Abstract
Killer-cell immunoglobulin-like receptors (KIRs) are expressed predominantly on natural killer cells, where they play a key role in the regulation of innate immune responses. Recent studies show that inhibitory KIRs can also impact adaptive T cell-mediated immunity. In mice and in human T cells in vitro, inhibitory KIR ligation enhanced CD8+ T cell survival. To investigate the clinical relevance of these observations, we conducted an extensive immunogenetic analysis of multiple, independent cohorts of HIV-1, hepatitis C virus (HCV) and human T cell leukemia virus (HTLV-1)-infected individuals in conjunction with in vitro assays of T cell survival, analysis of ex vivo KIR expression and mathematical modeling of host-virus dynamics. Our data suggest that functional engagement of inhibitory KIRs enhances the CD8+ T cell response against HIV-1, HCV and HTLV-1 and is a significant determinant of clinical outcome in all three viral infections.
Introduction
Killer-cell immunoglobulin-like receptors (KIRs) are a family of transmembrane glycoproteins with activating and inhibitory isoforms; their ligands include the HLA class I molecules (1, 2). KIRs are expressed predominantly on natural killer (NK) cells and are primarily associated with innate NK cell-mediated immunity. However, recent studies have shown that KIRs can also impact adaptive T cell-mediated immunity.
Two distinct routes whereby inhibitory KIRs (iKIRs) affect T cell responses have been described (3, 4). First, iKIR expression on CD8+ T cells directly affects their longevity and function. KIRs are expressed on T cells with an effector memory TEM(RA) phenotype (CD28− CD45RA+/–CD45RO–/+CCR7–); the frequency of KIR+ T cells increases with age and some persistent viral infections (5–9). In mice, inhibitory Ly49 receptors (the functional homolog of iKIRs) enhance CD8+ T cell survival (10). Similarly, in KIR-transgenic mice, ligation of iKIRs on T cells reduces activation-induced death (11, 12). Furthermore, iKIR expression on human T cells is associated with higher levels of the survival molecule Bcl-2, reduced cell death and impaired functionality (13–15). Second, KIRs indirectly modify the CD8+ T cell response via their expression on NK cells. NK cells regulate T cell immunity by cytokine secretion and killing activated CD4+ and CD8+ T cells; this regulation may be modified by the expression of inhibitory receptors on NK cells (4, 16, 17). In mice, lymphocytic choriomeningitis virus-specific CD8+ T cell responses are modulated via NK cell mediated-killing of activated CD4+ T cells (18) and in murine cytomegalovirus infection, IL-10 production by NK cells impairs the CD8+ T cell response (19). Additionally, human NK cells can lyse hepatitis B virus-specific CD8+ T cells in vitro (20).
The clinical relevance of an interaction between iKIRs and CD8+ T cells is difficult to infer from these earlier studies, because it is impossible to tell whether changes in CD8+ T cell lifespan in mice or in vitro alter the course of human disease in vivo. To investigate whether iKIRs have a clinically significant impact on the CD8+ T cell response, our starting point was the well-documented association between certain HLA class I alleles and disease outcome as this represents compelling evidence that CD8+ T cells are clinically relevant. We reasoned that if KIRs significantly affect the CD8+ T cell response then KIRs will affect HLA class I disease associations. In a previous study, we validated this approach by showing that KIR2DL2 exaggerates both protective and detrimental HLA class I associations with disease progression in individuals infected with hepatitis C virus (HCV) or human T cell leukemia virus type 1 (HTLV-1) (21). However, it remains unclear whether this constitutes a generalizable phenomenon that extends to other viruses and iKIRs. Furthermore, the mechanism underlying the effect was not investigated.
In this study we used epidemiological data from multiple, independent cohorts of HIV-1, HCV and HTLV-1-infected individuals; in vitro T cell survival assays; ex vivo analyses of KIR expression and mathematical modelling of host-virus dynamics. Our data indicate that both two-domain and three-domain iKIRs, together with their HLA class I ligands, enhance the CD8+ T cell response to HIV-1, HTLV-1 and HCV by prolonging CD8+ T cell survival and are a significant determinant of clinical outcome in all three viral infections.
Results
We studied a well-characterized cohort of HIV-1 seroconverters from sub-Saharan Africa who were identified when seronegative and followed under Protocol C of the International AIDS Vaccine Initiative (IAVI) (22). First we sought to identify KIR-HLA associations that could potentially confound our study. We define a “functional KIR gene” as a KIR gene together with a gene encoding its HLA class I ligand. Both functional KIR3DS1 (KIR3DS1 with the gene encoding its putative ligand HLA-Bw4-80I) and functional KIR3DL1 (KIR3DL1 with the gene for its ligand HLA-Bw4) have previously been associated with good NK cell-mediated control of HIV-I infection (23). In the IAVI cohort, the protective effect of functional KIR3DS1, but not functional KIR3DL1 was replicated, Table S1. To prevent confounding effects arising from strong linkage disequilibrium between the KIR genes we therefore excluded all individuals with functional KIR3DS1. As expected, HLA-B*57 was significantly associated with low early set-point viral load (Coefficient = –0.42, P = 0.004) and slow progression to low CD4 count (Hazard Ratio = 0.44, P = 0.02). Where the Coefficient (Coeff) is the change in log10[early viral load set point] associated with possession of HLA-B*57 and the Hazard Ratio (HR) is the relative risk of progression to low CD4 count associated with possession of HLA-B*57.
iKIR genes are associated with an enhancement of the protective effect of HLA-B*57
To determine whether HLA-B*57 was more protective in the presence of functional KIR2DL2, we stratified the cohort into individuals with functional KIR2DL2 and without functional KIR2DL2 and analyzed the protective effect of HLA-B*57 in each stratum. Two definitions of functional KIR2DL2 were considered: (i) strong functional KIR2DL2 (KIR2DL2 with genes encoding its strong HLA-C1 ligand); and (ii) weak functional KIR2DL2 (KIR2DL2 with genes encoding its weaker HLA-C2 ligand). For both early set-point viral load and time to low CD4 count, HLA-B*57 was significantly protective in the stratum with functional KIR2DL2 but weaker and non-significant in its absence; additionally, the magnitude of the protection was greater with the stronger ligand (Fig. 1A, Table 1). These differences were not attributable to cohort size because there were fewer HLA-B*57+ individuals in the stratum with functional KIR2DL2 than in the stratum without functional KIR2DL2. We then extended the analysis to other iKIRs with known ligands (KIR2DL1 with HLA-C2 and KIR2DL3 with HLA-C1). Functional KIR3DL1 could not be analysed because there were no individuals with HLA-B*57 who lacked functional KIR3DL1. iKIRs with disputed ligands were excluded from the analysis. Both functional KIR2DL1 and functional KIR2DL3 enhanced the protective effect of HLA-B*57 on both early set-point viral load and time to low CD4 count (Fig. 1A, Table 1). The odds of seeing this enhancement by chance are P = 2x10-6 (Permutation test).
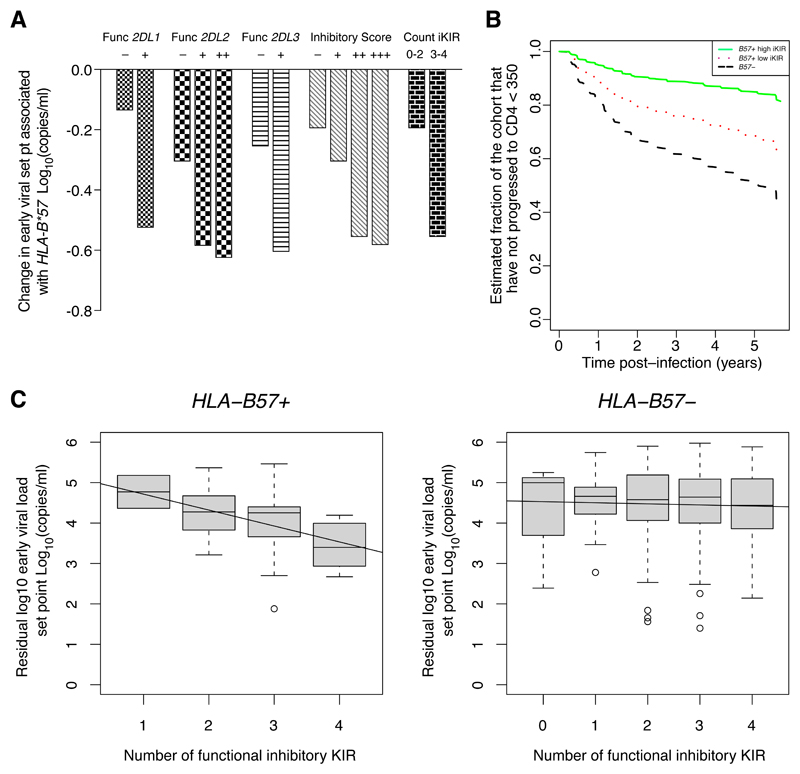
Figure 1. Inhibitory KIR genes enhance the protective effect of HLA-B*57 on early set-point viral load and time to low CD4 count.
A. The IAVI cohort (N=461) was stratified into individuals with or without the functional iKIR of interest, into individuals with different inhibitory scores and into individuals with a low or high count of functional iKIRs. In each stratum, the protective effect of HLA-B*57 on early set-point viral load was analyzed using multivariate linear regression. The coefficients (change in early set point viral load associated with HLA-B*57) are plotted. It can be seen that the protective effect of HLA-B*57 was enhanced by all 3 iKIR, by high inhibitory score and by high count of iKIRs with a dose effect with inhibitory score. Definitions. Func 2DL1+: KIR2DL1+C2+. Func 2DL2++: KIR2DL2+C1+. Func 2DL2+: KIR2DL2+C2+. Func 2DL3+: KIR2DL3+C1+. Func 2DL1-: KIR2DL1- or KIR2DL1+C2-. Func 2DL2-: KIR2DL2-. Func 2DL3-: KIR2DL3- or KIR2DL3+C1-. Inhibitory score: +++: >2.5, ++: >2.0, +: ≤2.5. -: ≤2.0.
B. Cox survival curves are plotted for HLA-B*57 − individuals (black dashed line), HLA-B*57+ individuals with a low inhibitory score (red dotted line) and HLA-B*57+ individuals with a high inhibitory score (green solid line). It can be seen that individuals with HLA-B*57 and a high inhibitory score (>2.5) progressed to low CD4 count (<350 cells per mm3) at the slowest rate (N = 491).
C. The cohort (N=461) was stratified into individuals with or without HLA-B*57 and the relationship between early set-point viral load and the count of functional iKIRs was analyzed. Early viral load is shown in grey boxes, and the corresponding linear regression is shown as a line. The number of functional iKIRs was significantly associated with decreasing viral load in HLA-B*57+ individuals (P = 0.018) but not HLA-B*57 − individuals (P = 0.59). This indicates that iKIRs enhance the HLA-B*57-associated reduction in early viral load in a dose-dependent manner; and that iKIRs in the absence of HLA-B*57 are not in themselves significantly protective (see also Supplementary Table S2).
Table 1. Functional iKIRs enhance the protective effect of HLA-B*57 on early set-point viral load and time to low CD4 count.
The IAVI cohort was stratified into individuals with or without the functional iKIR of interest, with or without a high inhibitory score (two cut-offs considered) and with or without a high count of functional iKIRs (only one cut-off possible). In each stratum, the protective effect of HLA-B*57 on early set-point viral load (A) and time to low CD4 (B) was analyzed using multivariate linear regression and the Cox proportional hazards model, respectively. The coefficients (Coeff), hazard ratios (HR), P values and cohort sizes are reported. The coefficient (Coeff) is the change in log10 viral load associated with possession of the HLA allele (a Coeff>0 represents a detrimental effect). The hazard ratio (HR) is the relative risk of progression associated with possession of the HLA allele (HR >1 represents a detrimental effect).
It can be seen that HLA-B*57 was consistently and significantly protective in the presence but not in the absence of functional iKIRs.
Definitions: Functional 2DL1+: KIR2DL1+C2+. Functional 2DL2+ (strong): KIR2DL2+C1+. Functional KIR2DL2+ (weak): KIR2DL2+C2+. Functional 2DL3+: KIR2DL3+C1+. Functional 2DL1-: KIR2DL1- or KIR2DL1+C2-. Functional 2DL2-: KIR2DL2-. Functional 2DL3-: KIR2DL3- or KIR2DL3+C1-. Inhibitory score (cut-off=2.0): inhibitory score>2.0. Inhibitory score (cut-off=2.5): inhibitory score>2.5. Count (cut-off=2.0): count of functional iKIRs>2.0. The column headings, “KIR+” and “KIR-” refer to the high and low strata. E.g. for Functional 2DL1, “KIR+” denotes the Functional KIR2DL1+ strata, “KIR-” the Functional KIR2DL1- strata; for inhibitory score (cut off=2.0), “KIR+” denotes the strata of individuals with inhibitory score>2.0, “KIR-” denotes the strata of individuals with inhibitory score<=2.0 etc.
Note the stratification provided by the count of iKIRs is the same as one of the stratifications provided by inhibitory score. We report both because the count benefits from being a more objective measure (independent of the choice of weight) whilst the inhibitory score benefits from a greater diversity of values, typically allowing for stratification at more than one cut-off.
All cut-offs that gave a sufficiently balanced analysis were considered.
| A. Outcome: Early viral load setpoint | ||||||||
| B57+ KIR− | B57+ KIR+ | Cohort numbers | ||||||
| Coeff | P value | Coeff | P value | B57+ KIR− | B57− KIR− | B57+ KIR+ | B57− KIR+ | |
| Functional 2DL1 | -0.14 | 0.6 | -0.53 | 0.003 ** | 10 | 125 | 25 | 301 |
| Functional 2DL2 (strong) | -0.31 | 0.1 | -0.63 | 0.02 * | 17 | 153 | 10 | 178 |
| Functional 2DL2 (weak) | -0.59 | 0.01 * | 15 | 208 | ||||
| Functional 2DL3 | -0.26 | 0.2 | -0.61 | 0.002 ** | 18 | 180 | 17 | 246 |
| Inhibitory score (cut-off=2.0) | -0.20 | 0.4 | -0.56 | 0.004 ** | 14 | 195 | 21 | 231 |
| Inhibitory score (cut-off=2.5) | -0.31 | 0.1 | -0.59 | 0.0096 ** | 21 | 258 | 14 | 168 |
| Count (cut-off=2.0) | -0.20 | 0.4 | -0.56 | 0.004 ** | 14 | 195 | 21 | 231 |
| B. Outcome: Time to low CD4 count (<350 cells/mm3) | ||||||||
| B57+ KIR− | B57+ KIR+ | Cohort numbers | ||||||
| HR | P value | HR | P value | B57+ KIR− | B57− KIR− | B57+ KIR+ | B57− KIR+ | |
| Functional 2DL1 | 0.73 | 0.6 | 0.36 | 0.01 * | 10 | 132 | 25 | 324 |
| Functional 2DL2 (strong) | 0.36 | 0.09 ⋅ | 0.19 | 0.03 * | 17 | 168 | 10 | 187 |
| Functional 2DL2 (weak) | 0.71 | 0.40 | 15 | 221 | ||||
| Functional 2DL3 | 0.56 | 0.18 | 0.32 | 0.05 ⋅ | 18 | 195 | 17 | 261 |
| Inhibitory score (cut-off=2.0) | 0.49 | 0.17 | 0.42 | 0.06 ⋅ | 14 | 214 | 21 | 242 |
| Inhibitory score (cut-off=2.5) | 0.57 | 0.15 | 0.22 | 0.04 * | 21 | 280 | 14 | 176 |
| Count (cut-off=2.0) | 0.49 | 0.17 | 0.42 | 0.06 ⋅ | 14 | 214 | 21 | 242 |
Having investigated the effect of the presence or absence of individual functional iKIR we next investigated the effect of the number of functional iKIRs. For each subject in the cohort we assigned two numbers: (i) a ‘count of functional iKIR’ (a count of the number of iKIR+ligand pairs); and (ii) an ‘inhibitory score’ (similar to the count but weighted to reflect the strength of iKIR signalling; Methods). We first stratified the cohort based on the inhibitory score (all cut-offs that gave balanced strata were considered) and analyzed the strength of HLA-B*57-mediated protection in each stratum. Again, we found that the protective effect of HLA-B*57 on both early set-point viral load and progression to low CD4 count was enhanced in the high inhibitory score stratum and that the size of the protective effect increased with the magnitude of the score (Fig. 1A & B, Table 1). Repeating the analysis using the count of functional iKIRs gave similar results indicating that our findings were not dependent on the choice of scoring system. Within HLA-B*57+ individuals (but not HLA-B*57– individuals), both the inhibitory score and the count of functional iKIRs were significantly associated with a decrease in early set-point viral load (Fig. 1C). This result indicates that iKIRs enhance the HLA-B*57 protective effect in a gene dose-dependent manner and shows that functional iKIRs alone (i.e. not in the context of HLA-B*57) are not significantly protective; this latter conclusion was confirmed by multivariate analysis (Table S2).
The iKIR-associated enhancement of HLA-B*57-mediated protection was wholly dependent on the presence of the KIR ligands. If we counted iKIRs in the absence of ligand, we found no enhancement of HLA-B*57-mediated protection (Fig. S1).
Repeating the analysis of the impact of iKIRs on HLA-B*57 using a different approach (introducing a covariate with different levels for HLA-B*57 -, HLA-B*57+ KIR– and HLA-B*57+KIR+ and performing multivariate regression and Cox survival analysis on the whole, unstratified cohort) yielded identical conclusions: the iKIRs KIR2DL1, KIR2DL2 and KIR2DL3 with genes encoding their HLA ligands, a high inhibitory score and a high count of functional iKIRs all enhanced the protective effect of HLA-B*57 on both outcome measures: early viral load and time to low CD4 count (Table S3).
To check that the iKIR-associated enhancement we observed was not driven by leukocyte immunoglobulin-like receptor subfamily B member 2 (LILRB2) binding (24) (which was significantly protective in our cohort, Table S4) or by functional KIR3DL1 (which was not protective in our cohort, Table S1, but has been reported elsewhere (23)), we included these terms as covariates in our model. In both cases, the conclusions were unchanged (Table S5), indicating that functional iKIR enhancement was not attributable to these potentially confounding factors. The conclusions were also unchanged when we included KIR3DL2 with genes for its classical HLA ligands (HLA-A3, -A11 and -B27) or genes for its classical and non-classical HLA ligands (HLA-A3, -A11, -B27 and -F (25)) in the count of functional iKIRs and the inhibitory score.
Which iKIR is responsible for the enhancement of HLA-B*57- mediated protection?
The KIR genes are in strong linkage disequilibrium so the enhancement of HLA-B*57-mediated protection by functional KIR2DL1, functional KIR2DL2 and functional KIR2DL3 are not independent. Establishing which iKIR gene(s) was/were associated with the enhancement was difficult because the cohort size limited the power of further sub-stratification. Across the cohort, functional KIR2DL1 and functional KIR2DL2 were both negatively associated with functional KIR2DL3 while functional KIR2DL1 and functional KIR2DL2 were unassociated. Given that all three functional iKIR enhanced HLA-B*57 and none of them were positively associated this suggests that more than one functional iKIR was enhancing HLA-B*57-mediated protection (the requirement for ligand makes enhancement via ‘absence of a KIR’ unlikely). Furthermore, among HLA-B*57+ subjects, both the count of functional iKIRs and the inhibitory score were better predictors of early viral load than any of the individual functional iKIRs, again suggesting that more than one iKIR was enhancing the protective effect of HLA-B*57. Henceforth, because of the difficulty in determining which iKIR(s) were responsible for the enhancement of HLA-B*57 together with the evidence that more than one iKIR was implicated we focus on the count of iKIRs and the inhibitory score rather than individual iKIR.
Maintenance of the protective effect of HLA-B*57 depends on iKIRs
Next, we investigated longitudinal viral load in the IAVI cohort (a total of 5197 person-visits between 0–2000 days post-infection). The cohort was stratified in three ways: (i) functional iKIR-negative and –positive; (ii) low and high functional iKIR count; or (ii) low and high inhibitory score. In each case, the local non-parametric regression lines were calculated (Fig. 2A, Fig. S2). A consistent picture emerged for each of the three functional iKIRs and for both the iKIR count and inhibitory score. In the presence of a strong inhibitory signal (functional iKIR-positive or a high inhibitory score/count), the protective effect of HLA-B*57 on viral load was sustained over time. Conversely, in the absence of a strong inhibitory signal (functional iKIR-negative or a low inhibitory score/count), the protective effect of HLA-B*57 on viral load was weak. Interestingly, in the absence of a strong inhibitory signal (Fig. 2A, left panel), HLA-B*57 appeared to protect at very early time points post-infection, but this protective effect was eroded with time. Bootstrap estimation of the correlation between HLA-B*57-mediated protection and time confirmed this observation: the protective effect of HLA-B*57 decreased significantly more rapidly in people with a low inhibitory score, Fig. 2B. These findings are consistent with our hypothesis that a strong inhibitory signal enhances T cell survival.
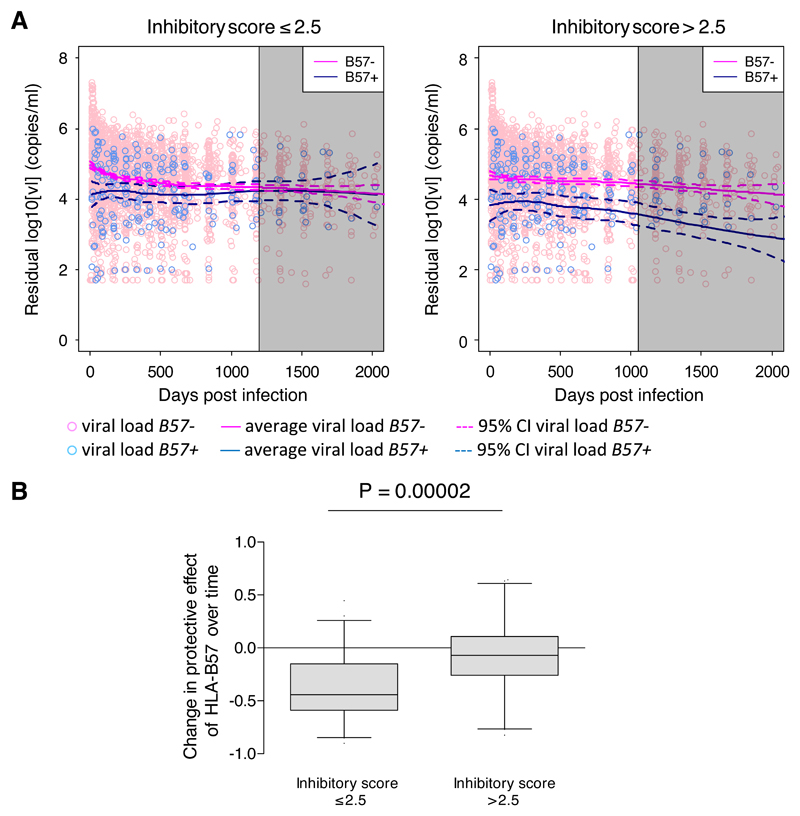
Figure 2. The HLA-B*57 protective effect on viral load is maintained in the presence of a strong inhibitory KIR signal but lost in its absence.
A. The IAVI cohort was stratified into individuals with a low inhibitory score (≤2.5, left) and individuals with a high inhibitory score (>2.5, right). In each stratum, the local nonparametric regression lines (loess) were calculated from the longitudinal viral load measurements (5197 person-visits). Pink circles denote viral load measurements in HLA-B*57 – individuals; summarised with the pink loess line. Blue circles denote viral load measurements in HLA-B*57+ individuals; summarised with the blue loess line. Dashed lines are the 95% confidence intervals around the loess lines. The vertical lines denote the time at which the number of individuals in either arm fell below 10 (loess lines to the right of this time point, i.e. within the grey shaded area, are unreliable and should be ignored). Viral load is residualized for gender; plots of raw viral load are indistinguishable.
B. Bootstrap estimation of the correlation between the HLA-B*57 protective effect and time in the low (≤2.5) and high (>2.5) inhibitory score strata. Analysis was restricted to timepoints <500 days to ensure large cohort size. The protective effect of HLA-B*57 decreased significantly more rapidly in people with a low inhibitory score (P=0.00002, Wilcoxon Mann Whitney two-tailed).
What is the impact of iKIRs on the detrimental effect of HLA-B*35Px?
In the context of HTLV-1 infection, KIR2DL2 has also been shown to enhance the detrimental effects of the susceptibility allele HLA-B*54 (21). We therefore investigated the impact of iKIRs on the well-established detrimental association between HLA-B*35Px and the outcome of HIV-1 infection (26). In the IAVI cohort, HLA-B*35Px was associated with an increase in set-point viral load (Coeff = +0.35, P = 0.0007); but this effect was not consistently enhanced by iKIRs (Table S6). However, when we studied longitudinal viral dynamics, it could again be seen that the detrimental impact of HLA-B*35Px on viral load was progressively eroded in individuals with a low inhibitory score and maintained in individuals with a high inhibitory score (Fig. S3). This observation explains why iKIR enhancement of the HLA-B*35Px effect was not visible in the original outcome measure, early set-point viral load, which focuses on the first 3–9 months post-infection. Furthermore, bootstrap estimates of the correlation between time and the detrimental effect of HLA-B*35Px showed that the detrimental effect weakened in individuals with a low inhibitory score but was maintained in individuals with a high inhibitory score (P=6x10-7, P=5x10-9, P=9x10-18 for inhibitory score cut-offs of 2.0, 2.5, 2.75 respectively, Fig. S3). HLA-B*35Px did not influence the time to low CD4 count in the IAVI cohort (HR = 1.04, P = 0.8), so we did not investigate the interplay between iKIRs and HLA-B*35Px for this outcome measure.
Results are replicated in independent cohorts
We considered two independent replication cohorts: IAVI-partners and US.
IAVI-partners
IAVI-partners is a cohort of 315 HIV-1+ individuals from sub-Saharan Africa (the partners of the incident cases in the main IAVI cohort). In this cohort, HLA-B*57 showed a trend to reduce median viral load (Coeff = – 0.33, P = 0.06) and HLA-B*35Px was non-significant. Pooling IAVI and IAVI-partners provided a much larger cohort (N = 776). In this pooled cohort, HLA-B*57 was strongly protective (Coeff = – 0.43, P = 0.00003) and HLA-B*35Px showed a trend to be detrimental (Coeff = +0.16, P = 0.06). In both the unpooled and the pooled cohorts, the effects of HLA-B*57 and HLA-B*35Px were enhanced in people with a high inhibitory score (Table 2). Strikingly, HLA-B*35Px, which only showed a trend for susceptibility in the pooled cohort and nothing in the unpooled cohort, was highly significantly detrimental in people with a high inhibitory score.
Table 2. Functional iKIRs enhance the protective effect of HLA-B*57 and the detrimental effect of HLA-B*35Px in two independent replication cohorts.
Two independent replication cohorts were considered (IAVI-partners and US). In a separate analysis IAVI and IAVI-partners were pooled to increase cohort size. Cohorts were stratified into individuals with a low or high inhibitory score (two cut-offs considered) and into individuals with a low or high count of functional iKIR (only one cut-off possible). In each stratum the effects of HLA-B*57 and HLA-B*35Px were analyzed. HLA-B*35Px was not analyzed in IAVI partners as the HLA gene alone had no detectable impact (Coeff=+0.04 P=0.8).
Definitions as in Table 1. In all cohorts, all variables which had a significant impact on outcome were included in the model as covariates. A: linear regression, covariates: gender. B&C: linear regression, covariates: gender, cohort. D&F: Cox regression, covariates: age, subcohort. E&G: Cox regression, covariates: KIR3DL1+Bw4-80I+, age, ethnicity, subcohort. In all cohorts where functional KIR3DS1 (KIR3DS1+Bw4-80I+) was protective individuals with functional KIR3DS1 were removed prior to analysis to give a clean background (cohorts reported in B, C, E, G).
Note, the only balanced stratification provided by the “count of functional iKIRs” (namely ≤2 vs. >2) is the same as the stratification provided by the inhibitory score with a cut-off of 2.0. The inhibitory score has more diversity than the count and so typically more stratifications by inhibitory score than by count are possible. All cut-offs that gave a sufficiently balanced analysis were considered.
| A. IAVI partners HLA-B*57 | Outcome: log viral load | |||||||
| B57+ KIR- | B57+ KIR+ | Cohort numbers | ||||||
| Coeff | P value | Coeff | P value | B57+ KIR− | B57− KIR− | B57+ KIR+ | B57− KIR+ | |
| Inhibitory score (cut-off=2.0) | -0.37 | 0.3 | -0.33 | 0.1 | 10 | 122 | 31 | 152 |
| Inhibitory score (cut-off=2.5) | -0.21 | 0.4 | -0.46 | 0.06 | 18 | 159 | 23 | 115 |
| Count iKIR (cut-off=2.0) | -0.37 | 0.3 | -0.33 | 0.1 | 10 | 122 | 31 | 152 |
| B. Pooled IAVI & IAVI partners HLA-B*57 | Outcome: log viral load | |||||||
| B57+ KIR− | B57+ KIR+ | Cohort numbers | ||||||
| Coeff | P value | Coeff | P value | B57+ KIR− | B57− KIR− | B57+ KIR+ | B57− KIR+ | |
| Inhibitory score (cut-off=2.0) | -0.33 | 0.08 ∙ | -0.47 | 0.0002 *** | 27 | 330 | 64 | 420 |
| Inhibitory score (cut-off=2.5) | -0.35 | 0.01 * | -0.51 | 0.0007 *** | 47 | 439 | 44 | 311 |
| Count iKIR (cut-off=2.0) | -0.33 | 0.08 ∙ | -0.47 | 0.0002 *** | 27 | 330 | 64 | 420 |
| C. Pooled IAVI & IAVI partners HLA-B*35Px | Outcome: log viral load | |||||||
| B35Px+ KIR− | B35Px+ KIR+ | Cohort numbers | ||||||
| Coeff | P value | Coeff | P value | B35Px+ KIR− | B35Px− KIR− | B35Px+ KIR+ | B35Px− KIR+ | |
| Inhibitory score (cut-off=2.0) | -0.12 | 0.4 | +0.31 | 0.003 ** | 49 | 308 | 104 | 380 |
| Inhibitory score (cut-off=2.5) | +0.08 | 0.5 | +0.27 | 0.04 * | 87 | 399 | 66 | 289 |
| Count iKIR (cut-off=2.0) | -0.12 | 0.4 | +0.31 | 0.003 ** | 49 | 308 | 104 | 380 |
| D. US HLA-B*57 | African Americans | Outcome: time to low CD4 count (<200 cells/mm3) | ||||||
| B57+ KIR− | B57+ KIR+ | Cohort numbers | ||||||
| HR | P value | HR | P value | B57+ KIR− | B57− KIR− | B57+ KIR+ | B57− KIR+ | |
| Inhibitory score (cut-off=2.0) | 0.54 | 0.42 | 0.21 | 0.04 * | 8 | 92 | 23 | 89 |
| Inhibitory score (cut-off=2.5) | 0.54 | 0.41 | 0.22 | 0.04 * | 10 | 110 | 21 | 71 |
| Count iKIR (cut-off=2.0) | 0.51 | 0.37 | 0.21 | 0.04 * | 8 | 92 | 23 | 88 |
| E. US HLA-B*57 | All ethnicities | Outcome: time to low CD4 count (<200 cells/mm3) | ||||||
| B57+ KIR− | B57+ KIR+ | Cohort numbers | ||||||
| HR | P value | HR | P value | B57+ KIR− | B57− KIR− | B57+ KIR+ | B57− KIR+ | |
| Inhibitory score (cut-off=2.0) | 0.29 | 0.24 | 0.25 | 0.004 ** | 8 | 236 | 36 | 210 |
| Inhibitory score (cut-off=2.5) | 0.34 | 0.15 | 0.25 | 0.01 * | 11 | 271 | 33 | 175 |
| Count iKIR (cut-off=2.0) | 0.36 | 0.32 | 0.27 | 0.006 ** | 8 | 235 | 36 | 211 |
| F. US HLA-B*35Px | African Americans | Outcome: time to low CD4 count (<200 cells/mm3) | ||||||
| B35Px+ KIR− | B35Px+ KIR+ | Cohort numbers | ||||||
| HR | P value | HR | P value | B35Px+ KIR− | B35Px− KIR− | B35Px+ KIR+ | B35Px− KIR+ | |
| Inhibitory score (cut-off=2.0) | 0.22 | 0.15 | 2.06 | 0.05 ∙ | 11 | 89 | 34 | 78 |
| Inhibitory score (cut-off=2.5) | 0.82 | 0.68 | 2.08 | 0.08 ∙ | 24 | 96 | 21 | 71 |
| Count iKIR (cut-off=2.0) | 0.29 | 0.13 | 2.04 | 0.06 ∙ | 11 | 89 | 34 | 77 |
| G. US HLA-B*35Px | All ethnicities | Outcome: time to low CD4 count (<200 cells/mm3) | ||||||
| B35Px+ KIR− | B35Px+ KIR+ | Cohort numbers | ||||||
| HR | P value | HR | P value | B35Px+ KIR− | B35Px− KIR− | B35Px+ KIR+ | B35Px− KIR+ | |
| Inhibitory score (cut-off=2.0) | 1.54 | 0.22 | 2.57 | 0.001 ** | 20 | 224 | 44 | 202 |
| Inhibitory score (cut-off=2.5) | 1.89 | 0.02 * | 2.42 | 0.01 * | 37 | 245 | 27 | 181 |
| Count iKIR (cut-off=2.0) | 1.81 | 0.09 ∙ | 2.74 | 0.0004 *** | 20 | 223 | 44 | 203 |
US
The US is a cohort of 548 HIV-1+ individuals with known seroconversion date. 57.8% of subjects were white, 38.7% African American and 3.5% Hispanic/”Other”; we therefore stratified by ethnicity. Among whites, but not African Americans, both functional KIR3DS1 (KIR3DS1+Bw4-80I+) and functional KIR3DL1 (KIR3DL1+Bw4+) were protective. We therefore excluded functional KIR3DS1+ individuals and included functional KIR3DL1 as a covariate in the white stratum (exclusion of functional KIR3DS1+ individuals was not possible due to their high frequency). Although neither of these steps were necessary in the African American stratum, we repeated the analysis with both steps to check that the weak, insignificant effects of functional KIR3DS1 or functional KIR3DL1 were not responsible for our results.
Among whites, 15 HLA-B*57+ and 16 HLA-B*35Px+ individuals remained after exclusion of functional KIR3DS1+ individuals. Stratification of these groups left at most 8 people in each arm, so analysis was not possible. In contrast, among African Americans, there were 31 HLA-B*57+ and 45 HLA-B*35Px+ individuals. Analysis showed that both the protective effect of HLA-B*57 and the detrimental effect of HLA-B*35Px on the time to CD4 count < 200 cells/mm3 were consistently enhanced by functional iKIRs (Table 2). On pooling white, Hispanic/”Other” and African Americans (with ethnicity as a covariate), the protective effect of HLA-B*57 and the detrimental effect of HLA-B*35Px were still enhanced, although the enhancement of HLA-B*57 appeared weaker (Table 2) suggesting that the iKIR effect on HLA-B*57 was largely driven by the African American population. These findings were replicated when high-expressing KIR3DL1 alleles were included as a covariate (Table S7).
We conclude that in HIV-1 infection, in three independent cohorts, the effects of both protective and detrimental HLA alleles on viral load and disease progression are stronger in the presence of genes coding for iKIRs with their ligands.
iKIRs enhance HLA associations with asymptomatic status in HTLV-I infection
We previously reported that KIR2DL2 enhances HLA class I associations in HTLV-1 infection, but found little evidence for enhancement by other iKIRs (21). In contrast, the findings reported here, show that all iKIRs amenable to analysis enhanced HLA-B*57 and HLA-B*35Px disease associations in HIV-1 infection. We therefore re-examined the HTLV-1 data.
We confirmed that, in the HTLV-1-infected Kagoshima cohort, functional KIR2DL2 strongly enhanced the protective effect of HLA-C*08 and the detrimental effect of HLA-B*54 on the odds of the inflammatory disease HTLV-I-associated myelopathy/tropical spastic paraparesis (HAM/TSP). Previously, we reported that, unexpectedly, functional KIR2DL2 did not enhance the protective effect of HLA-A*02 (21). However, we recently found that only HLA-A*02:07, and not the more frequent HLA-A*02:01 and HLA-A*02:06, associates with protection in the Kagoshima cohort (Table S8). We therefore repeated the analysis at the level of 4-digit HLA resolution. Although subject numbers were low, we found evidence that KIR2DL2 did indeed enhance the protective effect of HLA-A*02:07 (halving the risk of developing HAM/TSP). Investigating the other iKIRs, we found that the low frequency of HLA-C2 precluded a meaningful analysis of functional KIR2DL1 (too few functional KIR2DL1+ individuals) and functional KIR2DL3 (too few functional KIR2DL3- individuals), explaining why we had not previously seen an effect for these other iKIRs. However, for functional KIR3DL1 both the positive and negative stratum were of sufficient size to permit analysis and there we found good evidence that functional KIR3DL1 enhanced both the detrimental effect of HLA–B*54 and the protective effects of HLA–A*02:07 and HLA–C*08. Furthermore, the effects of HLA–B*54, HLA–A*02:07 and HLA–C*08 were, in each case, strongest in individuals with a high inhibitory score and were weak and non-significant in individuals with a low inhibitory score (Table 3).
Table 3. Functional iKIRs enhance HLA class I associations in HTLV-1 infection.
The Kagoshima cohort was stratified into individuals with and without the functional iKIR of interest. In each stratum the effect of (A) HLA-B*54, (B) HLA-A*02:07 and (C) HLA-C*08 on HAM/TSP status was analyzed using multivariate logistic regression (with covariates age & gender). The odds ratio (OR), P values and cohort sizes are reported. An OR <1 represents a protective effect (reduced odds of HAM/TSP).
The detrimental effect of HLA-B*54 and the protective effects of HLA-A*02:07 and HLA-C*08 on HAM/TSP status were enhanced in people with strong functional KIR2DL2, functional KIR3DL1 and a high inhibitory score. There was an insufficient number of people with HLA-C2 to analyse functional KIR2DL1, weak functional KIR2DL2 and functional KIR2DL3. Similar results were obtained if we use factors instead of stratification. The ligands of KIR3DL1 are contentious with one paper reporting that KIR3DL1 binds HLA-A*23, -A*24 and –A*32 as well as HLA-Bw4 molecules (58). If we amended our definition of functional KIR3DL1 to include these HLA-A alleles then the conclusions were unchanged with the exception of HLA-C*08 where our conclusion was strengthened (magnitude of enhancement of protective effect of HLA-C*08 in the functional KIR3DL1-positive stratum is greater).
All cut-offs that gave a sufficiently balanced analysis were considered. The definitions of ‘low’ and ‘high’ iKIR counts and scores were reduced compared with other cohorts in this study to compensate for lower overall iKIR counts and scores in the Japanese cohort.
| KAGOSHIMA COHORT. OUTCOME: ODDS OF DISEASE (HAM/TSP) | ||||||||
| A. HLA-B*54 | ||||||||
| B54+ KIR− | B54+ KIR+ | Cohort numbers | ||||||
| OR | P value | OR | P value | B54+ KIR− | B54− KIR− | B54+ KIR+ | B54− KIR+ | |
| Functional 2DL2 (strong) | 1.69 | 0.17 | 12.14 | 0.004 ** | 62 | 231 | 20 | 79 |
| Functional 3DL1 | 1.98 | 0.13 | 4.45 | 0.007 ** | 48 | 147 | 34 | 163 |
| Inhibitory score (cut-off=1.5) | 1.29 | 0.7 | 5.33 | 0.0007 *** | 36 | 94 | 46 | 216 |
| Inhibitory score (cut-off=1.75) | 2.19 | 0.03 * | 9.35 | 0.06 ∙ | 73 | 253 | 9 | 57 |
| Count functional iKIR (cut-off=1) | 1.31 | 0.6 | 5.31 | 0.0007 *** | 36 | 93 | 46 | 217 |
| B. HLA-A*02:07 | ||||||||
| A0207+ KIR− | A0207+ KIR+ | Cohort numbers | ||||||
| OR | P value | OR | P value | A0207+ KIR− | A0207− KIR− | A0207+ KIR+ | A0207− KIR+ | |
| Functional 2DL2 (strong) | 0.33 | 0.04 * | 0.15 | 0.1 | 23 | 270 | 7 | 92 |
| Functional 3DL1 | 0.54 | 0.3 | 0.07 | 0.006 ** | 21 | 174 | 9 | 188 |
| Inhibitory score (cut-off=1.5) | 0.81 | 0.8 | 0.10 | 0.003 ** | 16 | 114 | 14 | 248 |
| Inhibitory score (cut-off=1.75) | Insufficient number of A02:07+KIR+ individuals | |||||||
| Count functional iKIR (cut-off=1) | 0.83 | 0.8 | 0.10 | 0.003 ** | 16 | 113 | 14 | 249 |
| C. HLA-C*08 | ||||||||
| C08+ KIR− | C08+ KIR+ | Cohort numbers | ||||||
| OR | P value | OR | P value | C08+ KIR− | C08− KIR− | C08+ KIR+ | C08− KIR+ | |
| Functional 2DL2 (strong) | 0.66 | 0.4 | 0.16 | 0.02 * | 43 | 250 | 13 | 86 |
| Functional 3DL1 | 0.53 | 0.19 | 0.48 | 0.3 | 39 | 156 | 17 | 180 |
| Inhibitory score (cut-off=1.5) | 1.09 | 0.9 | 0.32 | 0.02 * | 28 | 102 | 28 | 234 |
| Inhibitory score (cut-off=1.75) | 0.62 | 0.2 | 0.08 | 0.04 * | 49 | 277 | 7 | 59 |
| Count functional iKIR (cut-off=1) | 1.11 | 0.9 | 0.32 | 0.02 * | 28 | 101 | 28 | 235 |
The high inhibitory score stratum was heavily enriched for people with strong functional KIR2DL2 (P < 10-14 cut-off = 1.5, P < 10-15 cut-off = 1.75; Chi squared) so the KIR2DL2 enhancement and high iKIR score enhancement of HLA–B*54, –A*02:07 and –C*08 are not independent. However, functional KIR3DL1 is not a surrogate for functional KIR2DL2 (P = 0.86 Chi-squared), so the enhancement due to KIR2DL2 is independent of the enhancement due to KIR3DL1. We conclude that, although most iKIRs cannot be analysed (due to the low frequency of HLA-C2 alleles in Japan) there is evidence that, independent of KIR2DL2, another iKIR, KIR3DL1 and a high inhibitory score enhance HLA class I disease associations in HTLV-1 infection.
iKIRs enhance HLA associations with spontaneous clearance of HCV
We previously reported that functional KIR2DL2 enhances the protective effect of HLA-B*57 on spontaneous clearance of hepatitis C virus (HCV). Again, the earlier analysis found no evidence that other iKIRs showed similar behaviour (21). Reanalysis of this cohort confirmed the KIR2DL2 enhancement. It was not possible to analyze any other individual iKIR (insufficient numbers of HLA-B57+ functional KIR2DL1- and HLA-B57+ functional KIR3DL1+ individuals; and functional KIR2DL3 alone is protective (27), Table S2). We also found a strong enhancement of the HLA-B*57 effect in people with a high inhibitory score (Table S9). However, the high inhibitory score stratum was heavily enriched for people with strong functional KIR2DL2 for all three cut-offs considered (P < 2.10-16 in each case; Chi squared) and so the functional KIR2DL2 enhancement and strong iKIR enhancement of HLA-B*57 are not independent. We conclude that functional KIR2DL2 and a high inhibitory score enhance the protective effect of HLA-B*57 in HCV infection but it is not possible to test whether or not other iKIRs show the same behaviour.
Primary HLA class I associations are unlikely to be attributable to NK cells
In theory, an HLA class I disease association could be attributable to the role of these molecules as recognition targets for either CD8+ T cells (via somatically rearranged T cell antigen receptors) or NK cells (via germline-encoded KIRs). If a given HLA association is attributable to NK cells rather than CD8+ T cells, then other HLA class I molecules with similar KIR-binding properties should be similarly protective/detrimental (in the context of the relevant KIRs). This pretext forms the basis of classical KIR-HLA analyses (28–31). We investigated this possibility for each of the HLA class I alleles included in this study (HLA-B*57 and HLA-B*35Px in the HIV-1-infected IAVI cohort, HLA-A*02:07, HLA-C*08 and HLA-B*54 in the HTLV-1-infected Kagoshima cohort, and HLA-B*57 in the HCV-infected cohort). The results are detailed in the Supplementary Material & Table S10. We found no evidence to suggest that any of the HLA class I associations could be explained by a ligand preference for certain KIRs. This is consistent with existing evidence that these HLA class I associations are attributable to CD8+ T cells (32–37). Furthermore, simply considering iKIR:ligand pairs (without also considering the protective or detrimental HLA class I alleles) finds no significant protective or detrimental effect of any individual iKIR:ligand or of the count of functional iKIR or of the Inhibitory score in HIV-1, HTLV-1 or HCV (Table S2).
iKIR ligation enhances CD8+ T cell survival in vitro
Studies in mice and in vitro have shown that inhibitory NK receptors on T cells reduce activation-induced cell death (10–12). An iKIR-mediated enhancement of CD8+ T cell survival may therefore explain our immunogenetic findings. However, whilst our immunogenetics findings show presence of the HLA ligand is crucial (Fig. S1), the role of ligand in the in vitro survival studies is less clear. Two studies reported that iKIRs are associated with CD8+ T cell survival independent of ligand (13, 38), while a third indicated that KIR-ligation is required for the increased survival (15). To clarify and extend this work we investigated the impact of blocking iKIR ligation on CD8+ T cell survival in vitro. Three iKIR-expressing CD8+ T cell lines were established: two expressing KIR2DL3 and one expressing KIR3DL1, (Fig. S4). The lines were co-cultured with the HLA class-I-deficient B cell line 721:221 (henceforth 221) that was either untransfected, transfected to express a non-cognate HLA molecule (HLA-B57:01 or HLA-C04:01 in the case of KIR2DL3 and HLA-C03:04 or HLA-C04:01 in the case of KIR3DL1) or transfected to express a cognate HLA molecule (HLA-C03:04 in the case of KIR2DL3 and HLA-B57:01 in the case of KIR3DL1). Activation was induced with staphylococcal enterotoxin E (SEE). T cell survival was quantified in the presence of antibodies that block KIR (GL183, Dx9 (39)), or block HLA class I (Dx17 (40)); or an isotype control. The gating strategy is illustrated in Fig. S5. We found that blocking the iKIR-HLA interaction either by blocking KIR or by blocking HLA class I significantly decreased the count of live T cells compared to isotype control (median decrease = 40% and 48% respectively; Fig. 3A & 3B). Survival was similarly impaired in the absence of ligand (221 transfected with non-cognate HLA or untransfected; median decrease =57% and 53% respectively; Fig. 3A & 3B).
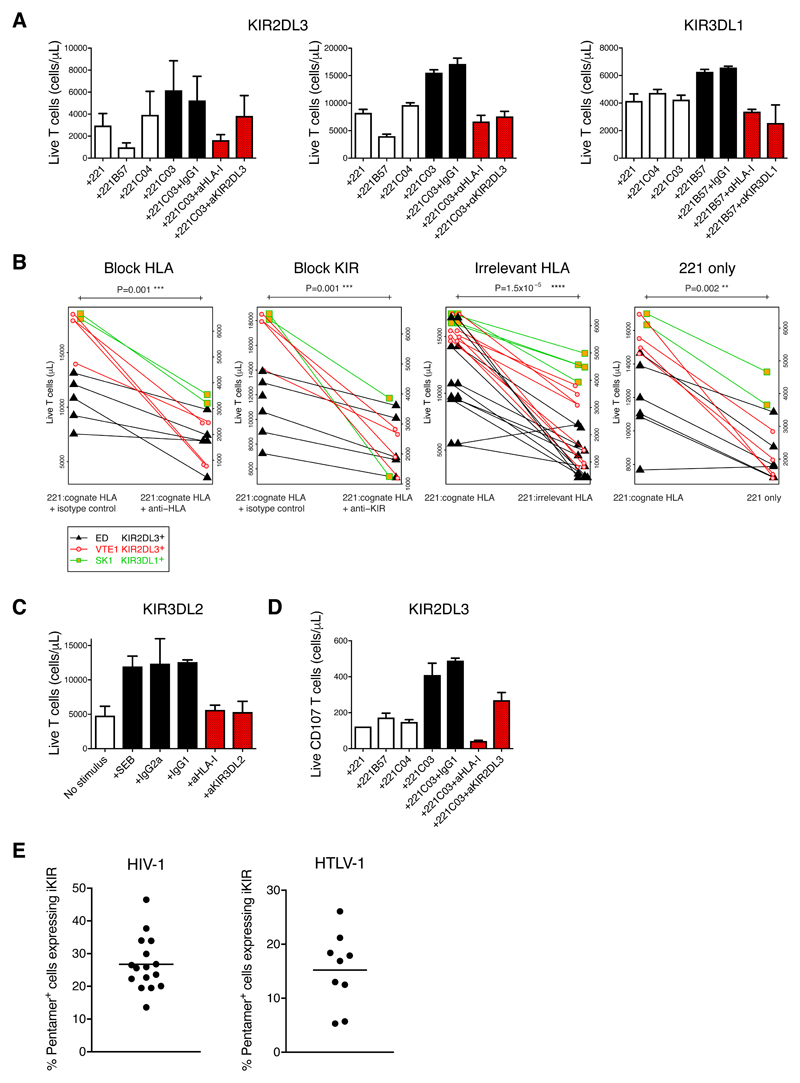
Figure 3. Impact of iKIR-HLA interaction on CD8+ T cell survival in vitro & iKIR expression ex vivo.
A. CD8+ T cell lines (plots from left ED, KIR2DL3+; VTE1 KIR2DL3+; SK1 KIR3DL1+) were cocultured with superantigen either without adding ligand for KIR i.e. with 221 cells or with 221 cells transfected to express irrelevant, non-cognate HLA (open bars); with ligand i.e. 221 transfected to express cognate HLA with or without isotype control antibody (black bars) and with KIR-HLA blocking i.e. 221 transfected to express cognate HLA with addition of antibody to block KIR (GL183, DX9) or to block HLA (DX17) (red bars). The count of live CD8+ T cells after 5 days is shown (mean± SE). ED: 5 experiments each consisting of 3 replicates; VTE1: 1 experiment, 4 replicates; SK1: 1 experiment, 2 replicates. It is consistently seen that there are greater numbers of viable T cells with isotype control than when the KIR-HLA interaction is blocked or when cognate ligand is absent.
B. Metaanalysis of survival of the 3 KIR-expressing CD8+ T cell lines. Block HLA. Coculture with 221 cells transfected to express cognate HLA with isotype control antibody compared to coculture with 221 cells transfected to express cognate HLA with anti-HLA antibody. Median difference = 48% P = 0.001. Block KIR. Coculture with 221 cells transfected to express cognate HLA with isotype control antibody compared to coculture with 221 cells transfected to express cognate HLA with anti-KIR antibody. Median difference = 40% P = 0.001. Irrelevant HLA. Coculture with 221 cells transfected to express cognate HLA compared to coculture with 221 cells transfected to express irrelevant, non-cognate HLA. Median difference = 57% P =1.5x10-5. 221 only. Coculture with 221 cells transfected to express cognate HLA compared to coculture with 221 cells. Median difference = 53% P = 0.002. Survival of VTE1 plotted on left hand y axis (for all conditions), ED and SK1 plotted on right hand y axis (for all conditions). We consistently found that absence of KIR:HLA ligation (either by blocking HLA, blocking KIR, adding a non-ligating HLA or not adding HLA) resulted in significantly decreased CD8+ T cell survival.
C. PBMCs from 3 HLA-B27+ individuals with ankylosing spondylitis were left untreated (open bars); cultured with superantigen (SEB) with or without isotype control antibody (black bars) or cultured with SEB with antibodies that block KIR3DL2 or homodimeric HLA-B27 (red bars). The count of live KIR3DL2+ CD4- T cells after 5 days is shown, (mean± SE). Blocking the iKIR-HLA interaction with specific antibodies decreased the count of live KIR3DL2+ T cells compared to isotype control (block KIR: median difference=63%; block HLA: median difference=50%).
D. The KIR2DL3-expressing CD8+ T cell line VTE1 was cultured as in A. The number of live CD8+ CD107a+ T cells was quantified by flow cytometry at day 5.
E. The fraction of pentamer+ CD8+ T cells expressing iKIRs (KIR2DL1, KIR2DL2/L3, KIR3DL1 or KIR3DL2) directly ex vivo is plotted for HLA-A2+ individuals infected with HIV-1 (N = 16) or HTLV-1 (N = 9).
Next, we extended this work to primary CD8+ T cells. Peripheral blood mononuclear cells (PBMCs) from three HLA-B*27+ individuals with ankylosing spondylitis, who have a high proportion of PBMCs expressing KIR3DL2 (6), were stimulated with staphylococcal enterotoxin B. KIR3DL2 binds HLA-B27 homodimers (6). Survival of KIR3DL2+ CD8+ T cells was quantified in the presence of antibodies that block KIR3DL2 (DX31), that block HLA-B27 homodimers (HD6 (41)) or an isotype control, Fig. S6. We found that blocking the iKIR-HLA interaction reduced the count of live KIR3DL2+ CD8+ T cells compared to isotype control (block KIR: median difference=63%; block HLA: median difference=50%; Fig. 3C).
Finally, iKIR ligation has been shown to inhibit CD8+ T cell killing under some conditions (42). To assess this possibility, which could mitigate the biological impact of enhanced survival, we quantified the impact of iKIR ligation on the mobilization of CD107a, a surrogate marker of degranulation. Greater numbers of live CD107a+ cells were observed in the presence of a functional KIR-HLA interaction after stimulation of the KIR2DL3-expressing CD8+ T cell line VTE1 with SEE (Fig. 3D). In summary, KIR ligation enhances the number of viable cells and the number of functional cells both for CD8+ T cell lines and for primary T cells in vitro.
Virus-specific CD8+ T cells express iKIR ex vivo
To probe the relevance of these findings, we quantified the fraction of virus-specific CD8+ T cells which expressed iKIRs (KIR2DL1, KIR2DL2/L3, KIR3DL1 and KIR3DL2) in PBMCs from HIV-1-positive and HTLV-1-positive donors. We found considerable proportions of iKIR-expressing pentamer+ CD8+ T cells in both groups (HIV-1: mean = 27%; HTLV-1: mean = 15%), Fig. 3E, Fig. S7, S8. On analysing the differentiation stage of virus-specific KIR-expressing CD8+ T cells we found, consistent with previous work on bulk CD8+ T cells (9), that approximately 40% of iKIR+ pentamer+ cells lay within the CD45RA+CD28- T effector memory revertant gate; the remainder were distributed approximately equally between CD45RA-CD28+ central memory T cells, CD45RA-CD28- effector memory T cells and CD45RA+CD28+ stem cell memory/naïve T cells, Fig. S9.
We did not investigate iKIR expression on pentamer+ CD8+ T cells from HCV-seropositive individuals because the frequency of HCV-specific CD8+ T cells in the blood is known to be extremely low (43, 44), due in part to T cell homing to the liver (45). Instead we investigated the relationship between functional iKIR expression and cell death ex vivo using bulk CD8+ and CD4+ T cells from HCV-seropositive individuals and healthy donors. We found that iKIR+ CD8+ and iKIR+ CD4+ T cells were both less likely to stain with annexin V in individuals where the cognate HLA ligand was present (Supplementary Results & Fig. S10).
Mathematical modelling can explain the enhancement of both protective and detrimental HLA associations
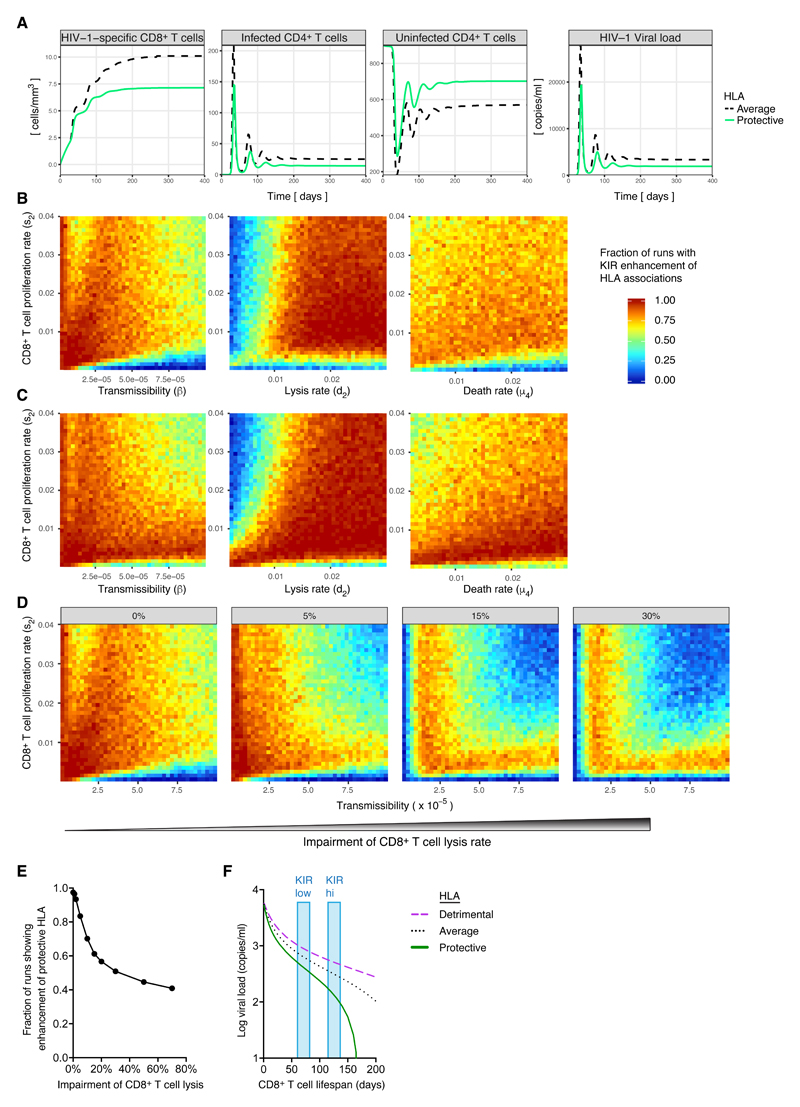
The hypothesis that iKIRs enhance HLA class I disease associations by enhancing T cell survival sounds reasonable, but closer inspection reveals a number of difficulties. First, iKIRs would be expected to increase the survival of all CD8+ T cells not just those restricted by protective HLA alleles. In an immunogenetic study, a protective HLA effect reflects the difference in viral control between average and protective HLA alleles, so it is not obvious how a non-specific increase in T cell survival would enhance protective HLA associations. Second, is it difficult to envisage how the same mechanism could also have the opposite effect and enhance detrimental HLA associations. Finally, there is evidence that iKIRs can suppress the lytic activity of CD8+ T cells under some conditions (14) (although this was not replicated in our work with CD107a). This raises another question: is it reasonable to suggest that in people with a high iKIR score, and potentially a higher level of functional inhibition, CD8+ T cell protection is actually enhanced? To address these questions we constructed a mechanistic mathematical model of host-virus dynamics for HIV-1. In this model, CD8+ T cells restricted by protective HLA class I molecules were assumed to lyse targets more efficiently than average, whereas CD8+ T cells restricted by detrimental HLA class I molecules were assumed to lyse targets less efficiently than average. In the first instance, we assumed that the only effect of iKIRs was to enhance T cell survival; in later analyses, we assumed that iKIRs also impaired CD8+ T cell lysis. We generated 100,000 random parameter combinations by sampling from the physiological range for each parameter (Table S11) and solved the system numerically (Fig. 4A). For each parameter combination we recorded whether the protective effect (decrease in set-point viral load associated with possession of a protective HLA class I molecule compared with an average HLA class I molecule) was enhanced in people with a high number of functional iKIRs. We found that the majority of runs (97%) showed iKIR-mediated enhancement of protective HLA class I associations (Fig. 4B). Next, we repeated the analysis for detrimental HLA class I molecules. This showed that detrimental HLA class I-associated effects were also enhanced across a wide range of parameter space (Fig. 4C). Comparison of the region of parameter space where protective HLA class I associations were enhanced and where detrimental HLA class I associations were enhanced showed that there was broad overlap; i.e. there exist parameter regions where both protective and detrimental HLA class I associations were simultaneously enhanced. Additionally, when iKIRs were assumed to inhibit T cell lysis, enhancement of protective and detrimental HLA class I effects was decreased, but the majority of runs still showed enhancement (Fig. 4D & E). Further analysis showed these results were robust to both structural and parametric changes in the model (Supplementary Methods).
Figure 4. Simulations of iKIR enhancement of HLA class I disease associations in HIV-1.
A. Dynamics of HIV-1-specific CD8+ T cells, infected and uninfected CD4+ T cells and viral load during HIV-1 infection, for a representative hypothetical in silico subject with average (dashed black line) or protective (green solid line) HLA class I alleles.
B&C. Fraction of runs that result in an iKIR-mediated enhancement of the protective (B) or detrimental (C) HLA effect as a function of virus transmissibility (β) and CD8+ T cell proliferation rate (s2) (left); as a function of CD8+ T cell lysis rate (d2) and proliferation rate (s2) (center) and as a function of CD8+ T cell death rate (μ4) and proliferation rate (s2) (right).
D&E. Distribution (D) and frequency (E) of runs with enhancement when we consider an impairment in the CD8+ T cell lysis rate due to iKIRs (percentage reduction from left to right: 0%, 5%, 15% and 30%).
F. Viral load as a function of CD8+ T cell lifespan (1/μ4) for a hypothetical person with detrimental (magenta dotted), average (black dashed) or protective (green solid) HLA class I alleles. It can be seen that as the survival of CD8+ T cells increases both the effect of protective HLA (difference in viral load between average and protective HLA) and the effect of detrimental HLA (difference in viral load between average and detrimental HLA) increases. Compare for example, a hypothetical person with a high number of iKIR-HLA pairs (blue bar marked KIR hi) and a hypothetical person with a low number of iKIR-HLA pairs (blue bar marked KIR low).
To understand how an increase in CD8+ T cell survival can enhance both protective and detrimental HLA class I associations, we simulated viral load as a function of CD8+ T cell lifespan for three representative individuals: one with average HLA class I molecules, one with a protective HLA class I molecule and one with a detrimental HLA class I molecule (Fig. 4F). On seeing this figure, the explanation immediately becomes obvious. In the limit, when CD8+ T cell lifespan is very short, it makes little difference whether an individual possesses average, protective or detrimental alleles; everyone has a similar viral load independent of their HLA genotype. In an immunogenetics study this would manifest as an absence of association between either protective or detrimental alleles and outcome. As CD8+ T cell lifespan is increased (for example by iKIRs) then the “quality” of the CD8+ T cell response becomes important, and a large difference in viral load emerges between people with average, protective and detrimental HLA class I alleles. In an immunogenetics study this would manifest as large protective and detrimental effects i.e. as CD8+ T cell lifespan increases both protective and detrimental effects are enhanced. Although this model does not prove the hypothesis it does show that the hypothesis predicts our findings and provides a plausible and intuitive explanation for a seemingly contradictory observation.
Discussion
In this study, we conducted an immunogenetic analysis of three independent cohorts of HIV-1-infected individuals to investigate the impact of iKIR genes on well-established HLA class I disease associations. We found that genes for iKIRs consistently enhanced the protective effect of HLA-B*57 and the detrimental effect of HLA-B*35Px on viral load and progression to low CD4 count. The result was critically dependent on the presence of the KIR ligand. A reanalysis of two other persistent viral infections, HCV and HTLV-1, found consistent results: all four HLA class I associations studied were enhanced in people with a high inhibitory score. We previously reported this effect as an enhancement associated with one particular iKIR (KIR2DL2) (21). In HCV infection, enhanced HLA class I associations were detected for individuals with KIR2DL2 and individuals with a high inhibitory score, but it was impossible to determine which effect was the driver. However, in HTLV-1 infection, new data made it possible to show that, independent of KIR2DL2, another iKIR (functional KIR3DL1) enhanced both protective and detrimental HLA class I disease associations. The probability of seeing this result by chance is P = 10-16; furthermore the HIV-1 findings were replicated in two independent cohorts. Collectively, these data suggest that all iKIRs behave in a similar way and that KIR2DL2 is not functionally unique (though its prevalence and distribution make its effects easier to detect in some cohorts; a hypothesis we suggested previously (21)). The iKIRs are closely related, being the product of gene duplication (46), so it is gratifying that we find a unifying picture in which all iKIRs enhance HLA class I associations in a similar fashion.
We investigated the underlying mechanism using three approaches: (i) analysis of the duration of HLA class I-mediated effects in the HIV-1-infected cohort; (ii) in vitro survival assays combined with ex vivo KIR expression analysis and (iii) mathematical modelling. In the HIV-1 cohort, the phenomenon of iKIR-mediated enhancement appears to be related to the longevity of the CD8+ T cell response as both the HLA-B*57 protective effect and the HLA-B*35Px detrimental effect were eroded over time in individuals with a low inhibitory score but maintained in individuals with a high inhibitory score; the difference in rates between these two groups was consistently highly significant. In vitro, we found that blocking the KIR:HLA interaction significantly reduced cultured and primary T cell survival and functional CD107a+ T cell survival following activation with superantigen. In addition, presence of the KIR ligand was associated with a decrease in annexin V binding on CD4+ and CD8+ T cells expressing iKIR directly ex vivo. We also found substantial levels of iKIR expression on virus-specific CD8+ T cells among PBMCs isolated from HIV-1 and HTLV-1-seropositive individuals. Finally, in a mathematical model of host-virus dynamics in HIV-1 infection, we predicted that if iKIRs increase CD8+ T cell lifespan (either directly or indirectly) then we would see an enhancement of HLA class I associations, consistent with the immunogenetic data. Further, the model provides an explanation for how increasing CD8+ T cell lifespan can simultaneously have two seemingly contradictory effects: enhancing both protective and detrimental associations. The modelling shows that if CD8+ T cells have longer lifespans (e.g. due to the presences of iKIRs) then the quality of the CD8+ T cell response becomes more important and so differences between protective, average and detrimental HLA alleles become more apparent i.e. both protective and detrimental effects are enhanced.
Our conclusions are that iKIRs enhance HLA class I associations and that this can be explained by an increase in CD8+ T cell survival in the presence of functional iKIRs. A number of ways in which iKIRs can increase CD8+ T cell survival have been described. These can be broadly divided into “direct” (iKIRs on T cells affect T cell lifespan) and “indirect” (iKIRs on other cell populations, e.g. NK cells, affect T cell lifespan); these pathways are summarised in Fig. S11. Our finding that, in vitro, iKIR expression on CD8+ T cells is directly associated with increased survival does not preclude a role for iKIR on NK cells in indirectly enhancing CD8+ T cell survival. Both effects may be occurring and increasing CD8+ T cell survival and both are consistent with our immunogenetics analysis, the longitudinal study and the mathematical modelling.
The iKIR-ligand binding groups which we use in our definition of “functional iKIR” are simplistic. Incomplete knowledge of how different alleles and different peptides affect binding and signalling precludes a more sophisticated definition. Nevertheless, these simple groupings have proved very powerful in other studies (23, 27, 29, 31, 47, 48). With the definition of inhibitory score used we saw clear and reproducible results. This suggests that the inhibitory score is a meaningful metric (indeed the odds of seeing all the results by chance is <10-23). It is worth noting that the score is used only to split the cohorts in half, so second order changes to the calculation of the score will not necessarily change the results; indeed we found that our conclusions were robust to quite dramatic changes to the score (e.g. whether KIR3DL2 with classical class I ligands was included or not). This is because a person’s score can change by a large amount and they will still remain in the same strata. Indeed, whether we simply count KIR-ligand pairs or use the inhibitory score has little impact on the conclusions. Another limitation of this work is that the survival assays were only conducted in vitro. The strong associations between possession of combinations of iKIR/HLA genes and clinical outcomes such as time to low CD4 count suggest that these effects are important in vivo but direct in vivo assays were not performed. Murine in vivo assays provide limited information since Ly49 receptors differ significantly from KIR (this includes profound differences in structure and in the pattern of expression on T cells). Human in vivo studies are limited by ethical considerations but would be an important next step in this work.
HCV, HIV-1 and HTLV-1 are all characterized by continuous high level viral replication and chronic immune activation which has been associated with CD8+ T cell exhaustion and activation-induced T cell death (49–52). Chronic immune cell activation is also associated with increased KIR expression on T cells (5–9, 42, 53). iKIR ‘rescuing’ of activated CD8+ T cells may be particularly important in this setting. Most KIR-HLA genetic associations are not replicated across viral infections e.g. KIR3DS1 with HLA-Bw4 is protective in the context of HIV-1 but not HTLV-1 (54); this is difficult to understand unless a very strong peptide dependence is postulated. In contrast, we report that all iKIRs enhance HLA class I associations in all three persistent virus infections we have studied (HIV-1, HCV and HTLV-1), which suggests a universality that is often missing from KIR studies.
The iKIR-HLA receptor-ligand system also has similarities to the programmed death 1 (PD-1)–programmed death-ligand 1/2 system. Both iKIR and PD-1 are inhibitory receptors which interfere with proximal T cell receptor signalling and are upregulated in the context of chronic viral infection and on tumour infiltrating lymphocytes. PD-1 and iKIR are both expressed on late stage differentiated memory T cells, typically in a mutually exclusive manner (55). Blockade or activation of iKIR would thus affect a different population of T cells compared with PD-1 activation/blockade. These observations, combined with the demonstration herein that the iKIR-HLA receptor-ligand system is clinically significant, suggest novel and synergistic targets for therapeutic immune checkpoint blockade.
Materials and Methods
(Full Methods available in Supplementary Information)
Study design
The aim of this study was to investigate whether iKIR have a clinically significant impact on the human CD8+ T cell response. We studied the impact of iKIR genotype on 6 well-documented HLA class I disease associations in 3 viral infections (Table 4) in 5 independent cohorts. The work was extended and interpreted by mechanistic mathematical modelling, in vitro T cell survival assays and ex vivo analysis of iKIR expression.
Table 4. HLA class I disease associations studied.
| Protective | Detrimental | |
|---|---|---|
| HIV-1 | HLA-B*57 | HLA-B*35Px |
| HTLV-1 | HLA-A*0207, HLA-C*08 | HLA-B*54 |
| HCV | HLA-B*57 |
Ethics statement
The immunogenetics study was approved by the NHS Research Ethics Committee (13/WS/0064) and the Imperial College Research Ethics Committee (ICREC_11_1_2). Informed consent was obtained at the study sites from all individuals. For the cellular work, written informed consent was obtained from all participants in accordance with the Declaration of Helsinki. Research was conducted under the governance of the Imperial College Healthcare NHS Trust Tissue Bank, approved by the UK National Research Ethics Service (09/H0606/106, 15/SC/0089).
Immunogenetics Cohorts
IAVI is a prospective cohort of treatment-naïve HIV-1 seroconverters from sub-Saharan Africa, identified and followed under Protocol C of the International AIDS Vaccine Initiative (IAVI) (22, 56). Individuals who possessed both KIR3DS1 and its putative ligand HLA-Bw4-80I were excluded from the analysis to eliminate the potentially confounding effects of functional KIR3DS1. The cohort comprised individuals with time to low CD4 count information (N=491) and individuals with early viral load information (N=461). IAVI partners is an HIV-1 seroprevalent cohort (N=315). US is a cohort of HIV-1 seroconverters (N=548). Kagoshima is a cohort of HTLV-1-infected subjects (N=392). HCV comprises four sub-cohorts of HCV-seropositive subjects (N=782).
Regression
The impact of genotype on outcome was analyzed using multivariate linear, logistic and Cox regression in R v3.0.1 (57). Potentially confounding covariates were identified and included in the analysis (listed in Supplementary Methods). All reported P values are two tailed.
In vitro T cell survival
CD8+ T Cell Lines
Three iKIR-expressing CD8+ T cell lines were established from the peripheral blood of healthy volunteers (Fig S4). The lines were co-cultured with the HLA class I-deficient B cell line 221, either untransfected; or transfected to express a non-cognate HLA molecule (HLA-B57:01 or HLA-C04:01 in the case of KIR2DL3, and HLA-C03:04 or HLA-C04:01 in the case of KIR3DL1); or transfected to express a cognate HLA molecule (HLA-C03:04 in the case of KIR2DL3, and HLA-B57:01 in the case of KIR3DL1). Cells were activated with superantigen staphylococcal enterotoxin E. The count of live T cells at day 5 was enumerated using CountBright Absolute Counting Beads (Invitrogen) in the presence of antibodies to block KIR (GL183, Dx9 (39)) or to block HLA class I (Dx17 (40)) or as an isotype control. The gating strategy is illustrated in Fig. S5.
Primary T cells
PBMC from three HLA-B*27+ individuals with ankylosing spondylitis were stimulated with staphylocococcal enterotoxin B. Survival of KIR3DL2+ CD4- T cells was quantified in the presence of isotype controls or antibodies that block KIR3DL2 (DX31) or homodimeric HLA-B27 (HD6 (41)).
Pentamer staining
Cryopreserved PBMC samples from HLA-A*02+ HIV-1+ (N=16) and HLA-A*02+ HTLV-I+ (N=9) subjects were thawed and stained sequentially with a fixable amine dye to exclude dead cells, fluorochrome-labelled peptide-HLA class I pentamers (ILKEPVHGV-A02:01, SLYNTVATL-A02:01 or LTFGWCFKL-A02:01 for HIV-1 and LLFGYPVYP-A02:01 for HTLV-1) and directly conjugated monoclonal antibodies specific for CD3, CD8, CD28, CD45RA, KIR2DL1, KIR2L2/L3, KIR3DL1 and KIR3DL2. The fraction of iKIR+ pentamer+ CD8+ T cells was enumerated within each phenotypic compartment defined by the expression of CD28 and CD45RA.
Supplementary Material
One sentence summary.
Inhibitory killer-cell immunoglobulin-like receptors (iKIRs) help maintain the CD8+ T cell response to chronic viral infections; in contrast to many reported iKIR-disease associations, these observations are remarkably wide-ranging, being seen for all iKIRs and all 3 viral infections studied.
Acknowledgements
We are grateful to Robert Busch, Aileen Rowan, Graham Taylor, Dimitra Peppa, Persephone Borrow and the Center for HIV/AIDS Vaccine Immunology AI067854.
Funding
B.A. is a Wellcome Trust (WT) Investigator (103865Z/14/Z) and is also funded by the Medical Research Council (MRC) (J007439 and G1001052), the European Union Seventh Framework Programme (317040, QuanTI) and Leukemia and Lymphoma Research (15012). M.S. received financial support from the Brazilian National Council for Scientific Development (206435/2014-2). C.h.R. is funded by the WT Institutional Strategic Support Fund (105609/Z/14/Z). E.C.Y.W is funded by the WT (090323/Z/09/Z) and the MRC (MR/L018373/L, MR/P001602/1). J.T. and J.A.T. are funded by the MRC, the National Institutes of Health (NIH), the Cambridge Biomedical Research Centre and the European Research Council under the European Union's Horizon 2020 Research and Innovation Programme (695551). C.L.T is supported by NIH R01 DA13324. D.A.P. is a WT Senior Investigator (100326Z/12/Z). The UK HCV cohort is funded by the MRC (MR/M019829/1). The Hemophilia Growth and Development Study is supported by the National Institute of Child Health and Human Development (R01-HD-41224). Data presented in this manuscript were collected by the ALIVE Study funded by the National Institute on Drug Abuse (U01-DA-036297, R01-DA-12568, K24-AI118591). This work was partially funded by IAVI with the generous support of USAID and other donors; a full list of IAVI donors is available at www.iavi.org. This project has been funded in whole or in part with federal funds from the Frederick National Laboratory for Cancer Research under contract HHSN261200800001E and by the Intramural Research Program of the NIH, Frederick National Laboratory, Center for Cancer Research. The content of this publication is the responsibility of the authors and does not necessarily reflect the views or policies of the Department of Health and Human Services, USAID or the US Government, nor does the mention of trade names, commercial products, or organizations imply endorsement by the US Government.
Footnotes
IAVI protocol C Investigators
Eduard J Sanders1,2, Omu Anzala3, Anatoli Kamali4, Etienne Karita5, William Kilembe6, Mubiana Inambao6, Shabir Lakhi6, Susan Allen7, Eric Hunter7, Vinodh Edward8, Pat Fast9, Matt A Price9,10, Jill Gilmour11, Jianming Tang12, Fran Priddy9, Mary H Latka8, Linda-Gail Bekker13
1Centre for Geographic Medicine–Coast/KEMRI, Kenya; 2University of Oxford, UK; 3Kenya AIDS Vaccine Institute–Institute of Clinical Research, Kenya; 4Medical Research Council/Uganda Virus Research Institute, Uganda Research Unit on AIDS, Uganda; 5Project San Francisco, Rwanda; 6Zambia Emory Research Project, Zambia; 7Emory University, Georgia, USA; 8The Aurum Institute, South Africa; 9IAVI, New York, USA; 10Department of Epidemiology and Biostatistics, University of California San Francisco, USA; 11IAVI Human Immunology Laboratory, Imperial College, UK; 12Ryals Public Health Building, University of Alabama, USA; 13Desmond Tutu HIV Foundation, South Africa.
Author Contributions
L.B. performed the analysis, contributed to interpretation, performed the statistical analysis and wrote the manuscript. B.D. and S.K. performed the experimental work. M.S. performed the mathematical modelling. C.h.R., J.J., J.A.T., J.T., J.M., A.S., K.T., K.L, K.L.M., J.G. and D.M. helped with experimental work. D.A.P. helped with experimental work and edited the manuscript. E.C.Y.W. and J.F contributed materials. Y.Q. and M.P.M. helped with statistical analysis. I.P.C.I, C.T., J.A., G.K., S.M.D., S. B., S.I.K. and J.J.G. provided cohort data. M.C. helped with analysis, contributed to interpretation and edited the manuscript. B.A. conceived and directed the project, obtained funding, performed the analysis, contributed to interpretation, performed the statistical analysis and wrote the manuscript.
Competing Interests
The authors declare that they have no competing interests.
References
(ref 59 onwards appear only in SI)
- 1.Parham P. Immunogenetics of killer cell immunoglobulin-like receptors. Mol Immunol. 2005;42:459–462. doi: 10.1016/j.molimm.2004.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 3.Cook LB, Elemans M, Rowan AG, Asquith B. HTLV-1: persistence and pathogenesis. Virology. 2013;435:131–140. doi: 10.1016/j.virol.2012.09.028. [DOI] [PubMed] [Google Scholar]
- 4.Crouse J, Xu HC, Lang PA, Oxenius A. NK cells regulating T cell responses: mechanisms and outcome. Trends Immunol. 2015;36:49–58. doi: 10.1016/j.it.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Anfossi N, Pascal V, Vivier E, Ugolini S. Biology of T memory type 1 cells. Immunol Rev. 2001;181:269–278. doi: 10.1034/j.1600-065x.2001.1810123.x. [DOI] [PubMed] [Google Scholar]
- 6.Bowness P, Ridley A, Shaw J, Chan AT, Wong-Baeza I, Fleming M, Cummings F, McMichael A, Kollnberger S. Th17 cells expressing KIR3DL2+ and responsive to HLA-B27 homodimers are increased in ankylosing spondylitis. J Immunol. 2011;186:2672–2680. doi: 10.4049/jimmunol.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonorino P, Leroy V, Dufeu-Duchesne T, Tongiani-Dashan S, Sturm N, Pernollet M, Vivier E, Zarski JP, Marche PN, Jouvin-Marche E. Features and distribution of CD8 T cells with human leukocyte antigen class I-specific receptor expression in chronic hepatitis C. Hepatology. 2007;46:1375–1386. doi: 10.1002/hep.21850. [DOI] [PubMed] [Google Scholar]
- 8.Poon K, Montamat-Sicotte D, Cumberbatch N, McMichael AJ, Callan MF. Expression of leukocyte immunoglobulin-like receptors and natural killer receptors on virus-specific CD8+ T cells during the evolution of Epstein-Barr virus-specific immune responses in vivo. Viral Immunol. 2005;18:513–522. doi: 10.1089/vim.2005.18.513. [DOI] [PubMed] [Google Scholar]
- 9.Bjorkstrom NK, Beziat V, Cichocki F, Liu LL, Levine J, Larsson S, Koup RA, Anderson SK, Ljunggren HG, Malmberg KJ. CD8 T cells express randomly selected KIRs with distinct specificities compared to NK cells. Blood. 2012;120:3455–3465. doi: 10.1182/blood-2012-03-416867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roger J, Chalifour A, Lemieux S, Duplay P. Cutting edge: Ly49A inhibits TCR/CD3-induced apoptosis and IL-2 secretion. J Immunol. 2001;167:6–10. doi: 10.4049/jimmunol.167.1.6. [DOI] [PubMed] [Google Scholar]
- 11.Ugolini S, Arpin C, Anfossi N, Walzer T, Cambiaggi A, Forster R, Lipp M, Toes RE, Melief CJ, Marvel J, Vivier E. Involvement of inhibitory NKRs in the survival of a subset of memory-phenotype CD8+ T cells. Nat Immunol. 2001;2:430–435. doi: 10.1038/87740. [DOI] [PubMed] [Google Scholar]
- 12.Ugolini S, Vivier E. Regulation of T cell function by NK cell receptors for classical MHC class I molecules. Curr Opin Immunol. 2000;12:295–300. doi: 10.1016/s0952-7915(00)00090-x. [DOI] [PubMed] [Google Scholar]
- 13.Young NT, Uhrberg M, Phillips JH, Lanier LL, Parham P. Differential expression of leukocyte receptor complex-encoded Ig-like receptors correlates with the transition from effector to memory CTL. J Immunol. 2001;166:3933–3941. doi: 10.4049/jimmunol.166.6.3933. [DOI] [PubMed] [Google Scholar]
- 14.Alter G, Rihn S, Streeck H, Teigen N, Piechocka-Trocha A, Moss K, Cohen K, Meier A, Pereyra F, Walker B, Altfeld M. Ligand-independent exhaustion of killer immunoglobulin-like receptor-positive CD8+ T cells in human immunodeficiency virus type 1 infection. J Virol. 2008;82:9668–9677. doi: 10.1128/JVI.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gati A, Guerra N, Gaudin C, Da Rocha S, Escudier B, Lecluse Y, Bettaieb A, Chouaib S, Caignard A. CD158 Receptor Controls Cytotoxic T-Lymphocyte Susceptibility to Tumor-Mediated Activation-Induced Cell Death by Interfering with Fas Signaling. Cancer Res. 2003;63:7475–7482. [PubMed] [Google Scholar]
- 16.Cerboni C, Zingoni A, Cippitelli M, Piccoli M, Frati L, Santoni A. Antigen-activated human T lymphocytes express cell-surface NKG2D ligands via an ATM/ATR-dependent mechanism and become susceptible to autologous NK-cell lysis. Blood. 2007;110:606–615. doi: 10.1182/blood-2006-10-052720. [DOI] [PubMed] [Google Scholar]
- 17.Soderquest K, Walzer T, Zafirova B, Klavinskis LS, Polic B, Vivier E, Lord GM, Martin-Fontecha A. Cutting edge: CD8+ T cell priming in the absence of NK cells leads to enhanced memory responses. J Immunol. 2011;186:3304–3308. doi: 10.4049/jimmunol.1004122. [DOI] [PubMed] [Google Scholar]
- 18.Waggoner SN, Cornberg M, Selin LK, Welsh RM. Natural killer cells act as rheostats modulating antiviral T cells. Nature. 2011;481:394–398. doi: 10.1038/nature10624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SH, Kim KS, Fodil-Cornu N, Vidal SM, Biron CA. Activating receptors promote NK cell expansion for maintenance, IL-10 production, and CD8 T cell regulation during viral infection. J Exp Med. 2009;206:2235–2251. doi: 10.1084/jem.20082387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peppa D, Gill US, Reynolds G, Easom NJ, Pallett LJ, Schurich A, Micco L, Nebbia G, Singh HD, Adams DH, Kennedy PT, et al. Up-regulation of a death receptor renders antiviral T cells susceptible to NK cell-mediated deletion. J Exp Med. 2013;210:99–114. doi: 10.1084/jem.20121172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seich Al Basatena NK, Macnamara A, Vine AM, Thio CL, Astemborski J, Usuku K, Osame M, Kirk GD, Donfield SM, Goedert JJ, Bangham CR, et al. KIR2DL2 enhances protective and detrimental HLA class I-mediated immunity in chronic viral infection. PLOS Pathog. 2011;7:e1002270. doi: 10.1371/journal.ppat.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.https://www.iavi.org/.
- 23.Martin MP, Qi Y, Gao X, Yamada E, Martin JN, Pereyra F, Colombo S, Brown EE, Shupert WL, Phair J, Goedert JJ, et al. Innate partnership of HLA-B and KIR3DL1 subtypes against HIV-1. Nat Genet. 2007;39:733–740. doi: 10.1038/ng2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bashirova AA, Martin-Gayo E, Jones DC, Qi Y, Apps R, Gao X, Burke PS, Taylor CJ, Rogich J, Wolinsky S, Bream JH, et al. LILRB2 interaction with HLA class I correlates with control of HIV-1 infection. PLOS Genet. 2014;10:e1004196. doi: 10.1371/journal.pgen.1004196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goodridge JP, Burian A, Lee N, Geraghty DE. HLA-F and MHC class I open conformers are ligands for NK cell Ig-like receptors. J Immunol. 2013;191:3553–3562. doi: 10.4049/jimmunol.1300081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, Kaslow R, Buchbinder S, Hoots K, O'Brien SJ. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 27.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, Cox S, et al. HLA and NK Cell Inhibitory Receptor Genes in Resolving Hepatitis C Virus Infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 28.Hirayasu K, Ohashi J, Kashiwase K, Hananantachai H, Naka I, Ogawa A, Takanashi M, Satake M, Nakajima K, Parham P, Arase H, et al. Significant association of KIR2DL3-HLA-C1 combination with cerebral malaria and implications for co-evolution of KIR and HLA. PLOS Pathog. 2012;8:e1002565. doi: 10.1371/journal.ppat.1002565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vince N, Bashirova AA, Lied A, Gao X, Dorrell L, McLaren PJ, Fellay J, Carrington M. HLA class I and KIR genes do not protect against HIV type 1 infection in highly exposed uninfected individuals with hemophilia A. J Infect Dis. 2014;210:1047–1051. doi: 10.1093/infdis/jiu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ayo CM, Frederico FB, Siqueira RC, Brandao de Mattos CC, Previato M, Barbosa AP, Murata FH, Silveira-Carvalho AP, de Mattos LC. Ocular toxoplasmosis: susceptibility in respect to the genes encoding the KIR receptors and their HLA class I ligands. Sci Rep. 2016;6 doi: 10.1038/srep36632. 36632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin MP, Gao X, Lee J-H, Nelson GW, Detels R, Goedert JJ, Buchbinder S, Hoots K, Vlahov D, Trowsdale J, Wilson M, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat Genet. 2002;31:429–434. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 32.Bangham CR. CTL quality and the control of human retroviral infections. Eur J Immunol. 2009;39:1700–1712. doi: 10.1002/eji.200939451. [DOI] [PubMed] [Google Scholar]
- 33.Altfeld M, Kalife ET, Qi Y, Streeck H, Lichterfeld M, Johnston MN, Burgett N, Swartz ME, Yang A, Alter G, Yu XG, et al. HLA Alleles Associated with Delayed Progression to AIDS Contribute Strongly to the Initial CD8(+) T Cell Response against HIV-1. PLOS Med. 2006;3:e403. doi: 10.1371/journal.pmed.0030403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Borghans JAM, Mølgaard A, de Boer RJ, Keşmir C. HLA Alleles Associated with Slow Progression to AIDS Truly Prefer to Present HIV-1 p24. PLOS One. 2007;2:e920–e920. doi: 10.1371/journal.pone.0000920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, Ndhlovu ZM, Liu D, Porter LC, Fang JW, Darko S, Brockman MA, Miura T, Brumme ZL, Schneidewind A, Piechocka-Trocha A, et al. TCR clonotypes modulate the protective effect of HLA class I molecules in HIV-1 infection. Nat Immunol. 2012;13:691–700. doi: 10.1038/ni.2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kosmrlj A, Read EL, Qi Y, Allen TM, Altfeld M, Deeks SG, Pereyra F, Carrington M, Walker BD, Chakraborty AK. Effects of thymic selection of the T-cell repertoire on HLA class I-associated control of HIV infection. Nature. 2010;465:350–354. doi: 10.1038/nature08997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim AY, Kuntzen T, Timm J, Nolan BE, Baca MA, Reyor LL, Berical AC, Feller AJ, Johnson KL, Schulze zur Wiesch J, Robbins GK, et al. Spontaneous control of HCV is associated with expression of HLA-B 57 and preservation of targeted epitopes. Gastroenterology. 2011;140:686–696.e681. doi: 10.1053/j.gastro.2010.09.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chwae Y-J, Chang MJ, Park SM, Yoon H, Park H-J, Kim SJ, Kim J. Molecular Mechanism of the Activation-Induced Cell Death Inhibition Mediated by a p70 Inhibitory Killer Cell Ig-Like Receptor in Jurkat T Cells. J Immunol. 2002;169:3726–3735. doi: 10.4049/jimmunol.169.7.3726. [DOI] [PubMed] [Google Scholar]
- 39.Binyamin L, Alpaugh RK, Hughes TL, Lutz CT, Campbell KS, Weiner LM. Blocking NK Cell Inhibitory Self-Recognition Promotes Antibody-Dependent Cellular Cytotoxicity in a Model of Anti-Lymphoma Therapy. J Immunol. 2008;180:6392–6401. doi: 10.4049/jimmunol.180.9.6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Andrea A, Chang C, Phillips JH, Lanier LL. Regulation of T cell lymphokine production by killer cell inhibitory receptor recognition of self HLA class I alleles. J Exp Med. 1996;184:789–794. doi: 10.1084/jem.184.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Payeli SK, Kollnberger S, Marroquin Belaunzaran O, Thiel M, McHugh K, Giles J, Shaw J, Kleber S, Ridley A, Wong-Baeza I, Keidel S, et al. Inhibiting HLA-B27 homodimer-driven immune cell inflammation in spondylarthritis. Arthritis Rheum. 2012;64:3139–3149. doi: 10.1002/art.34538. [DOI] [PubMed] [Google Scholar]
- 42.van der Veken LT, Campelo MD, van der Hoorn MA, Hagedoorn RS, van Egmond HM, van Bergen J, Willemze R, Falkenburg JH, Heemskerk MH. Functional analysis of killer Ig-like receptor-expressing cytomegalovirus-specific CD8+ T cells. J Immunol. 2009;182:92–101. doi: 10.4049/jimmunol.182.1.92. [DOI] [PubMed] [Google Scholar]
- 43.Willberg C, Barnes E, Klenerman P. HCV immunology-Death and the maiden T cell. Cell Death Differ. 2003;10:S39–S47. doi: 10.1038/sj.cdd.4401122. [DOI] [PubMed] [Google Scholar]
- 44.Björkström NK, Gonzalez VD, Malmberg K-J, Falconer K, Alaeus A, Nowak G, Jorns C, Ericzon B-G, Weiland O, Sandberg JK, Ljunggren H-G. Elevated Numbers of FcγRIIIA+ (CD16+) Effector CD8 T Cells with NK Cell-Like Function in Chronic Hepatitis C Virus Infection. The Journal of Immunology. 2008;181:4219–4228. doi: 10.4049/jimmunol.181.6.4219. [DOI] [PubMed] [Google Scholar]
- 45.Sung PS, Racanelli V, Shin E-C. CD8(+) T-Cell Responses in Acute Hepatitis C Virus Infection. Front Immunol. 2014;5:266. doi: 10.3389/fimmu.2014.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Norman PJ, Parham P. Complex interactions: The immunogenetics of human leukocyte antigen and killer cell immunoglobulin-like receptors. Semin Hematol. 2005;42:65–75. doi: 10.1053/j.seminhematol.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 47.Ahlenstiel G, Martin MP, Gao X, Carrington M, Rehermann B. Distinct KIR/HLA compound genotypes affect the kinetics of human antiviral natural killer cell responses. J Clin Invest. 2008;118:1017–1026. doi: 10.1172/JCI32400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nakimuli A, Chazara O, Hiby SE, Farrell L, Tukwasibwe S, Jayaraman J, Traherne JA, Trowsdale J, Colucci F, Lougee E, Vaughan RW, et al. A KIR B centromeric region present in Africans but not Europeans protects pregnant women from pre-eclampsia. Proc Natl Acad Sci U S A. 2015;112:845–850. doi: 10.1073/pnas.1413453112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Day CL, Kaufmann DE, Kiepiela P, Brown JA, Moodley ES, Reddy S, Mackey EW, Miller JD, Leslie AJ, DePierres C, Mncube Z, et al. PD-1 expression on HIV-specific T cells is associated with T-cell exhaustion and disease progression. Nature. 2006;443:350–354. doi: 10.1038/nature05115. [DOI] [PubMed] [Google Scholar]
- 50.Bengsch B, Seigel B, Ruhl M, Timm J, Kuntz M, Blum HE, Pircher H, Thimme R. Coexpression of PD-1, 2B4, CD160 and KLRG1 on exhausted HCV-specific CD8+ T cells is linked to antigen recognition and T cell differentiation. PLOS Pathog. 2010;6:e1000947. doi: 10.1371/journal.ppat.1000947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kozako T, Yoshimitsu M, Fujiwara H, Masamoto I, Horai S, White Y, Akimoto M, Suzuki S, Matsushita K, Uozumi K, Tei C, et al. PD-1/PD-L1 expression in human T-cell leukemia virus type 1 carriers and adult T-cell leukemia/lymphoma patients. Leukemia. 2009;23:375–382. doi: 10.1038/leu.2008.272. [DOI] [PubMed] [Google Scholar]
- 52.Asquith B, Zhang Y, Mosley AJ, de Lara CM, Wallace DL, Worth A, Kaftantzi L, Meekings K, Griffin GE, Tanaka Y, Tough DF, et al. In vivo T lymphocyte dynamics in humans and the impact of human T-lymphotropic virus 1 infection. Proc Natl Acad Sci U S A. 2007;104:8035–8040. doi: 10.1073/pnas.0608832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huard B, Karlsson L. KIR expression on self-reactive CD8+ T cells is controlled by T-cell receptor engagement. Nature. 2000;403:325–328. doi: 10.1038/35002105. [DOI] [PubMed] [Google Scholar]
- 54.O'Connor GM, Seich Al Basatena NK, Olavarria V, MacNamara A, Vine A, Ying Q, Hisada M, Galvao-Castro B, Asquith B, McVicar DW. In contrast to HIV, KIR3DS1 does not influence outcome in HTLV-1 retroviral infection. Hum Immunol. 2012;73:783–787. doi: 10.1016/j.humimm.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Duraiswamy J, Ibegbu CC, Masopust D, Miller JD, Araki K, Doho GH, Tata P, Gupta S, Zilliox MJ, Nakaya HI, Pulendran B, et al. Phenotype, function, and gene expression profiles of programmed death-1(hi) CD8 T cells in healthy human adults. J Immunol. 2011;186:4200–4212. doi: 10.4049/jimmunol.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kamali A, Price MA, Lakhi S, Karita E, Inambao M, Sanders EJ, Anzala O, Latka MH, Bekker LG, Kaleebu P, Asiki G, et al. Creating an African HIV clinical research and prevention trials network: HIV prevalence, incidence and transmission. PLOS One. 2015;10:e0116100. doi: 10.1371/journal.pone.0116100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.R: A language and environment for statistical computing. 2014 URL http://www.R-project.org/
- 58.Stern M, Ruggeri L, Capanni M, Mancusi A, Velardi A. Human leukocyte antigens A23, A24, and A32 but not A25 are ligands for KIR3DL1. Blood. 2008;112:708–710. doi: 10.1182/blood-2008-02-137521. [DOI] [PubMed] [Google Scholar]
- 59.Goedert JJ, Biggar RJ, Winn DM, Mann DL, Byar DP, Strong DM, DiGioia RA, Grossman RJ, Sanchez WC, Kase RG, et al. Decreased helper T lymphocytes in homosexual men. I. Sexual contact in high-incidence areas for the acquired immunodeficiency syndrome. Am J Epidemiol. 1985;121:629–636. doi: 10.1093/aje/121.5.629. [DOI] [PubMed] [Google Scholar]
- 60.Goedert JJ, Kessler CM, Aledort LM, Biggar RJ, Andes WA, White GC, Drummond JE, Vaidya K, Mann DL, Eyster ME. A prospective study of Human Immunodeficiency Virus type 1 infection and the development of AIDS in subjects with hemophilia. N Engl J Med. 1989;321:1141–1148. doi: 10.1056/NEJM198910263211701. [DOI] [PubMed] [Google Scholar]
- 61.Buchbinder SP, Katz MH, Hessol NA, O'Malley PM, Holmberg SD. Long-term HIV-1 infection without immunologic progression. AIDS. 1994;8:1123–1128. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 62.Vlahov D, Graham N, Hoover D, Flynn C, Bartlett JG, Margolick JB, Lyles CM, Nelson KE, Smith D, Holmberg S, Farzadegan H. Prognostic indicators for AIDS and infectious disease death in HIV-infected injection drug users: plasma viral load and CD4+ cell count. JAMA. 1998;279:35–40. doi: 10.1001/jama.279.1.35. [DOI] [PubMed] [Google Scholar]
- 63.Jeffery KJ, Usuku K, Hall SE, Matsumoto W, Taylor GP, Procter J, Bunce M, Ogg GS, Welsh KI, Weber JN, Lloyd AL, et al. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-I-associated myelopathy. Proc Natl Acad Sci U S A. 1999;96:3848–3853. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vlahov D, Anthony JC, Munoz A, Margolick J, Nelson KE, Celentano DD, Solomon L, Polk BF. The ALIVE study, a longitudinal study of HIV-1 infection in intravenous drug users: description of methods and characteristics of participants. NIDA Res Monogr. 1991;109:75–100. [PubMed] [Google Scholar]
- 65.Hilgartner MW, Donfield SM, Willoughby A, Contant CF, Evatt BL, Gomperts ED, Hoots WK, Jason J, Loveland KA, McKinlay SM. Hemophilia growth and development study. Design, methods, and entry data. The American Journal of Pediatric Hematology/Oncology. 1993;15:208–218. doi: 10.1097/00043426-199305000-00009. [DOI] [PubMed] [Google Scholar]
- 66.Thio CL, Gao X, Goedert JJ, Vlahov D, Nelson KE, Hilgartner MW, O'Brien SJ, Karacki P, Astemborski J, Carrington M, Thomas DL. HLA-Cw*04 and hepatitis C virus persistence. J Virol. 2002;76:4792–4797. doi: 10.1128/JVI.76.10.4792-4797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prentice HA, Price MA, Porter TR, Cormier E, Mugavero MJ, Kamali A, Karita E, Lakhi S, Sanders EJ, Anzala O, Amornkul PN, et al. Dynamics of viremia in primary HIV-1 infection in Africans: insights from analyses of host and viral correlates. Virology. 2014;449:254–262. doi: 10.1016/j.virol.2013.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jiang W, Johnson C, Simecek N, Lopez-Alvarez MR, Di D, Trowsdale J, Traherne JA. qKAT: a high-throughput qPCR method for KIR gene copy number and haplotype determination. Genome Med. 2016;8:99. doi: 10.1186/s13073-016-0358-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Therneau TM. A Package for Survival Analysis. 2015 http://CRAN.R-project.org/package=survival.
- 70.Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; New York: 2000. [Google Scholar]
- 71.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27:361–368. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 72.Shimizu Y, DeMars R. Production of human cells expressing individual transferred HLA-A,-B,-C genes using an HLA-A,-B,-C null human cell line. J Immunol. 1989;142:3320–3328. [PubMed] [Google Scholar]
- 73.Li MY, Shu H. Multiple stable periodic oscillations in a mathematical model of CTL response to HTLV-I infection. Bull Math Biol. 2011;73:1774–1793. doi: 10.1007/s11538-010-9591-7. [DOI] [PubMed] [Google Scholar]
- 74.Lim AG, Maini PK. HTLV-I infection: a dynamic struggle between viral persistence and host immunity. J Theor Biol. 2014;352:92–108. doi: 10.1016/j.jtbi.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 75.Gomez-Acevedo H, Li MY, Jacobson S. Multistability in a model for CTL response to HTLV-I infection and its implications to HAM/TSP development and prevention. Bull Math Biol. 2010;72:681–696. doi: 10.1007/s11538-009-9465-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ, Ge D, De Luca A, Martinez-Picado J, Wolinsky SM, Martinson JJ, et al. Copy number variation of KIR genes influences HIV-1 control. PLOS Biol. 2011;9:e1001208. doi: 10.1371/journal.pbio.1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Martin MP, Naranbhai V, Shea PR, Qi Y, Ramsuran V, Vince N, Gao X, Thomas R, Brumme ZL, Carlson JM, Wolinsky SM, et al. Killer cell immunoglobulin-like receptor 3DL1 variation modifies HLA-B*57 protection against HIV-1. J Clin Invest. 2018 doi: 10.1172/JCI98463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Saunders PM, Pymm P, Pietra G, Hughes VA, Hitchen C, O'Connor GM, Loiacono F, Widjaja J, Price DA, Falco M, Mingari MC, et al. Killer cell immunoglobulin-like receptor 3DL1 polymorphism defines distinct hierarchies of HLA class I recognition. J Exp Med. 2016;213:791–807. doi: 10.1084/jem.20152023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.O'Connor GM, Vivian JP, Gostick E, Pymm P, Lafont BA, Price DA, Rossjohn J, Brooks AG, McVicar DW. Peptide-Dependent Recognition of HLA-B*57:01 by KIR3DS1. J Virol. 2015;89:5213–5221. doi: 10.1128/JVI.03586-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Taner SB, Pando MJ, Roberts A, Schellekens J, Marsh SGE, Malmberg K-J, Parham P, Brodsky FM. Interactions of NK Cell Receptor KIR3DL1*004 with Chaperones and Conformation-Specific Antibody Reveal a Functional Folded State As Well As Predominant Intracellular Retention. J Immunol. 2011;186:62–72. doi: 10.4049/jimmunol.0903657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Norman PJ, Hollenbach JA, Nemat-Gorgani N, Guethlein LA, Hilton HG, Pando MJ, Koram KA, Riley EM, Abi-Rached L, Parham P. Co-evolution of human leukocyte antigen (HLA) class I ligands with killer-cell immunoglobulin-like receptors (KIR) in a genetically diverse population of sub-Saharan Africans. PLOS Genet. 2013;9:e1003938. doi: 10.1371/journal.pgen.1003938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Althaus CL, De Boer RJ. Dynamics of immune escape during HIV/SIV infection. PLOS Comput Biol. 2008;4:e1000103. doi: 10.1371/journal.pcbi.1000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Conway JM, Perelson AS. Post-treatment control of HIV infection. Proc Natl Acad Sci U S A. 2015;112:5467–5472. doi: 10.1073/pnas.1419162112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li F, Ma W, Jiang Z, Li D. Stability and Hopf Bifurcation in a Delayed HIV Infection Model with General Incidence Rate and Immune Impairment. Comput Math Methods Med. 2015;2015 doi: 10.1155/2015/206205. 206205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.van Deutekom HW, Wijnker G, de Boer RJ. The rate of immune escape vanishes when multiple immune responses control an HIV infection. J Immunol. 2013;191:3277–3286. doi: 10.4049/jimmunol.1300962. [DOI] [PubMed] [Google Scholar]
- 86.Batorsky R, Sergeev RA, Rouzine IM. The route of HIV escape from immune response targeting multiple sites is determined by the cost-benefit tradeoff of escape mutations. PLOS Comput Biol. 2014;10:e1003878. doi: 10.1371/journal.pcbi.1003878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muller V, Maree AF, De Boer RJ. Small variations in multiple parameters account for wide variations in HIV-1 set-points: a novel modelling approach. Proc Biol Sci. 2001;268:235–242. doi: 10.1098/rspb.2000.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kaslow RA, Carrington M, Apple R, Park L, Munoz A, Saah AJ, Goedert JJ, Winkler C, O'Brien SJ, Rinaldo C, Detels R, et al. Influence of combinations of human major histocompatibility complex genes on the course of HIV-1 infection. Nat Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 89.Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med. 2003;54:535–551. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 90.Perelson AS. Modelling viral and immune system dynamics. Nat Rev Immunol. 2002;2:28–36. doi: 10.1038/nri700. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.