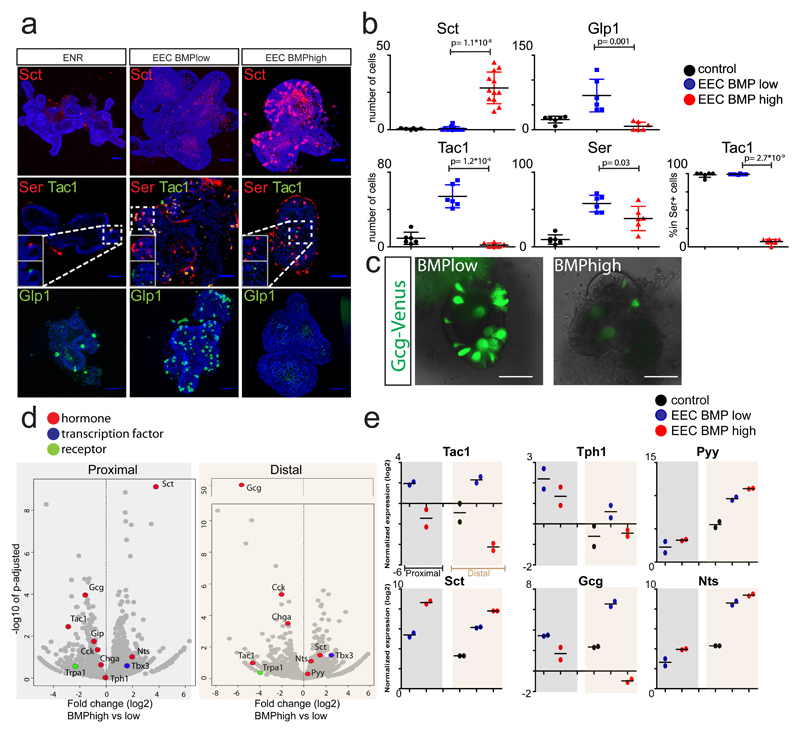
Multiple signaling gradients exist along the crypt-villus axis, e.g. Wnt levels are high at the crypt base, while BMP is highest at the villus tips13,14. Induction of EECs in murine intestinal organoids can be achieved through inhibition of the Wnt, MAPK and Notch signaling pathways, in the presence of the BMP inhibitor Noggin8. We used this differentiation system as a starting point to modulate selected signaling pathways8, while monitoring Sct and Gcg as a proxy of the villus- and crypt-hormone signatures respectively. Strikingly, we observed that all ECs in this culture co-expressed Serotonin and Tac1, while Sct was absent (Fig. 2a). This suggests niche signals acting on EECs are dominant over a default, temporal differentiation process. Manipulation of the Wnt, TGFβ and Hedgehog pathways did either reduce both Gcg and Sct transcripts, or had no significant on any assessed hormone (Fig. S2a). We then replaced Noggin by BMP4 in this EEC differentiation cocktail ('EEC BMPhigh' medium), generating cells immunoreactive for Sct, as well as ECs lacking Tac1 (Fig. 2a-b). Glp1+ cell numbers and total levels of secreted Glp1 were greatly diminished (Fig. 2a-c, S2b). We next performed bulk RNA sequencing on duodenal and ileal organoids stimulated with EEC BMPhigh or EEC BMPlow media (Fig. 2d), and validated the expression of selected genes by qPCR (Fig. 2e). EEC markers which are homogenously distributed on crypts and villi (Chga, Tph1 (enzyme catalyzing Serotonin production), Cck and duodenal Gip) are only mildly affected by BMP activation (Fig. 2d-e). We did observe an increase in Sct and a minor upregulation of Pyy and Nts (Fig. 2d-e), which are expressed highest in the villus10. Sct is reported to be enriched in the proximal part of the SI1, but we observe that our EEC differentiation protocol generates Sct+ cells equally well in the proximal and distal SI organoids. Trpa1, an irritant receptor enriched in EECs of the intestinal crypt15, decreased with BMP activation (Fig. 2d). BMP4-mediated changes in hormone expression could be overridden by the addition of BMPR1a inhibitor LDN193189, confirming involvement of the BMPR1/2 axis (Fig. S2c-d).
Fig.2. Activation of BMP signaling induces Villus-like hormone signature in mouse EECs.
a, Organoids are differentiated for 4 days to EECs in the absence (EEC BMPlow) or presence of BMP4 (EEC BMPhigh). ENR is used as a control. Activation of BMP signaling induces expression of Sct, while repressing Tac1 and Glp1. Images are presented as maximum projections. b, Quantification of a. The number of positive cells for each hormone were quantified and are displayed per mm of organoid epithelium. The percentage of Serotonin+ cells that co-express Tac1 are presented. Sample size represents n=2 biologically independent experiments, in which at least 3 organoids were quantified per replicate and staining. Statistics were derived comparing all organoids (n=12 for Sct, n=6 for other hormones) in BMP low and high conditions using a two-sided t-test. Mean values per treatment are shown, and error bars present SD. c, Overlay of brightfield and Venus image of organoids derived from GcgVenus mice after a 4 day treatment with a BMPlow or BMPhigh EEC differentiation cocktail. BMP activation represses expression of Gcg, without inducing morphological alterations. Experiment was repeated independently 10 times. d, Volcano plots showing results from RNA sequencing of organoids stimulated for 4 days with BMP low or high EEC differentiation cocktails, from proximal (left) and distal (right) SI organoids. Gene expression fold change (log2) of BMPhigh versus BMPlow is shown on the x-axis and significance on the y-axis. Each grey dot represents a gene, and dots representing relevant genes are highlighted in different colors, according to their function. Sample size represents n=2 biologically independent experiments, and p-adjusted values were calculated with a Wald test using the DESeq2 package. e, qPCR analysis of selected hormones from d. Expression levels are shown relative to control organoids in ENR medium. Experiment was performed in n=2 biologically independent experiments, and the mean expression is depicted. Scale bar is 50 µm.
To address whether BMP signaling can switch hormone expression in individual mature EECs rather than selectively depleting subtypes of EECs, we followed the fate of Gcg- or Tac1-expressing cells using cultures derived from GcgVenus and Tac1iresCre /Rosa26tdTomato mice11,16. Live-cell imaging of GcgVenus organoids demonstrated that BMP activation induced a decrease in Venus levels, suggesting downregulation of Gcg (Fig. S2e-f). We did not observe BMP-induced apoptosis of Venus+ or tdTomato+ cells (Fig. S2g).
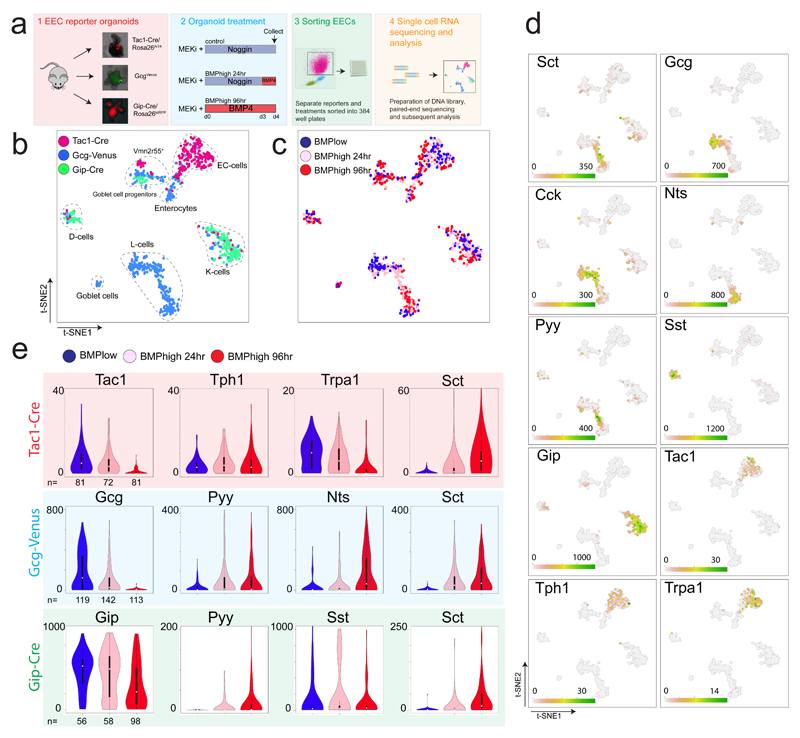
Changes in hormone expression in individual EECs might be caused by dynamics in transcriptional networks, and accompanied by production of other sensory receptors. To identify dynamics at a single cell level, we performed single cell RNA sequencing of traced Tac1-, Glp1- or Gip-expressing murine cells16. Gip+ K-cells exist both in crypts and villi and were isolated from organoids derived from a GipCre /Rosa26tdRFP mouse17. Guided by their regional in vivo abundance, we isolated organoids from the proximal SI of the Gip-, from the distal SI of Gcg- and from the whole SI of Tac1 reporter mice. Organoids were treated with a MEK inhibitor to limit new EEC generation and either exposed to Noggin (control) or BMP4 for 24 or 96 hours (Fig. 3a). Next, EECs derived from the reporters/treatments were sorted for the reporter fluorescence and single cell RNA sequencing was performed using SORTseq18, an automated version of CELseq2 (Fig. 3a, S3a)19. K-medoids clustering by RaceID2 algorithm20 showed that Tac1-, Gcg- and Gip-traced cells, classically defined as ECs, L- and K- cells respectively, clustered accordingly to their cell-type and mostly independently of the treatment in a t-SNE space (Fig. 3b-c). We identified Alpi+ enterocytes and Muc2+ Goblet cells derived from the GcgVenus reporter that displayed the lowest Venus fluorescence intensity (Fig. S3b). We also detected a cluster of unknown identity with expression of the vomeronasal receptor Vmn2r55, which was identified previously7. BMP-stimulated ECs displayed lower Sct expression compared to L- and K-cells (Fig. 3c-e). Within ECs, expression of Tph1 remained unchanged during BMP treatment, while Tac1 decreased (Fig. 3c-e). Cells clustering as classical L-cells dramatically reduced their Gcg (Glp1) expression, while activating Nts and Pyy transcription following BMP treatment (Fig. 3d and S3c). Some GcgVenus-sorted cells from ileal organoids expressed Gip, combined with low levels of Gcg (Fig. 3b-d). These cells clustered together with K-cells and could not be induced to express Nts, indicating that these cells represent K-cells and not L-cells. (Fig. 3d). L-cells and to a lesser extent K-cells express Cck independent of treatment (Fig. 3d). Within Gip-traced K-cells, we observed a separate population of Sst-producing cells. BMP activation had no effect on Sst expression and caused a mild reduction in Gip expression in these clusters, while activating Sct expression only in Gip+ but not in Sst+ cells (Fig. 3d-e, S3c). Pyy expression could be induced in both Gip+ and in Sst+ cells, but to a lesser extent than in L- cells (Fig. S3c). We did observe low Cck, Gcg and no Nts expression in K-cells, indicating that these represent a separate lineage from ileal L-cells (Fig. 3d-e).
Fig.3. Single cell RNA sequencing reveals BMP regulated plasticity among different EEC subtypes.
a, Experimental paradigm. Different EEC reporter organoids were treated with a MEK inhibitor, while receiving Noggin or BMP4. After 4 days, organoids were dissociated and traced EECs sorted and processed for single cell RNA sequencing. b and c, t-SNE map of single cell RNA sequenced EECs using the RaceID2 algorithm. Different colors, as indicated in the legend, highlight cells isolated from different reporter organoids (b) and treatments (c). d, Expression levels of selected hormones and receptors in the tSNE space of (b-c). e, Expression of individual hormones within different EEC reporter sorted cells are presented in violin plots, with different colors for the different treatments (as indicated in the legend). Violin plots depict median values (white dot), 50% of the values (within thick black line) and 95% of the values (within thin black line). The number of cells per treatment and reporter is depicted. Different dynamics of hormone expression were observed over the course of BMP treatment in subtypes of EECs.
We identified uniquely expressed genes in the various clusters, that corresponded to known expression or function along the crypt-to-villus axis in vivo. The irritant receptor Trpa1 (proposed to be involved in serotonin release) has been shown to be enriched in the crypt 15,21. Concordantly, it decreases during BMP treatment within ECs (Fig. 3d-e). We find the orphan receptor Asic5 to be expressed by Sst+ cells (Fig. S3d). This same population also expresses the islet amyloid polypeptide (Iapp) Amylin (Fig. S3d), a peptide previously found in pancreatic β-cells and known to have a wide range of metabolic effects22. The LIM homeobox factor Lmx1a occurs in ECs, as suggested recently23. The homeobox protein Hhex -not previously observed in the gut- was expressed by Sst-producing cells (Fig. S3d). Interestingly, it has been described as an essential factor for Sst-producing δ cells in the pancreas24. The T-box transcription factor Tbx3 is produced in BMP-activated EECs (Fig. S3d), and is enriched in the villus25. Finally, we find specific activation of classical BMP target genes in BMP-treated cells, such as Id1, Id2 and Id3, confirming pathway activation (Fig. S3d)26.
Transcript dynamics might not be fully predictive for changes at the peptide hormone level. Therefore, we repeated the same experimental strategy as for the single cell RNA sequencing and assessed co-expression of relevant peptide hormones in the Tac1iresCre /Rosa26tdTomato- and GcgVenus- reporter organoids. Over the course of 4 days, BMP-inhibited tdTomato+ cells remained immunoreactive for Tac1 and Serotonin, while only rarely expressing Sct (Fig S4a). Strikingly, BMP-activated tdTomato+ cells lost Tac1 immunoreactivity, maintained Serotonin and gained Sct positivity (Fig S4a). Glp1 positivity was strongly correlated with Venus expression in GcgVenus organoids in BMP-untreated conditions, while this correlation is lost in BMP-treated samples (Fig. S4a). Conversely, Venus positivity was increasingly predictive for Sct expression after BMP treatment (Fig. S4a). Pyy peptide positivity remained unchanged irrespective of BMP treatment (Fig S4a), in line with the constant peptide levels between crypt and villus in vivo10.
We performed live cell imaging of the Tac1iresCre /Rosa26tdTomato- and GcgVenus- reporter organoids in BMP-untreated and -treated conditions. tdTomato+ cells that existed at the beginning of the BMP treatment continued to persist over the course of 60 hours, while losing Tac1 and gaining Sct expression (Fig. S4b). Untreated cells retained Tac1 positivity (Fig. S4b). In GcgVenus- reporter organoids, we observed a similar increase in Sct peptides (Fig. S4b). Collectively, these data indicated that individual EECs can rewire their peptide hormone profile upon activation of BMP signaling.
The number of tdTomato+ and Venus+ EECs increased significantly more over 4 days when BMP signaling is inhibited compared to when activated (Fig. S4a). We found that the first transcription factor expressed by and defining the EEC lineage, Neurogenin 327, is inhibited by BMP activation (Fig. S4c). To circumvent a bias that occurs at the bulk population level due to this inhibition of EEC specification, we first generated a large pool of ileal EECs using our differentiation protocol for 3 days. Next, we switched to BMP-high conditions for 24 hours (Fig. S4d). Increases in Sct, Pyy and Nts expression were more pronounced compared to a continuous BMP inhibition (Fig. 2 and Fig. S4c). These data imply that initial EEC specification requires BMP low conditions, such as exists at the bottom of the crypt.
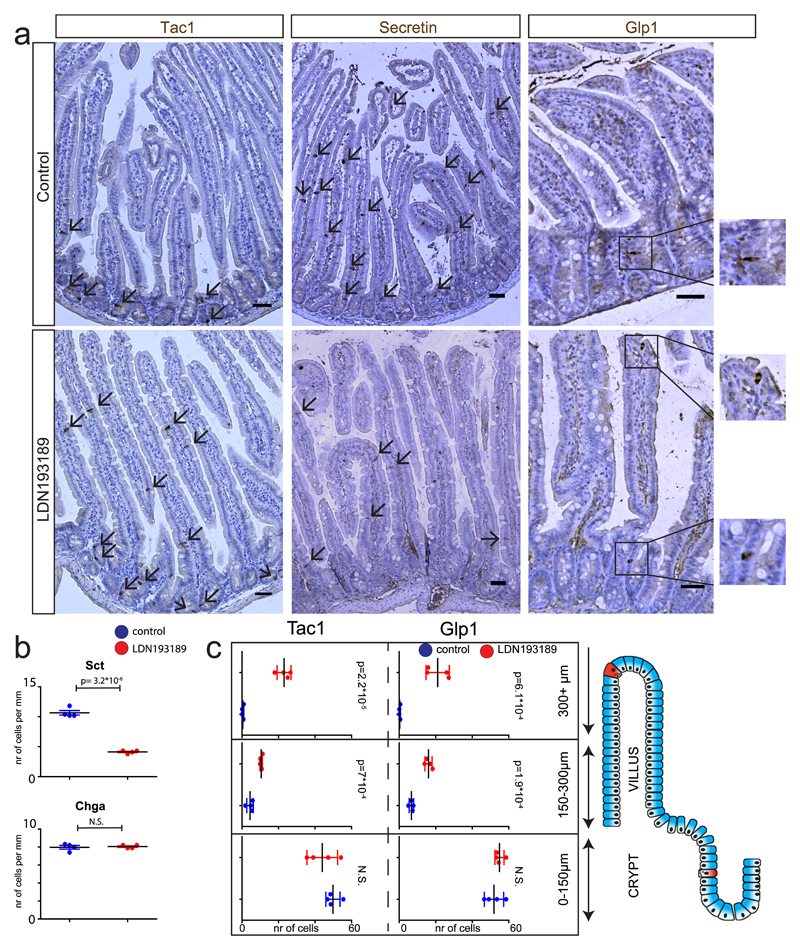
To investigate whether villus-produced BMP controls hormone expression in vivo, we analyzed intestines from mice that ectopically express BMP inhibitor Noggin in the intestinal epithelium (VillinNoggin)13. As expected, we observed an increased expression of Tac1 and Glp1 in the villi of these mice, while Sct was reduced (Fig S5a-d). We next tested the feasibility of influencing hormone expression by targeting the BMP gradient with the BMPR1a inhibitor LDN19318928. An 80-hour oral treatment caused a reduction in Sct+ cell numbers. Overall histology of the intestine was unaffected, and the number of Chga+ EECs did not change significantly (Fig. 4a-b), consistent with a previous study showing no change in Chga after BMP inhibition29. We quantified the numbers of Tac1+ and Glp1+ along different segments of the crypt-villus axis, assuming that BMP inhibition would not increase these in the BMP-low crypt. BMP inhibition did not cause significant changes in the lowest crypt-villus segment in the number of Tac1+ or Glp1+ EECs (Fig. 4a, 4c). However, the increase in cells immunoreactive for Tac1 or Glp1 was very pronounced higher up in the villus (Fig. 4a, 4c).
Fig. 4. Manipulation of BMP gradient alters hormone expression in mice.
a, Mice were treated for 80 hours with BMPr1a inhibitor LDN193819. Immunohistochemical analysis of the intestine shows a repression of Sct and induction of Glp1 and Tac1 expression upon BMP inhibition. Glp1+ and Tac1+ expressing cells are mostly restricted to the crypt in control mice, but expand into the upper villus region upon BMP inhibition. b, Quantification of a. Number of Chga+ and Sct+ positive cells per treatment. c, Quantification of a. Number of Tac1+ or Glp1+ cells are displayed for each segment of the crypt-villus axis in the different treatments. Cells positive for Tac1 and Glp1 increase in the higher villus segments upon LDN193189 treatment. Tac1+ cells are counted and displayed in 10mm of the proximal small intestine. Glp1+ cells are counted and displayed in 30mm of the distal small intestine. Results presented are derived from n=4 mice per treatment, and statistics were calculated using a two-sided t-test. Mean values per staining and treatment are shown, and error bars present SD. Scale bar is 50 µm.
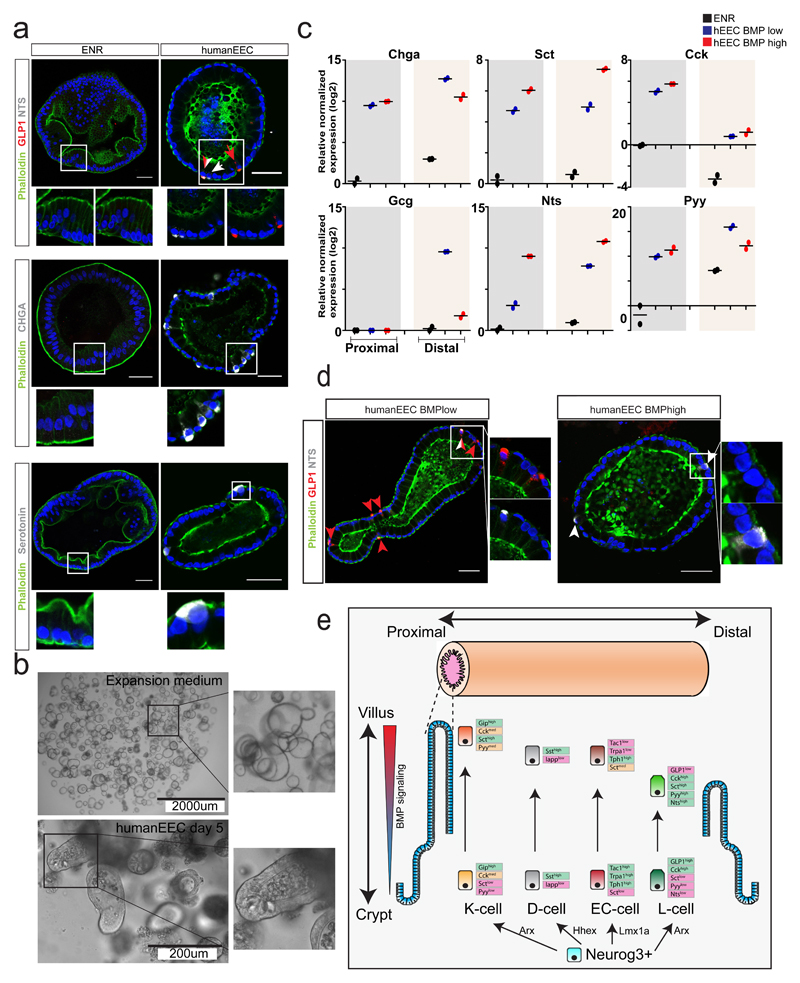
Finally, we pursued the establishment of a differentiation platform for induction of EECs in human intestinal organoids30. Best results were obtained with dual inhibition of Notch and MEK signaling (Fig. 5a-b). This allowed us to generate all subtypes of EECs in organoids either derived from human duodenal or ileal tissue. ENR differentiated organoids did not contain EECs but mostly enterocytes, as evidenced by their extensive brush border (Fig. 5a). In our EEC differentiation protocol, BMP activation induced similar trended alterations in EEC hormone repertoires as it did in murine organoids (Fig. 5c). NTS and SCT transcripts increased upon BMP stimulation, while we observed a dramatic reduction in GCG (Fig. 5c). BMP activation had a neutral effect on total CHGA expression, and in contrast to the mouse, PYY was not increased upon BMP stimulation (Fig. 5c). Although cells positive for both GLP1 and NTS peptides were observed in control conditions, we only observed NTS single positive cells in BMP-treated conditions (Fig. 5d). This implies that BMP-control of the expressed EEC hormone repertoire is a generalizable phenomenon. In line with our observations in the murine organoid system8, we find that human intestinal organoids maintain their regional identity in terms of representation of EEC subtypes. GCG, NTS and PYY were highly enriched in distal gut organoids, while CCK displays a much higher bias towards the duodenum (Fig. 5c).
Fig. 5. New human EEC differentiation protocol implies conserved BMP-controlled hormone expression.
a, Human small intestinal organoids were induced to differentiate either for 5 days by withdrawing Wnt signals (ENR) or to EECs through additional MEK and Notch inhibition. Immunofluorescence indicates the presence of different subtypes of EECs in human organoids. White arrows indicate NTS+ cells, red arrows Glp1+ cells. Experiment has been repeated four times independently with similar results. b, Brightfield images of human intestinal organoids in expansion medium or after a 5 day differentiation towards EECs. Experiment was repeated independently four times. c, The 5-day differentiation protocol of EECs (‘humanEEC’) was performed in the presence and absence of BMP4. Expression levels of hormones were determined by qPCR and are shown relative to duodenal ENR control. Gcg expression is shown relative to ileal ENR control, as it was not detected in duodenal organoids. Sample size represents n=2 biologically independent experiments, and the mean expression values are shown. d, Human EECs were produced in the absence (BMPlow) or presence (BMPhigh) of BMP4. NTS overlaps with GLP1 in BMP low conditions, while only NTS single positive cells are observed in BMPhigh conditions. Experiment was repeated independently four times. Scale bar is 50 µm. e, Model of EEC differentiation. K-cells are enriched in the proximal and L-cells in the distal part of the SI respectively, whereas ECs and D-cells are uniformly distributed. A crypt-to-villus BMP signaling gradient drives alterations in hormone repertoires in these EEC lineages.
Taken together, these data provide two main insights into EEC biology. First, the observations support that BMP controls hormone expression of EECs. EECs that are born in crypts from Lgr5 stem cells encounter increasing levels of BMP signaling when migrating towards the villus tips31, and can change their hormone profile during this journey. Second, this insight in combination with the single cell sequencing data proposes a simplification of EEC taxonomy (Fig. 5e). Confusingly, previous high resolution imaging and single cell RNA sequencing has suggested that almost every combination of EEC hormones can occur in individual EECs32. Our current data indicate that there might be fewer unrelated differentiation pathways of EECs ('lineages') than previously anticipated and that some of the marker-hormones indeed are not hard-wired. Our data imply that there might exist no EECs uniquely dedicated to the production of Sct or Nts (the so called S- or N-cells) or Pyy, and that most EECs initiate expression of Sct when entering the BMP-high villus domain. Indeed, we observe that all BMP-activated EECs except D-cells upregulate Sct to different degrees, while L-cells (but not Sst- or Serotonin-producing cells) increase Pyy and Nts. Gip-expressing cells can be induced to express lower levels of Pyy, but not Nts. Importantly, we find that Chga is a marker of Serotonin-positive cells but not of other EECs, and using it as a generic marker would not allow for identifying EEC regulators such as the BMP pathway.
Pulse-chase labeling using BrdU has indicated that EECs do not necessarily follow the conveyor-belt migration pattern in which cells follow a constant flow from the bottom of the crypt to the tip of the villus12,33. Recently it has been shown that EECs can physically interact with enteric neurons through synapses, which potentially could alter cellular migration 34,35. Subpopulations of EECs can be retained in the crypt for 2 weeks while maintaining expression of Tac1 or Glp1. Cells that migrate onto the villus are destined to lose Tac1 or Glp19,12,33. This ultimately suggests that controlling EEC migration along the crypt-villus axis and the Wnt and BMP gradients would be a way to influence hormone expression patterns.
Methods
Mouse strains and experiments
Primary organoid cultures used in this culture were derived from GcgVenus, Tac1iresCre /Rosa26tdTomato and GipiresCre /Rosa26tdRfp mice11,16, and established as described before6. All mice were bred on a C57BL/6 background. All animal procedures and experiments were performed in accordance with national animal welfare laws under a project license obtained from the Dutch Government, and were reviewed by the Animal Ethics Committee of the Royal Netherlands Academy of Arts and Sciences (KNAW). All rodents are housed in a barrier facility in conventional cages and are changed without using a change stations. All personnel entering the barrier must wear protective clothing (including head caps, specials clogs). All animals are received directly from approved vendors (Charles River) or generated in house. Animals arriving from other sources must pass the GDL –quarantine for screening or by embryo-transfer. After screening these SPF mice are housed in micro isolator cages and are transferred to the Hubrecht laboratory.
For the BMPR inhibition experiment, LDN193189 (Selleckchem) was dissolved in citric buffer (pH3-3.1) at 2mg ml-1. 12 week old mice (n=4) were given two oral doses of LDN193189 at 17.5 mg per kg bodyweight per day. Citric buffer was given to control mice. The total treatment was maintained for 80 hours. All mouse experiments were conducted under a project license granted by the Central Committee Animal Experimentation (CCD) of the Dutch government and approved by the Hubrecht Institute Animal Welfare Body (IvD), with project license number AVD8010020151.
Murine and human intestinal organoid culture
The basic culture medium (advanced Dulbecco’s modified Eagle’s medium/F12 supplemented with penicillin/streptomycin, 10 mM HEPES, Glutamax, B27 [Life Technologies, Carlsbad, CA] and 1 mM N-acetylcysteine [Sigma]) was supplemented with 50 ng/ml murine recombinant epidermal growth factor (EGF; Peprotech, Hamburg, Germany), R-spondin1 (conditioned medium, 5% final volume), and Noggin (conditioned medium, 5% final volume), called ‘’ENR” medium. Conditioned media were produced using HEK293T cells stably transfected with HA-mouse Rspo1-Fc (gift from Calvin Kuo, Stanford University) or after transient transfection with mouse Noggin-Fc expression vector. Advanced Dulbecco’s modified Eagle’s medium/F12 supplemented with penicillin/streptomycin, and Glutamax was conditioned for 1 week.
Human duodenal and ileal tissues were obtained from the UMC Utrecht with informed consent of each patient. The study was approved by the UMC Utrecht (Utrecht, The Netherlands) ethical committee and was in accordance with the Declaration of Helsinki and according to Dutch law. Patients were diagnosed with a small or large intestinal cancer and from the resected intestinal segments, a sample was taken from normal mucosa for this study. Human small intestinal cells were isolated, processed and cultured as described previously30.
Organoids were plated in BME (Trevigen). MEK signaling was inhibited using PD0325901 (1 μM for murine, 100nM for human organoids; Sigma Aldrich). Wnt secretion was inhibited with IWP-2 (5 μM; Stemgent) and Notch with DAPT (10uM, Sigma Aldrich). BMP signaling was activated by treating with human recombinant BMP4 (20ng/ml, Peprotech) and withdrawal of Noggin from the culture medium. Hedgehog signaling was inhibited with Vismodegib (10uM, Selleckchem). TGF beta signaling was activated using recombinant mouse TGF beta-1 (3ng ml-1, R&DSystems, MAB7666TGF beta-1. TGF beta type1 receptor signaling was inhibited using A83 (500nM, Tocris). All control organoids were treated with similar concentrations of the compound dissolvent, dimethyl sulfoxide (DMSO) or 0.1%BSA in PBS0. During treatments, cells were imaged using an EVOS microscope (Electron Microscopy Sciences).
For the induction of enteroendocrine differentiation in murine organoids, cells were cultured in standard culture conditions (ENR). 4-7 days after plating in BME, medium was removed and organoids were treated with different regimes. The cocktail for mouse EEC differentiation included: IWP2 (5 μM; Stemgent), DAPT (10 μM, Sigma Aldrich) and MEK inhibitor PD0325901 (1 μM; Sigma Aldrich), while BMP4 (20ng/ml, Peprotech) was added for activation of BMP signaling. In human organoids, differentiation was achieved by withdrawing p38 MAPK inhibitor SB202190, TGFbeta inhibitor A83, Nicotinamide and Wnt conditioned medium from the culture medium as described previously30. Differentiation into EECs was performed by on top treating with DAPT (10 μM, Sigma Aldrich) and MEK inhibitor PD0325901 (500nM; Sigma Aldrich), BMP4 (20ng/ml, Peprotech) was added for activation of BMP signaling.
Immunostainings
Whole organoids were collected by gently dissolving the BME in ice-cold medium, and subsequently fixed at RT in 4% formalin (Sigma) for at least 6 hours. Next, organoids were permeabilized and blocked in PBS containing 0,5% Triton X-100 (Sigma) and 2% normal donkey serum (Jackson ImunoResearch) for 30 min at room temperature. Organoids were incubated for 2 hr at room temperature in blocking buffer containing primary antibodies. Primary antibodies used were goat anti-Chromogranin A (1:500; Santa Cruz), goat anti-Cholestocystokin (sc-21617,1:100; Santa Cruz), rabbit anti-Neurotensin (sc-20806,1:100; Santa Cruz), goat anti-Secretin (sc-26630,1:100; Santa Cruz), goat anti-Somatostatin (sc-7819, 1:100; Santa Cruz), goat anti-Serotonin (ab66047, 1:1000, Abcam), rabbit anti-Gastric inhibitory polypeptide (ab22624-50, 1:500;Abcam), goat anti-GLP1 (sc-7782, 1:100; Santa Cruz), rabbit anti-GLP1 (ab22625, 1:200; Abcam), rabbit anti-Peptide YY (ab22663, 1:500; Abcam) and guinea pig anti-Substance P (1:200, ab10353; Abcam). Organoids were incubated with the corresponding secondary antibodies Alexa488, 568 and 647 conjugated anti-rabbit and anti-goat (1:1000; Molecular Probes) in blocking buffer containing DAPI (1;1000, Invitrogen). Sections were embedded in Vectashield (Vector Labs) and imaged using a Sp8 confocal microscope (Leica). Image analysis was performed using ImageJ software.
For immunohistochemistry of organoids within the BME (Figure S4), medium was removed from the wells and replaced with 4% formalin for 1 hour. Next, organoids were washed with PBS, permeabilized and blocked in PBS containing 0,5% Triton X-100 (Sigma) and 2% normal donkey serum (Jackson ImunoResearch) for 30 min at room temperature. The wells were incubated for 2 hr at room temperature in blocking buffer containing primary antibodies. After washing, secondary antibodies were added for 1 hr at room temperature in blocking buffer. Organoids were subsequently imaged within the plate using a Sp8 confocal microscope (Leica). For immunohistochemistry of mouse intestinal tissue, intestines were first flushed with 4% formaldehyde. Next, intestines were fixed for 6 hours at RT in 4% formalin. The tissue was either embedded in paraffin or Tissue-Tek O.C.T. for cryosectioning, and stained as described previously6,36.
Quantification of number/location of EECs on intestinal section images were performed in ImageJ software, as well as the intensity of Venus levels in the live cell imaging experiment in Figure S2e-f. Analysis of Glp1+ cell numbers and/or position in Figure 1a-c, Figure 4a-c and Figure S5 were performed in the ileum, and all other hormones were counted along the whole SI tract.
All quantifications were performed on the raw, unprocessed images.
RNA isolation and quantitative PCR
For qPCR analysis and bulk RNA sequencing, RNA was isolated from organoids using the RNAeasy kit (QIAGEN) as instructed in the manufacturer’s protocol. PCR analysis was performed using the SYBR-Green and Bio-Rad systems as described37. PCR reactions were performed in duplicate with a standard curve for every primer. Changes in expression were calculated using CFX manager software (Bio-Rad). Primers were designed using the NCBI primer design tool. Primers used in this study are presented in Supplementary Table 1.
Glp1 and Secretin secreted peptide
The supernatant from organoids was collected after 2-hour stimulation with Forskolin. GLP-1 concentration in the supernatant was measured with a GLP-1 EIA Kit (Rab0201, Sigma, detects both full length and N-terminal cleaved GLP-1) using the manufacturer’s protocol. Secretin concentration was measured with a Secretin EIA kit (EK-067-04, Phoenix Pharmaceuticals) using the manufacturer’s protocol.
Statistical analysis
Two sided t-tests were performed for all statistical analyses. Precise p-values are mentioned in the corresponding figures, and significance level was set at p < 0.05. In each figure legend, the number of biology replicates is mentioned for the corresponding experiment (n=x). For figures where representative images are shown, the number the experiment has been repeated is mentioned in the legend.
For immunohistochemistry, we counted the following number of cells, of organoids or length of intestine; at least 50 cells per hormone and replicate intestine (Fig. 1b-e, n=4 mice), at least 150 tdTomato+ cells per co-staining and replicate intestine (Fig. 1f-g, n=4 mice), at least 3 organoids per hormone and replicate (Fig. 2a-b, n=2 biologically independent experiments), 10 organoids per hormone and replicate (Fig. S4a, n=2 biologically independent experiments), 10mm of the proximal small intestine for Tac1 (Fig. 4b-c, n=4 mice per treatment), 30mm of the distal small intestine for Glp1 (Fig. 4b-c, n=4 mice per treatment). mice per genotype) and at least 50 cells per hormone and replicate intestine (Fig. S5b, n=2 mice per genotype),
Bulk and single cell RNA sequencing
For bulk RNA sequencing analysis, organoids stimulated with EEC BMPhigh or EEC BMPlow media for 4 days were collected and dissociated in RTL buffer (RNeasy Mini kit, Quiagen). Total RNA was isolated accordingly to manufacturer’s instruction (RNeasy Mini kit, Quiagen). Sequencing libraries were prepared based on a modified CELseq2 method19. Briefly, 1 ng of RNA was reverse transcribed using the Ambion kit and in vitro transcription was performed using 1 ng of cDNA as template. The aRNA was then used to prepare sequencing libraries. Samples were sequenced with sequenced paired-end at 75 bp read-length the on Illumina NextSeq.
For single cell RNA sequencing, organoids were first dissociated into single cells through mechanical disruption, after 15 min of Trypsin treatment at 37°C (TrypLE Express; Life Technologies). Next, cells were immediately sorted using a BD FACS Aria (BD Biosciences). For single cell sequencing experiment, cells were sorted as single cells into 384-well plates containing ERCC spike-ins (Agilent), RT primers and dNTP (Promega) as described before. Plates were prepared using Mosquito® HTS (TTPlabtech). Single cell RNA-sequencing libraries were prepared following the SORT-seq protocol 18, which is based on CEL-seq2 method19. Briefly, cells were first lysed 5 min at 65°C, and RT and second strand mixes were dispensed by the Nanodrop II liquid handling platform (GC biotech). Single cell double stranded cDNAs were pooled together and in vitro transcribed for linear amplification. Illumina sequencing libraries were prepared using the TruSeq small RNA primers (Illumina) and sequenced paired-end at 75 bp read length the Illumina NextSeq.
RNA-sequencing data analysis
Paired-end reads from Illumina sequencing were aligned to the mouse transcriptome genome by BWA38. For RNA-seq bulk data, normalization and differential gene expression analyses were performed using the DESeq2 package39 and visualized as volcano plots. For single cell RNA-seq data, read counts were first corrected for UMI barcode by removing duplicate reads that had identical combinations of library, cell-specific, and molecular barcodes and were mapped to the same gene. For each cell barcode the number of UMIs for every transcript was counted, and transcript counts were then adjusted to the expected number of molecules based on counts, 256 possible UMI’s and poissonian counting statistics40. Samples were then normalized by downsampling to a minimum number of 3000 transcripts/cell. Cells with fewer transcripts were excluded from the analyses. RaceID2 was used to cluster cells based on k-medoid method 20. All data analyses, quantification and data visualization were run on Rstudio. In total, we sequenced 2880 cells and, after applying a filtering criteria of 3000 expressed transcripts/cell, 820 cells were retained for further analysis.
Supplementary Material
Acknowledgements
We thank Stefan van der Elst, Reinier van der Linden and Yotam Bar-Ephraim, for their help with FACS experiments. B.A. is supported by NWO/VENI 863.15.015.
Footnotes
Data availability
Bulk and single-cell RNA–seq data that support the findings of this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE114988.
Source data for Fig. 1c, 1e, 1g, 2b, 2e, 4a, 5b, 5c, 6b and Supplementary Fig. 1a, 2a, 2b, 2d, 3b, 4b, 4c, 5a, 5b and 6 have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Competing financial interests
The authors disclose no conflicts of interest.
Author contributions
J.B. and H.C. conceived and designed the project. J.B. designed and performed all experiments. B.A. performed analysis of RNA sequencing data. F.R. and F.G. generated the Gcg and Gip reporter mice. H.Z. generated the Tac1 reporter mouse, and T.N.N. assisted in providing the tissue. J.H.v.E supervised and performed the mouse experiments, with the help of M.v.d.B. Y.P. assisted in histology preparation. J.B., B.A. and H.C. wrote the manuscript with input from all other authors.
References
- 1.Furness JB, Rivera LR, Cho H-J, Bravo DM, Callaghan B. The gut as a sensory organ. Nat Rev Gastroenterol Hepatol. 2013;1010:729–740. doi: 10.1038/nrgastro.2013.180. [DOI] [PubMed] [Google Scholar]
- 2.Connor TMO, et al. The Role of Substance P in Inflammatory Disease. doi: 10.1002/jcp.20061. [DOI] [Google Scholar]
- 3.Egerod KL, et al. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology. 2012;153:5782–5795. doi: 10.1210/en.2012-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 6.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 7.Grün D, et al. Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature. 2015;525:251–5. doi: 10.1038/nature14966. [DOI] [PubMed] [Google Scholar]
- 8.Basak O, et al. Induced Quiescence of Lgr5+ Stem Cells in Intestinal Organoids Enables Differentiation of Hormone-Producing Enteroendocrine Cells. Cell Stem Cell. 2017;20:177–190.e4. doi: 10.1016/j.stem.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 9.Roth Ka, Gordon JI. Spatial differentiation of the intestinal epithelium: analysis of enteroendocrine cells containing immunoreactive serotonin, secretin, and substance P in normal and transgenic mice. Proc Natl Acad Sci U S A. 1990;87:6408–6412. doi: 10.1073/pnas.87.16.6408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grunddal KV, et al. Neurotensin is coexpressed, coreleased, and acts together with GLP-1 and PYY in enteroendocrine control of metabolism. Endocrinology. 2016;157:176–194. doi: 10.1210/en.2015-1600. [DOI] [PubMed] [Google Scholar]
- 11.Harris JA, et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits. 2014;8 doi: 10.3389/fncir.2014.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aiken KD, Roth KA. Temporal differentiation and migration of substance P, serotonin, and secretin immunoreactive enteroendocrine cells in the mouse proximal small intestine. Dev Dyn. 1992;194:303–310. doi: 10.1002/aja.1001940406. [DOI] [PubMed] [Google Scholar]
- 13.Haramis A-PG. De novo crypt formation and Juvenile Polyposis on BMP inhibition in mouse intestine. Science (80-. ) 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- 14.Beumer J, Clevers H. Regulation and plasticity of intestinal stem cells during homeostasis and regeneration. Development. 2016;143:3639–3649. doi: 10.1242/dev.133132. [DOI] [PubMed] [Google Scholar]
- 15.Camacho S, et al. Anti-obesity and anti-hyperglycemic effects of cinnamaldehyde via altered ghrelin secretion and functional impact on food intake and gastric emptying. Sci Rep. 2015;5 doi: 10.1038/srep07919. 7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Habib AM, et al. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology. 2012;153:3054–3065. doi: 10.1210/en.2011-2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svendsen B, et al. GLP1- and GIP-producing cells rarely overlap and differ by bombesin receptor-2 expression and responsiveness. J Endocrinol. 2016;228:39–48. doi: 10.1530/JOE-15-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muraro MJ, et al. A Single-Cell Transcriptome Atlas of the Human Pancreas. Cell Syst. 2016;3:385–394. doi: 10.1016/j.cels.2016.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimshony T, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grün D, et al. De Novo Prediction of Stem Cell Identity using Single-Cell Transcriptome Data. Cell Stem Cell. 2016;19:266–277. doi: 10.1016/j.stem.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nozawa K, et al. TRPA1 regulates gastrointestinal motility through serotonin release from enterochromaffin cells. Proc Natl Acad Sci U S A. 2009;106:13. doi: 10.1073/pnas.0805323106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Z X-X, P Y-H, H Y-M. Neuroendocrine hormone amylin in diabetes. World J Diabetes. 2016;7:189–197. doi: 10.4239/wjd.v7.i9.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gross S, et al. The novel enterochromaffin marker Lmx1a regulates serotonin biosynthesis in enteroendocrine cell lineages downstream of Nkx2.2. Development. 2016;143:2616–2628. doi: 10.1242/dev.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J, McKenna LB, Bogue CW, Kaestner KH. The diabetes gene Hhex maintains ??-cell differentiation and islet function. Genes Dev. 2014;28:829–834. doi: 10.1101/gad.235499.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaaij LT, et al. DNA methylation dynamics during intestinal stem cell differentiation reveals enhancers driving gene expression in the villus. Genome Biol. 2013;14:R50. doi: 10.1186/gb-2013-14-5-r50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hollnagel A, Oehlmann V, Heymer J, Rüther U, Nordheim A. Id genes are direct targets of bone morphogenetic protein induction in embryonic stem cells. J Biol Chem. 1999;274:19838–19845. doi: 10.1074/jbc.274.28.19838. [DOI] [PubMed] [Google Scholar]
- 27.Jenny M, et al. Neurogenin3 is differentially required for endocrine cell fate specification in the intestinal and gastric epithelium. EMBO J. 2002;21:6338–6347. doi: 10.1093/emboj/cdf649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whissell G, et al. The transcription factor GATA6 enables self-renewal of colon adenoma stem cells by repressing BMP gene expression. Nat Cell Biol. 2014;16:695–707. doi: 10.1038/ncb2992. [DOI] [PubMed] [Google Scholar]
- 29.Qi Z, et al. BMP restricts stemness of intestinal Lgr5+ stem cells by directly suppressing their signature genes. Nat Commun. 2017;8:13824. doi: 10.1038/ncomms13824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 31.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154:274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 32.Fothergill LJ, Callaghan B, Hunne B, Bravo DM, Furness JB. Costorage of enteroendocrine hormones evaluated at the cell and subcellular levels in male mice. Endocrinology. 2017;158:2113–2123. doi: 10.1210/en.2017-00243. [DOI] [PubMed] [Google Scholar]
- 33.Aiken KD, Kisslinger JA, Roth KA. Immunohistochemical studies indicate multiple enteroendocrine cell differentiation pathways in the mouse proximal small intestine. Dev Dyn. 1994;201:63–70. doi: 10.1002/aja.1002010107. [DOI] [PubMed] [Google Scholar]
- 34.Bellono NW, et al. Enterochromaffin Cells Are Gut Chemosensors that Couple to Sensory Neural Pathways. Cell. 2017;170:185–198.e16. doi: 10.1016/j.cell.2017.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bohorquez DV, et al. Neuroepithelial circuit formed by innervation of sensory enteroendocrine cells. J Clin Invest. 2015;125:782–786. doi: 10.1172/JCI78361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Farin HF, et al. Visualization of a short-range Wnt gradient in the intestinal stem-cell niche. Nature. 2016;530:340–343. doi: 10.1038/nature16937. [DOI] [PubMed] [Google Scholar]
- 37.Muñoz J, et al. The Lgr5 intestinal stem cell signature: robust expression of proposed quiescent ‘+4’ cell markers. EMBO J. 2012;31:3079–3091. doi: 10.1038/emboj.2012.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grün D, Kester L, van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.