Abstract
An outstanding challenge of Epigenome-Wide Association Studies (EWAS) performed in complex tissues is the identification of the specific cell-type(s) responsible for the observed differential DNA methylation. Here, we present a novel statistical algorithm, called CellDMC, which is able to identify not only differentially methylated positions, but also the specific cell-type(s) driving the differential methylation. We provide extensive validation of CellDMC on in-silico mixtures of DNA methylation data generated with different technologies, as well as on real mixtures from epigenome-wide-association and cancer epigenome studies. We demonstrate how CellDMC can achieve over 90% sensitivity and specificity in scenarios where current state-of-the-art methods fail to identify differential methylation. By applying CellDMC to a smoking EWAS performed in buccal swabs, we identify differentially methylated positions occurring in the epithelial compartment, which we validate in smoking-related lung cancer. CellDMC may help towards the identification of causal DNA methylation alterations in disease.
Introduction
Somatic DNA methylation (DNAm) alterations have been shown to reflect cumulative exposure to environmental disease risk factors 1, and may contribute to disease risk by modifying cellular phenotypes 2,3. One major source of DNAm variation which may hamper the identification of DNAm alterations predisposing or driving disease in Epigenome-Wide Association Studies (EWAS) 4, is cell-type heterogeneity 5,6. While statistical methods for identifying differentially methylated cytosines (DMCs) in heterogeneous tissues have been developed 7–14, none allow the identification of the specific cell-types responsible for the observed differential methylation 10. Indeed, the only existing tool that can help pinpoint differentially methylated cell-types is an enrichment analysis method for cell-type specific DNase hypersensitive sites that is performed on a relatively large list of DMCs 15, not allowing for individual CpGs to be ranked according to their likelihood of differential methylation (DM) in individual cell-types. Here, we present and validate CellDMC, a novel statistical algorithm that can identify interactions between phenotype and the proportions of underlying cell-types in the tissue, thus allowing for the detection of differentially methylated cytosines in individual cell-types (DMCTs).
Results
Detection of DMCTs with CellDMC: rationale and statistical framework
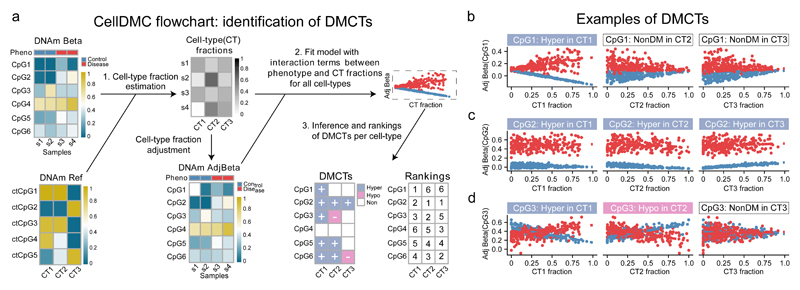
We reasoned that identification of DMCTs is possible within the same linear regression framework normally used to identify DMCs, by further inclusion of statistical interaction terms between phenotype and estimated cell-type fractions (Fig.1a, Supplementary Fig.1): intuitively, if a DMC is specific to one of the cell-types in the mixture, the observed differential methylation (DM) should be most prominent when the DM analysis is restricted to samples that contain the highest fraction of that cell-type (Fig.1b). CellDMC analyses the DNAm patterns of interactions of all cell-types in the mixture to infer DMCTs and their directionality of change (i.e. hyper or hypomethylation) (Fig.1, Online Methods, Supplementary Fig.1). Importantly, CellDMC also works in scenarios where all cell-types are uni-directionally differentially methylated to a similar degree (Fig.1c). CellDMC can also handle more complex scenarios, where a DMC occurs in two cell-types with opposite directionality (i.e. hypomethylated in one and hypermethylated in another) (Fig.1d), and which may not be identifiable by current state-of-the-art DMC calling algorithms (see later).
Figure 1. Identification of differentially methylated cell-types (DMCTs) using CellDMC.
a) For a given DNAm data matrix, CellDMC uses a reference DNAm matrix encompassing major cell-types (CTs) in the tissue of interest, to estimate cell-type fractions in each sample, subsequently adjusting the DNAm data matrix for these estimated fractions. It then fits statistical models adjusting for cell-type fractions, that include interaction terms between the phenotype and estimated cell-type fractions to identify DMCs in specific cell-types (DMCTs). These can then be ranked according to statistical significance in each cell-type. b,c,d) Scatterplots of adjusted beta-values against cell-type fraction for 3 different types of DMCTs. b) A DMCT (CpG1) which is hypermethylated in cell-type CT1 but not in cell-types CT2 and CT3. c) A DMCT (CpG2) hypermethylated in all three cell-types. d) A DMCT (CpG3) occurring in two cell-types (CT1 & CT2) but with DNAm changes occurring in opposite direction (hypermethylated in CT1, hypomethylated in CT2).
To estimate cell-type fractions, CellDMC applies our previously validated EpiDISH algorithm 16 in an iterative hierarchical procedure, called HEpiDISH 17, which leads to improved cell-type fraction estimates in complex tissues by recognizing that cell-types are naturally arranged along a developmental tree (Online Methods, Supplementary Data 1). In the context of epithelial tissues, HEpiDISH accomplishes this by using two distinct DNAm reference matrices, a primary reference matrix for the estimation of total epithelial, total fibroblast and total immune-cell (IC) fractions, and a separate, secondary, non-overlapping DNAm reference for the estimation of underlying IC cell subtype fractions. Justification and proof that this procedure works is given elsewhere 17, while additional proof that HEpiDISH works is provided here in the context of identifying DMCTs. In total, HEpiDISH can estimate cell-type fractions for 9-10 different cell-types commonly found in epithelial tissues, including epithelial cells, fibroblasts and 7 major types of immune-cells (Online Methods) 17. For tissues like breast, which contain a relatively large fraction of adipocytes 18, the primary reference also includes a representative DNAm profile for fat cells 17.
Validation of CellDMC on in-silico mixtures
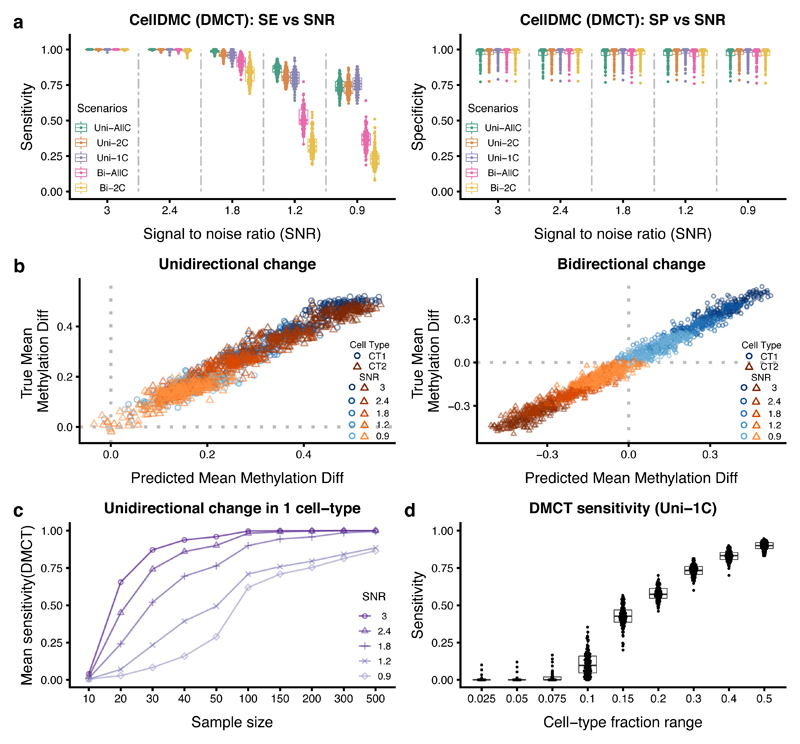
To test CellDMC’s ability to detect DMCTs, we first considered a number of different simulation scenarios, where we mixed together real DNAm profiles representing epithelial, fibroblast and immune cell types, in known mixing proportions, introducing DMCs in one, two or all cell-types, with parallel or opposite directionality, and over a range of different signal-to-noise ratios (SNRs) (Online Methods, Supplementary Table 1). Our choice of SNRs are conservative, corresponding to differences in average DNAm within an affected cell-type of approximately 0.4 to 0.5 (i.e. 40 to 50%) in the high SNR regime (SNR~3) down to changes as small as 0.1 (i.e. 10%) for the low SNR regime (SNR<1) (Supplementary Fig.2). In effect, the low SNR regime corresponds to a scenario where only 10% of say single epithelial cells exhibit a common DNAm change. Considering first scenarios where the underlying cell-type fractions do not change appreciably between cases and controls, CellDMC obtained a high sensitivity and specificity to detect DMCTs (Fig.2a). For instance, even when SNR<1, CellDMC obtained a sensitivity above 0.75 for all scenarios where the differential methylation is unidirectional, regardless of the number of affected cell-types (Fig.2a). For scenarios with bi-directional changes, CellDMC could detect DMCTs with a sensitivity above 0.75 as long as the SNR was approximately 1.8 or above (Fig.2a). In line with these high sensitivity values, we observed excellent agreement between the predicted DNAm difference in the underlying DMCTs with the true DNAm difference, irrespective of SNR and whether DNAm alterations were uni-or-bi-directional (Fig.2b). Importantly, CellDMC allows DMCTs to be ranked in each cell-type according to their statistical significance with the ranking highly correlated with true effect size, as required (Supplementary Fig.2). All of the above results were largely unchanged if cell-type fractions were allowed to vary between cases and controls (Supplementary Fig.3), or if DMCs only occurred in one of the underlying immune cell subtypes (compared to the previous case when the DMCs were common to all ICs) (Supplementary Fig.4, 5, 6). CellDMC also attained high sensitivity and specificity if in-silico mixtures were generated purely from IC subtypes (without admixture by epithelial and fibroblast cells), representing the more common scenario of EWAS performed in whole/peripheral blood (Supplementary Fig.6). CellDMC performed similarly if the phenotype is continuously valued as opposed to binary, or for binary heterogeneous phenotypes with cases defined by a bi-modal distribution, as observed for instance in cancer (Supplementary Data 2). We further verified that CellDMC is robust to the choice of cell-type fraction estimation algorithm (Supplementary Data 3). Underlying this robustness, we observed that results were also unchanged if cell-type fraction point estimates were perturbed using either theoretical or empirical confidence intervals (Supplementary Data 4 and 5).
Figure 2. Validation of CellDMC on simulated data.
a) Sensitivity (SE) and specificity (SP) (y-axis) of CellDMC to detect DMCTs in five different scenarios and for five different SNRs (x-axis). The scenarios correspond to uni-directional DNAm changes being unique to one cell-type (Uni-1C), shared by two cell-types (Uni-2C) or occurring in all cell-types (Uni-AllC), and to bi-directional changes (i.e hypermethylated and hypomethylated) in two cell-types (Bi-2C) or all cell-types (Bi-AllC). SNR values range from SNR~3 (mean DNAm difference in affected cell-types of approx. 0.4 to 0.5) to SNR < 1 (mean DNAm difference ~ 0.1). Each box contains 100 data points from 100 Monte-Carlo runs. Boxes indicate 25th and 75th centiles. Whiskers extend to the largest values no further than 1.5IQR from these centiles where IQR=inter-quartile range. b) Scatterplots of the true DNAm difference in affected cell-types (y-axis) versus the predicted DNAm difference from CellDMC (x-axis) for the uni-directional (left panel) and bi-directional (right panel) cases. Data points are shown for 100 DMCs, 2 affected cell-types (CT1 and CT2) and for five SNR levels. c) Sensitivity to detect DMCTs as a function of total sample size (numbers of cases and controls are each 100) for five different SNR values in the Uni-1C scenario. Each data point represents the mean over 100 Monte-Carlo runs. d) Sensitivity to detect DMCTs as a function of the cell-fraction range exhibited by the affected cell-type in a mixture of three cell-types. Each boxplot derives from 100 data points (100 Monte-Carlo runs). Boxes indicate 25th and 75th centiles. Whiskers extend to the largest values no further than 1.5IQR from these centiles where IQR=inter-quartile range.
We also assessed CellDMC’s power and specificity to detect DMCTs as a function of sample size, cell-type complexity and missing cell-types. At a sample size of 100 controls and 100 cases, power of CellDMC was always greater than 0.75 if the SNR was larger than 1.8 (corresponding to DNAm changes greater than 0.2 in individual cell-types) (Fig.2c, Supplementary Fig.7). Of note, as long as the fraction of the affected cell-type is distributed uniformly across samples (thus exhibiting a fairly large range), CellDMC’s sensitivity was robust to cell-type complexity, i.e. to the number K of cell-types in the mixture, although due to limitations on data availability, we could only assess this up to a value of K=7 (Supplementary Fig.8). We also assessed performance as a function of the variance of the altered cell-type fraction, estimating that sensitivity of CellDMC would remain over 50% as long as the range of fractions exhibited by the affected cell-type is higher than 0.2 (Fig.2d, Supplementary Fig.8). CellDMC was also robust if one major unaffected cell-type was missing from the reference DNAm matrix, while reduction in sensitivity was relatively marginal if an altered cell-type was missing from the reference (Supplementary Fig.9). In the case of blood tissue we simulated a realistic scenario, where cell-type fractions were modelled as observed in real blood EWAS but with one cell-type (CD8+ T-cells) missing from our reference: sensitivities to detect DMCTs in CD4+ T-cells were reduced at most by only 15% (Supplementary Fig.9). Thus, these data support the view that CellDMC can reliably detect DMCTs in a wide range of different realistic scenarios. However, we also expect CellDMC’s power to be strongly influenced by cell-type complexity, in line with the fact that DNAm references become less reliable as K increases, and also because specific cell-type proportions may exhibit small variation or fall below detection levels.
CellDMC outperforms state-of-the-art reference-based and reference-free DMC calling methods
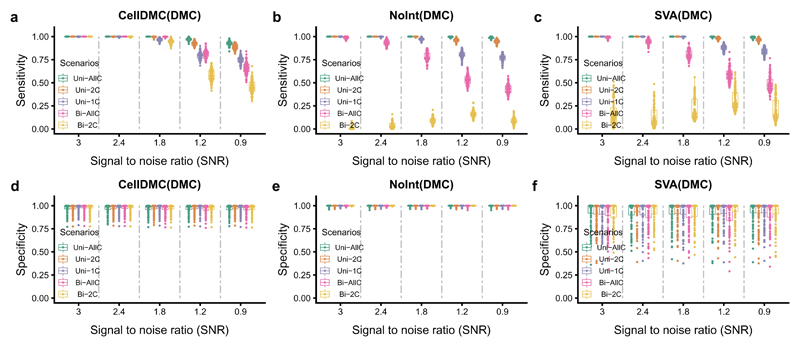
Next, we compared CellDMC to state-of-the-art reference-based and reference-free DMC calling methods. Although CellDMC outperformed competing methods in effectively all considered scenarios, differences were especially striking when DNAm changes were bi-directional (Fig.3, Supplementary Fig.10, 11). For instance, when DNAm changes occurred in two of the underlying cell-types with opposite directionality (denoted “Bi-2C”), conventional reference-based DMC calling that only includes the estimated cell-type fractions as covariates 7 would not resolve the underlying DMCs, resulting in sensitivities well below 0.25 (Fig.3, Supplementary Fig.10). In contrast, CellDMC attained sensitivities that ranged from over 0.95 for high SNRs to around 0.5 for low SNRs. If bi-directional changes occurred in more than 2 cell-types (denoted “Bi-AllC”), CellDMC also attained significantly higher sensitivity values than conventional non-interaction based DMC calling (Fig.3, Supplementary Fig.10). Similar improvements in sensitivity of CellDMC over state-of-the-art reference-free methods (SVA & RefFreeEWAS) 8,19,20 were also observed, which were again particularly prominent in bi-directional scenarios affecting two or more cell-types (Fig.3, Supplementary Fig.11). In line with the sensitivity results, CellDMC’s specificity was also higher and more stable than that of competing methods (Fig.3). Thus, these data demonstrate that CellDMC can attain sensitivity and specificity values close to 100% in scenarios where current state-of-the-art methods would fail to detect DMCs.
Figure 3. Benchmarking of CellDMC to state-of-the-art reference based and reference-free DMC calling algorithms.
a-c) From left to right, plots of the sensitivity of CellDMC, of conventional reference-based DMC calling (which includes cell-type fractions only as covariates without interaction-terms, denoted “NoInt”), and of the reference-free method SVA, against the SNR for five different differentially methylated cell-type scenarios where mean cell-type fractions are not different between cases and controls. Because the competing methods can only detect DMCs, and not DMCTs, we compare the methods in terms of their sensitivity to detect DMCs. Results are for 100 Monte-Carlo runs, where in each run 200 in-silico mixture samples were simulated (100 “control” and 100 “disease”). d-f) As a-c), but now for the specificity. Boxes indicate 25th and 75th centiles. Whiskers extend to the largest values no further than 1.5IQR from these centiles where IQR=inter-quartile range.
Validation of CellDMC on whole-genome bisulfite sequencing data
All previous simulation models mixed together DNAm profiles generated with Illumina 450k/EPIC technology 21,22, a platform matched to the one used to generate the DNAm references. Thus, to demonstrate applicability of CellDMC to samples generated with a different technology, we devised in-silico mixture simulation models using whole-genome bisulfite sequencing (WGBS) profiles of purified cell-types, generated as part of IHEC 23 (Supplementary Data 6). Although CellDMC’s performance measures dropped upon application to WGBS data, the reduction was only relatively marginal, with sensitivity and specificity values remaining high (sensitivity and specificity was still over 90% for the higher SNR values, Supplementary Fig.12). Thus, we conclude that profiling technology is not a major limiting factor for CellDMC.
Validation of CellDMC in a blood EWAS
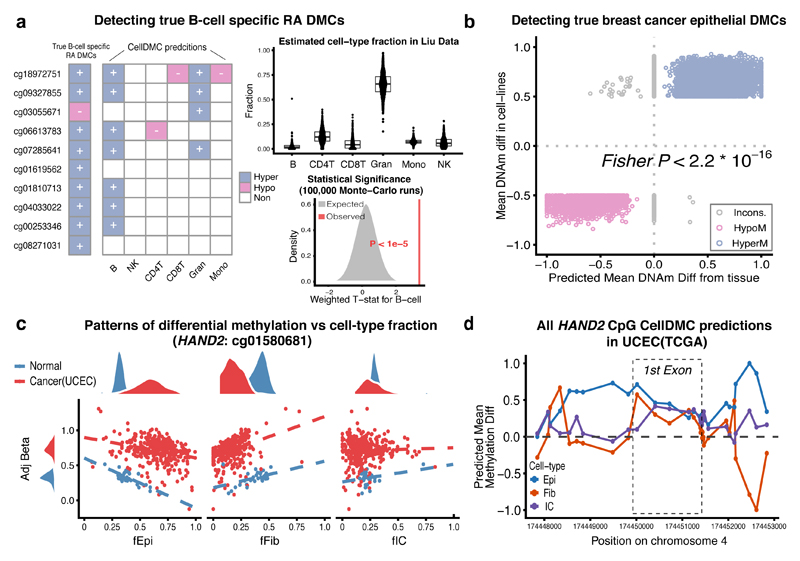
Next, we tested the ability of CellDMC to detect known DMCTs in a real EWAS. A recent EWAS for Rheumatoid Arthritis (RA) conducted on purified B-cells identified a total of 10 DMCs, which were validated in two independent EWAS cohorts which also profiled purified B-cells 24. Thus, we reasoned that application of CellDMC to an independent RA EWAS performed on 689 blood samples 5, should be able to identify these 10 RA-DMCs as being differentially methylated specifically in B-cells. CellDMC predicted 8 out of these 10 to be differentially methylated, and 7 out of 10 to be B-cell DMCTs (FDR<0.05, Fig.4a). Of note, CellDMC predicted the DNAm difference between RA cases and controls to be occurring primarily in B-cells, and not in the other immune cell subtypes (Fig.4a), despite the fact that on average B-cells only accounted for 2% of blood cell subtypes in the samples, and despite exhibiting relatively little variation (Fig.4a). Although the median absolute difference in B-cell fractions between the 689 samples was only 1.5%, 5 samples exhibited fractions larger than 10%, and therefore the ability of CellDMC to detect validated B-cell DMCTs is consistent with our previous simulation result (Fig.2d). Importantly, we estimated the chance that CellDMC would call 7 out of 10 randomly selected CpGs to be B-cell DMCTs at their observed significance levels to be less than 0.00001 (Fig.4a). We verified that results were robust to the choice of cell-type fraction estimation algorithm, or if one of the other minor blood cell subtypes was removed from the DNAm reference matrix (Supplementary Fig.13). Of note, had we applied a standard non-interaction based reference model to identify DMCs, 6 of the 10 validated RA-DMCTs would have been detected as DMCs using the same FDR<0.05 threshold but without knowledge of the underlying DMCT being B-cells. We also note that CellDMC led to a larger than 4-fold reduction of DMCs compared to a method that did not adjust for cell-type fractions, and a larger than 2-fold reduction of DMCs compared to a non-interaction based model, with relatively little overlap between DMCs called by CellDMC and the standard non-interaction model (Supplementary Table 2). Thus, like the standard non-interaction based model, CellDMC is able to remove large numbers of associations caused by changes in the granulocyte/lymphocyte ratio 5, whilst also identifying different DMCs to those found using the standard model.
Figure 4. Validation of CellDMC in real EWAS and cancer epigenome data.
a) Validation of CellDMC in a Rheumatoid Arthritis (RA) EWAS performed on 689 blood samples (Liu et al 5). The bar to the left indicates 10 CpG sites that have been validated to be RA-associated DMCs in 3 independent purified B-cell cohorts, with their directionality of DNAm change as indicated 24. The central heatmap displays the predicted DMCTs from CellDMC, based on a two-tailed t-test for the interaction term. Statistical significance of this result was assessed using 100,000 Monte-Carlo runs, where in each run 10 CpGs were selected at random and their weighted averaged t-statistic (grey curve and area) compared to the observed value (vertical red line). Boxplots display the cell-type fraction estimates across the 689 samples. Boxes indicate 25th and 75th centiles. Whiskers extend to the largest values no further than 1.5IQR from these centiles where IQR=inter-quartile range. b) Validation of CellDMC to detect true breast cancer epithelial DMCs. Scatterplot of the DNAm difference between breast cancer cell lines and normal mammary epithelial cell-lines (y-axis) against the predicted DNAm difference from CellDMC between the epithelial component of breast cancer tissue (n=305 samples) and that of normal breast (n=92), for a total of approximately 19,000 true positive breast cancer epithelial DMCs. One-tailed Fisher-test P-value is given. c) Validation of CellDMC in the endometrial TCGA cancer study. Scatterplots of the beta value (adjusted for cell-type fractions) of one of the HAND2 CpGs against the epithelial, fibroblast and total immune-cell (IC) fractions, with samples labeled according to endometrial cancer (red) or normal endometrium (blue). d) CellDMC predictions for DNAm differences between cancer and normal in each of the 3 cell-types, for all CpGs mapping to HAND2.
Validation of CellDMC in cancer epigenome studies
To demonstrate the ability of CellDMC to detect DMCTs in solid epithelial tissues, we applied it to the breast cancer EWAS setting, for which we had previously constructed a gold-standard set of 19,379 true positive breast cancer epithelial DMCs (bcDMCs) 9,17. This set was obtained by intersecting DMCs from a comparison of breast cancer epithelial to normal mammary epithelial cell-lines (thus representing relatively pure epithelial cell populations), with a corresponding list of DMCs derived from the TCGA breast cancer study 25. We reasoned that applying CellDMC on our independent breast cancer tissue EWAS 18 should not only validate the breast cancer epithelial DMCs, but should also predict the epithelial compartment to be the main DMCT. Confirming this, mean methylation differences between the epithelial cells found in breast cancer and those found in the normal tissue as predicted by CellDMC, exhibited an excellent correlation with the DNAm differences observed in the actual cell-lines (Fig.4b). Overall, the magnitude of DNAm changes were still larger in the cell-lines possibly owing to cell-culture effects or their longer proliferative history (Fig.4b). CellDMC exhibited approximately 75% sensitivity and 70% specificity to detect DMCs and over 60% sensitivity to detect the DMCTs as occurring in the epithelial compartment (Supplementary Fig.14). Although it is unknown if the 19,379 true positive bcDMCs are also altered in fibroblasts, fat and immune cells, CellDMC predicted much smaller fractions of these loci to be altered in fat and ICs (Supplementary Fig.14).
As a third validation on real data, we considered the case of the HAND2 gene in endometrial cancer 26,27. We had previously discovered and validated hypermethylated DMCs in endometrial cancer, mapping to the 1st exon region of the HAND2 gene, which is a main target of the progesterone receptor tumor suppressor pathway 26,27. HAND2 is expressed in normal endometrial tissue including stromal fibroblasts, but lacks expression in endometrial cancer and the stromal fibroblasts of the cancer tissue 26, suggesting that the observed hypermethylation occurs in both epithelial and fibroblast cells. Indeed, the relatively large difference in DNAm (Δβ~0.5) observed between endometrial cancer and normal endometrial tissue 26 is a strong indication that the unidirectional DNAm change is occurring in both epithelial and fibroblast compartments. Application of CellDMC to the TCGA endometrial cancer study 28 confirmed that the DNAm change is occurring in the epithelial and fibroblast cells of the tissue (Fig.4c-d), and that the alteration in fibroblasts is local to the 1st exon region (Fig.4d, Supplementary Data 7). Interestingly, CellDMC also predicted the immune-cell compartment to be differentially methylated, albeit less strongly so (Fig.4c-d). This is also consistent with the observation that these HAND2 CpG sites become hypermethylated with age in blood 29, and that the unmatched TCGA endometrial cancers were derived from older women. Thus, the prediction of CellDMC that these specific HAND2 CpGs define DMCs in all 3 major cell-types within the endometrial tissue is entirely consistent with previous knowledge and data.
CellDMC identifies smoking-associated DNAm changes in squamous epithelial cells
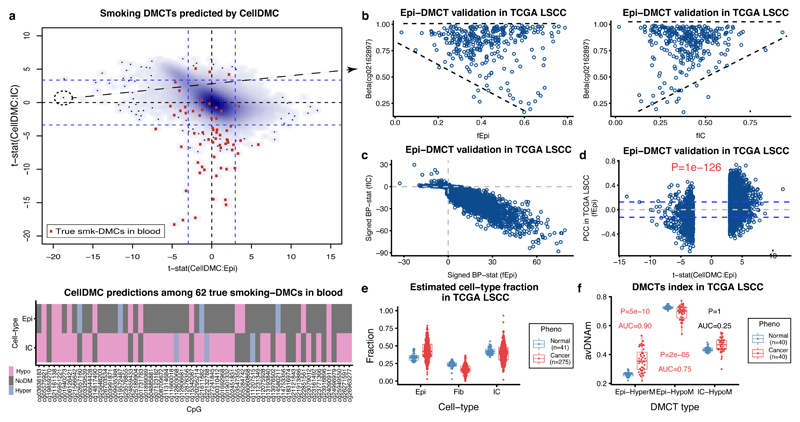
Finally, to demonstrate how CellDMC may help gain novel biological insight, we applied it to the largest EWAS cohort performed in buccal swabs 30, consisting of 790 samples from women all aged 53 but with a wide range of different lifetime levels of smoking exposure. As expected, buccal swabs contained mainly epithelial and immune cells (Supplementary Data 8), and thus we used CellDMC to predict smoking-associated DMCTs in these two cell-types. By comparison to a gold-standard list of 62 true smoking-associated DMCs which have been validated in at least three independent whole blood EWAS studies 31, we confirmed CellDMC’s sensitivity to detect true DMCTs: most of the gold-standard DMCs were predicted to be hypomethylated specifically in the immune cell compartment of the buccal tissue (Fig.5a), in line with their observed hypomethylation in blood EWAS 31. Confirming CellDMC’s specificity, the top ranked DMCTs in immune cells have all been previously validated as being smoking-associated DMCs in blood (Fig.5a). Although most of the 62 gold-standard smoking-DMCs in blood were not predicted to be altered in the epithelial cells of the buccal swabs, the great majority (i.e over 90%) of DMCTs were, however, predicted to occur in the epithelial compartment, with a strong skew towards hypermethylation (Supplementary Fig.15). To validate these epithelial DMCTs, we posited that their variance in DNAm levels would increase with the fraction of epithelial cells in lung squamous cell carcinoma 32, a cancer strongly associated with smoking, whilst also exhibiting a concomitant decrease in variance in samples with a higher immune-cell content. We were able to confirm this pattern for the top-ranked hypomethylated epithelial DMCT (Fig.5b), and for most of the other epithelial DMCTs, at high statistical significance (Fig.5c, Online Methods). Moreover, predicted hyper-and-hypo methylated epithelial DMCTs exhibited a highly significant trend towards positive and negative correlations between their DNAm levels and the estimated epithelial cell-type fractions, as required (Fig.5d). To confirm the biological and clinical significance of these findings, we posited that the DNAm alterations happening in the epithelial cells would exhibit increased deviations in lung cancer compared to the normal adjacent tissue, owing to an increase in the epithelial fraction in the tumors (Fig.5e). Confirming our expectation, hypermethylated and hypomethylated epithelial DMCTs exhibited increased and decreased levels of DNAm in lung cancer, respectively (Fig.5f). Consistent with the total immune cell fraction not being altered between normal and cancer, the top ranked DMCTs specifically hypomethylated in immune cells did not exhibit decreased levels in cancer, but increased levels, probably due to shifts in the specific immune cell subtype proportions (Fig.5f) Thus, the epithelial smoking-DMCTs identified here represent epigenetic alterations occurring in the squamous cell of origin of smoking related lung cancer and therefore may mark cells that are being selected for during lung carcinogenesis.
Figure 5. Identification of epithelial specific smoking DMCTs and their relevance in lung cancer.
a) Smoothed scatterplot of CellDMC t-statistics in epithelial cells (x-axis) vs immune cells (IC) (y-axis) derived from a 790 buccal swab EWAS with smoking as the phenotype. Calling of DMCTs was based on a two-tailed t-test. Blue dashed lines indicate level of statistical significance (FDR<0.05). Red points label CpGs from a gold-standard list of 62 true smoking-DMCs derived in blood. Lower panel displays a heatmap of CellDMC predictions (significance of signed adjusted P-values) for the 62 true smoking-DMCs. b) Scatterplot of the DNAm level (beta-value) of the top-ranked smoking-associated epithelial-specific DMCT against the epithelial cell-type fraction (left panel) and total immune cell fraction (right panel) across the 275 lung squamous cell carcinoma (LSCC) samples from the TCGA. c) Scatterplot of the signed Breusch-Pagan (BP) differential variance statistic of DNAm against immune-cell fraction (y-axis) versus the corresponding statistic for the epithelial fraction (x-axis), as computed across the 275 lung cancer samples, for all predicted epithelial-specific smoking-DMCTs derived from buccal swab EWAS. d) Scatterplot of the Pearson Correlation Coefficients (PCC, y-axis) between DNAm and epithelial cell-type fraction, as computed over the 275 lung cancer TCGA samples, against the CellDMC t-statistics for predicted epithelial-specific DMCTs derived in the buccal swab smoking EWAS (x-axis). Dashed lines indicate level of statistical significance (FDR<0.05). P-value is from a one-tailed Fisher’s exact test on the CpGs passing significance in each quadrant. e) Boxplots of cell-type fractions estimated with EpiDISH for epithelial, fibroblast and immune cells in the normal and lung squamous cell carcinoma (LSCC) samples from the TCGA. Number of data points in each boxplot is indicated within panel. f) Average DNAm levels of three categories of DMCTs derived with CellDMC from the buccal smoking EWAS, in the matched (n=40) normal-adjacent and LSCC sample pairs from the TCGA. The DMCT categories are the 50 top-ranked hypomethylated, 50 top-ranked promoter hypermethylated epithelial DMCTs, and the 50 top-ranked hypomethylated immune-cell DMCTs. P-values are from a one-tailed Wilcoxon rank sum test. AUC discriminatory values derived from Wilcox-test statistic are given. Number of data points in each boxplot is 40 corresponding to the number of matched normal-adjacent cancer pairs. In e-f), boxes indicate 25th and 75th centiles. Whiskers extend to the largest values no further than 1.5IQR from these centiles where IQR=inter-quartile range.
Discussion
We have presented a novel algorithm aimed at identifying DMCTs, and have extensively validated the method on simulated as well as real data, encompassing common EWAS and cancer epigenome scenarios, and different technologies. Results obtained on real data are in line with those derived from simulation. For instance, we have shown how CellDMC was able to identify validated DMCTs in a minor cell subpopulation (B-cells) within a large whole blood EWAS for Rheumatoid Arthritis, as predicted from our simulation analysis. The application of CellDMC to a smoking EWAS performed in buccal swabs is also noteworthy since it suggests that many of the top-ranked smoking associated CpGs detected in these buccal swabs are the result of DNAm changes in the infiltrating immune cells, but that there are also many other DMCs occurring specifically in the epithelial compartment. As shown here, these alterations and not those seen in the immune cells, become aggravated in lung cancer. It is plausible that these specific DNAm changes in the lung epithelium affect or reflect deregulation of transcription factor regulatory networks that contribute causally to lung cancer development in addition to those that occur in immune cells 33. Thus, we expect that widespread application of CellDMC to existing and future EWAS will help elucidate the role of DNAm changes and cell-types in disease. In this regard, we point that the main limitations of our method are only extrinsic factors such as the need for a large sample size and the availability of DNAm reference profiles for the main cell-types in the tissue of interest.
In summary, the ability of CellDMC to rank CpGs according to their probability of being DM in each of the underlying cell-types will be invaluable for EWAS and cancer epigenome studies to help improve biological interpretation, for prioritizing candidates that require functional validation, and to help elucidate causal pathways to disease.
Online Methods
Estimation of cell-type fractions in complex tissues using EpiDISH/HEpiDISH
Estimation of the fractions of cell-types in a given sample constitutes the first step of the CellDMC algorithm and uses our previously validated EpiDISH 16 and HEpiDISH 17 methods. We use these methods to estimate cell-type fractions for in-silico generated mixtures as well as for real samples from EWAS and cancer epigenome studies. Which method we use, i.e. EpiDISH or a hierarchical iterative version of it called HEpiDISH depends on the desired cell-type resolution, as we explain next.
In the case of whole blood, peripheral blood or in-silico mixtures of blood cell subtypes, we use the EpiDISH procedure with a DNAm reference matrix defined over 333 IC cell subtype specific DMCs and 7 IC cell subtypes 16. Briefly, EpiDISH models the DNAm profile of any given sample as a linear mixture over the DNAm profiles for the individual cell-types making up the sample. The associated multivariate linear regression is run using only the 333 IC cell subtype specific DMCs, and is performed using a robust M-estimator, as implemented in the rlm function of the MASS R-package 34. We refer to this implementation as Robust Partial Correlations (RPCs) 16. In this framework, the non-negativity and normalization constraints on the cell-type fractions (i.e. the estimated regression coefficients) are imposed a-posteriori, i.e. negative weights are set to zero and all other non-zero weights are normalized so that their sum adds to 1. As shown by us 16, RPCs improves inference, albeit marginally so, over the alternative approach which is to impose these constraints during inference via quadratic programming or constrained projection (CP) optimization 7. RPCs also performs similarly to another non-constrained approach that uses a penalized support vector regression model called CIBERSORT 16,35. In this work we use EpiDISH with RPCs throughout and only use CP for comparative purposes.
In the case of solid epithelial tissues and in-silico mixtures of epithelial, fibroblast and various IC cell subtypes, and when we only need to estimate cell-type fractions for the total epithelial, total fibroblast and total IC fractions, we once again use EpiDISH with RPCs, but now with a DNAm reference matrix defined over 716 DMCs and 3-cell types (generic Epithelial, generic Fibroblast, and a generic IC, denoted as the EpiFibIC reference matrix), as derived and validated previously by us in Zheng et al 17. The 716 DMCs are highly discriminative of the 3 main cell-types, exhibiting large differences in mean DNAm between epithelial, fibroblast and ICs 17. In some of the simulation scenarios, we also estimate fractions for individual IC cell subtypes, in which case we use our validated HEpiDISH algorithm 17. As explained in Zheng et al, HEpiDISH works by implementing EpiDISH in an iterative hierarchical fashion, first estimating total epithelial, total fibroblast and total IC fractions using the EpiFibIC reference, and then estimating proportions for the 7 individual IC cell subtypes using a 188 DMC subset of the original 333 DMC reference used to estimate fractions in blood. A key point is that the 188 DMCs do not share any overlap with the 716 DMCs of the EpiFibIC reference, and that all 188 DMCs have been selected to ensure that their median DNAm levels do not vary appreciably (most differences are less than 0.05 and the maximum difference is 0.3, in absolute terms) between the epithelial, fibroblast and the collection of ICs. This latter criterion helps ensure that the error or noise induced by inferring IC cell subtype fractions in the mixture using a reference matrix that does not include epithelial and fibroblast references, is less than the effect size or signal of the 188 DMCs (since any of the original 333 DMCs in the blood reference exhibit big differences in DNAm, i.e. at least +/-0.5 with the great majority exhibiting over +/-0.8 differences between at least one IC cell subtype and all others. See Zheng et al 17 for further justification and validation.
The CellDMC algorithm
CellDMC consists of 3 basic steps: (1) Estimation of cell-type fractions in a given sample using EpiDISH 16 or Hierarchical EpiDISH (HEpiDISH) 17, depending on the cell-type resolution desired (as described in previous section), (2) Estimation of differentially methylated cell-types (DMCTs) for a given phenotype of interest (binary or continuous), (3) Ranking of DMCTs per cell-type.
The key idea behind CellDMC is that a DNAm alteration occurring in a specific cell-type will exhibit a significant interaction between the phenotype and the corresponding cell-type fraction variable. To introduce relevant notation, we shall assume that we have a DNA methylation beta-valued matrix xcs, with c (c = 1,..,C) labeling the CpGs and s (s = 1,…,S) labeling the samples. For each sample s, let ys denote the phenotype (e.g. a 0 or 1 variable for binary phenotypes, or a continuously-valued number for continuous phenotypes), and let denote the vector of phenotype values over all samples. Correspondingly, we define to be the vector of beta (DNAm) values for CpG c across the S samples. We further assume that the fractions for the main cell-types, indexed by k (k=1…K and with K the number of cell-types), in the given samples have already been estimated using EpiDISH/HEpiDISH (or another algorithm the user wishes to use). We denote the estimated fractions for cell-type k over the S samples with the vector . CellDMC is then based on the following statistical model, which is run separately for each CpG c:
where are regression coefficients to be estimated, * denotes the interaction term and where we assume that errors are Gaussianly distributed with a variance that may depend on the specific CpG c. We note that the above summations don’t run over all K cell-types, because cell-type fractions are normalized and must add to 1. Because of this normalization constraint, the intercept term and the phenotype main effects term can be absorbed into the corresponding summations, so that an equivalent model is
The regression coefficients inform us as to whether there is a significant interaction between the phenotype and the corresponding fraction for cell-type k. We note that if differential methylation associated with the phenotype occurs at a CpG c and in cell-type k that the observed differential methylation should be larger in samples with high fractions for that cell-type k compared to samples with low content for cell-type k, and should be detectable via a statistically significant interaction term . We solve the above model using least squares with the lm function in R, which provides estimates for the regression coefficients and their statistical significance via P-values . The P-values for each cell-type k are adjusted for multiple hypothesis testing using Benjamini-Hochberg (BH) False Discovery Rate (FDR) estimation. For those CpGs with BH-FDR values less than a predefined significance threshold (i.e. typically BH FDR<0.05), we call it a DMCT in the given cell-type. Finally, CpGs can be ranked within each cell-type according to the associated P-value of significance.
Finally, we note that additional covariates representing other biological (e.g. age, gender, ethnicity) or technical factors (batch) can be included in the above model. For instance, for a set Q of such factors, let (q=1…Q) denote the factor values across the S samples, then the model above becomes
DNA methylation datasets used for in-silico mixture simulation experiments
The following lists the DNAm datasets used in this manuscript to generate the in-silico mixtures, with their GEO (www.ncbi.nlm.nih.gov/geo) accession numbers or download links. We note that all these samples were not used in the construction of the DNAm references used in EpiDISH and HEpiDISH and therefore represent truly independent samples: Illumina 450k data of 4 epithelial cell-lines and 10 fibroblasts cell-lines from Stem-Cell Matrix Compendium-2 (SCM2) 36 (GSE31848), Illumina 450k data of 8 B-cells, 71 CD4+ T-cells, and 28 monocytes from Absher et al 37 (GSE59250), Illumina 450k data of 31 CD4+ T-cells from Limbach et al 38 (GSE71955), Illumina 450k data of 4 B-cells, 6 CD4+ T-cells, and 5 monocytes from Mamrut et al (GSE71244), Illumina 450k data of 36 monocytes from Marabita et al 39 (GSE43976), Illumina 450k data of 8 CD4+ T-cells from Nestor et al 40 (GSE50222), Illumina 450k data of 214CD4+ T-cells, and 1202 monocytes from Reynolds et al 41 (GSE56047), Illumina 450k data of 6 B-cells, 6 CD4+ T-cells, and 6 monocytes from Zilbauer et al 42 (https://www.ebi.ac.uk/arrayexpress/experiments/E-MTAB-2145/). Two fibroblasts cell-lines were removed from SCM2 dataset after purity check. All of the above Illumina 450k data together made the purified cells pool for generating in-silico mixtures. In total, we have 4 epithelial cell lines, 8 fibroblast lines, 66 B-cells, 336 CD4+ T-cells, and 1277 monocytes.
We also downloaded processed WGBS bed files from IHEC (http://epigenomesportal.ca/ihec) samples that we expected to be of relatively high purity, representing epithelial, fibroblast and immune cell-subtypes. Samples of low WGBS coverage / sequencing depth were removed. A total of 6 epithelial (4 podocytes and 2 breast epithelial), 2 fibroblast/stromal, 8 B-cells, 11 CD4+ T-cells, and 9 monocyte samples were included. After merging them together, the number of common CpGs was 220351. Only 381 out of the 716 cell-type CpGs in EpiFibIC reference could be found. Thus, purity estimates were checked using 381 CpGs in the EpiFibIC reference using EpiDISH. One podocyte sample was predicted to have a high fibroblast fraction and was removed. For the in-silico mixtures, we thus used 5 epithelial, 2 fibroblast, 8 B-cells, 11 CD4+ T-cells, and 9 monocyte WGBS samples.
Generation of in-silico mixtures
To test performance of CellDMC in terms of sensitivity and specificity, we generated 200 in-silico mixtures with half of them representing controls and the other half representing cases (disease). For cases, we altered the DNAm levels of 150 CpGs in specific cell-types, thus defining true DMCTs (and DMCs). For each of the 200 mixtures, we randomly sampled an epithelial cell, fibroblast, B-cell, CD4+ T-cell, and monocyte from the corresponding pools of purified cells, and mixed them together with 5 weights drawn from a Dirichlet distribution. Since for each sample, DNAm changes were made to all immune cells (B-cells, CD4+ T-cells, and monocytes) simultaneously and equally, we could treat this as a three cell-type mixture (Epi, Fib, and IC) problem, where we inferred the total IC fraction alongside the total epithelial and fibroblast fractions. We considered 5 different DMCT scenarios, including unidirectional DNAm changes in all 3 cell-types, unidirectional changes in 2 cell-types, change in 1 cell-type, bidirectional changes in 3 cell-types, and bidirectional changes in 2 cell-types. For each scenario, we also had 5 different signal to noise ratios (SNR) with values of 3, 2.4, 1.8, 1.2, and 0.9, corresponding to approximate absolute DNAm differences in individual cell-types (between cases and controls) of 0.42, 0.32, 0.22, 0.12, and 0.12, respectively. The latter two SNR values differ because we increased the noise/variance in the case where SNR=0.9. For every scenario, we ran 100 Monte-Carlo runs.
We note that we also considered scenarios, where a DNAm change was only induced in a particular IC cell-subtype (and not in all ICs simultaneously), in which case we used HEpiDISH to estimate the cell-type fractions for all IC cell subtypes, in addition to the total epithelial and fibroblast fractions. We also considered a scenario mimicking a blood EWAS, where we only mixed together purified IC subtypes.
The overall strategy used to generate in-silico mixtures with a continuous phenotype is similar to that used for a binary phenotype. For each sample, the continuous phenotype y was assumed to take a value between 0 and 1. In the case where y=1, the corresponding beta value was sampled as described previously for disease samples in the binary phenotype context, whereas for samples with values y <1, the altered beta-values were correspondingly scaled by y. The SNR values were defined at y=1.
In the scenario where we allow for a bi-modal distribution within the disease phenotype, we use the same parameters as for the binary phenotype case. The only difference is that for each DMCT, only 70% of the disease samples were randomly selected to be altered, whereas the other 30% disease samples were modelled as control/normal samples.
Perturbations of cell-type fraction point estimates
We used two different approaches to assess the robustness of CellDMC to the use of cell-type fraction point estimates. Briefly, in one approach, instead of using the point estimates, we randomly sampled a number from the interval given by point estimate ± standard deviation*3. In the second approach, we bootstrapped DMCs making up the reference matrix, thus generating a new reference DNAm matrix with the same number of DMC but with several of these appearing multiple times. Inference of cell-type fractions was then performed using this bootstrapped DNAm reference matrix, with the new point estimates subsequently used in CellDMC.
Simulations involving missing cell-types in DNAm reference matrix
To study the impact of missing cell-types, we first considered a scenario where there are 3 cell-types with one cell-type being differentially methylated (Uni-1C scenario) and another cell-type missing. In detail, we only altered epithelial cells introducing 150 true epithelial DMCTs. Cell-type fractions were then estimated using the EpiFibIC reference matrix but with one cell-type (fibroblasts) missing from the reference DNAm matrix. Correspondingly, CellDMC was subsequently run with only 2 cell-types. We note that when generating the in-silico mixtures, the cell-type fractions for all 3 cell-types were drawn from a uniform Dirichlet distribution, so that each cell-type exhibited the same underlying mean and variance for the fraction.
We also ran a second simulation but now with 2 of the 3 cell-types being altered (Uni-2C scenario), and with one of the altered cell-types missing from the DNAm reference.
In a third analysis, we focused on blood tissue, considering the Uni-1C scenario. In more detail, we generated in-silico mixtures of 5 blood cell subtypes (B-cells, CD4+ and CD8+ T-cells, Monocytes and Neutrophils) using our pools of purified blood cell subtypes constructed earlier, drawing cell proportions from a weighted Dirchlet distribution, with weights determined to yield realistic cell-type fractions in blood, as inferred from a blood EWAS 5. As before, 150 DMCTs were generated within one cell-type only (we chose CD4+ T-cells) with sensitivity and specificity computed by CellDMC using a DNAm reference matrix that did not include another cell-type (we chose CD8+ T-cells). As in all other simulations, total of 100 Monte-Carlo runs were performed, each run simulating 100 cases and 100 controls. We benchmarked results against the scenario where none of the 5 cell subtypes was missing from the DNAm reference matrix.
Implementation of CellDMC on in-silico mixtures and definition of sensitivity and specificity
We estimated cell-type fractions of all in-silico mixtures using EpiDISH (RPC mode) with a reference 17, which consists of 716 cell-type specific DMCs, to estimate fractions of 3 major cell-types (generic epithelial cells, generic fibroblasts, and total immune cells). Next, CellDMC was run with the estimated fractions, multiple hypothesis correction method set to “FDR”, and FDR (adjusted P-value) threshold 0.05, which resulted in a matrix containing predicted DMCT(s) and a matrix of coefficients for each cell-type including predicted DNAm change, raw and adjusted P-values, ranked by selected cell-type. As long as a CpG in one cell-type was predicted to be a DMCT, it was counted as a predicted DMC. Sensitivity to detect DMCs was defined as the number of predicted true DMCs divided by 150 (true number of DMCs). Sensitivity to detect DMCT was defined as the mean sensitivity to correctly predict DMCTs (considering directionality of change) averaged over all relevant cell-types. In the case where we induced DNAm changes in individual IC cell subtypes, we estimated cell-type fractions for all cell-types using our HEpiDISH framework, but all subsequent analysis proceeded as for the 3 cell-type scenario above.
Power calculations
To estimate the appropriate sample size to achieve a certain sensitivity and specificity, we performed corresponding power calculations for a DNAm change in one cell-type (Uni-1C) and unidirectional changes in all cell-types (Uni-AllC) scenarios. For each scenario, we did 100 Monte-Carlo runs for 5 SNR(s) as described earlier. We varied sample sizes from 10 to 500 (including 10, 20, 30, 40, 50, 100, 150, 200, 300, and 500), in each case with half of these as controls and the other half as cases. Mean sensitivities for DMCT detection over 100 Monte-Carlo runs were calculated.
Simulation analysis of cell-type complexity
For K=3, we mixed epithelial, fibroblast and monocytes and used EpiDISH with centEpiFibIC DNAm reference to estimate cell-type fractions. For larger K values, we iteratively added one more IC cell subtype to the mix in the order of neutrophils (K=4), CD4+ T-cells (K=5), B-cells (K=6) and CD8+ T-cells (K=7). For K>3, we used HEpiDISH. For each choice of K and in each Monte-Carlo run, there were 150 true DMCTs (75 hyper, and 75 hypo) occurring specifically in monocytes.
Application of CellDMC to a blood rheumatoid arthritis EWAS to detect B-cell specific rheumatoid arthritis DMCs
We obtained Illumina 450k data profiling peripheral blood for 335 controls and 354 rheumatoid arthritis cases from Liu et al (GSE42861) 5. Data was normalized as described elsewhere 29. We estimated cell-type fractions of all 7 major immune cell subtypes (B-cells, CD4T-cells, CD8T-cells, NK cells, monocytes, neutrophils, eosinophils) using EpiDISH with a DNAm reference consisting of 333 IC cell-type specific DMCs 16. All parameters of CellDMC were the default choices. We tested CellDMC’s predictions against 10 B-cell specific RA CpGs, which had been validated in 3 independent purified B-cell EWAS datasets, as reported in Julià et al 24. To assess the statistical significance of CellDMC’s DMCT predictions, we randomly picked 10 CpGs 100,000 times, and compared the weighted t-statistics of the estimated B-cell specific regression coefficients to the observed one. P value was calculated as the number of runs, where the statistic was larger than the original observed one, divided by the number of runs.
Application of CellDMC to endometrial cancer
We downloaded and processed level 3 Illumina 450k data of uterine corpus endometrial carcinoma (UCEC) from TCGA 28. This dataset contains 374 cancer samples and 34 normal adjacent control samples. We estimated cell-type fractions for the total epithelial, total fibroblast and total immune cell components, using our previously validated EpiFibIC DNAm reference (716 cell-type specific DMCs).
Application of CellDMC to a breast cancer EWAS
To test CellDMC’s ability to detect DMCTs occurring in breast cancer epithelial cells, we used a breast cancer tissue DNAm 450k dataset, which included 92 normal controls and 305 breast cancer cases, from Teschendorff et al (GSE69914) 18. We estimated cell-type fractions using EpiDISH in conjunction with a previously validated DNAm reference for breast tissue, consisting of 491 cell-type specific DMCs and 4 cell-types (generic epithelial, generic fibroblast, fat and total IC) 17. All parameters choices in CellDMC were the default ones. The true positive breast cancer epithelial DMCTs/DMCs and a corresponding list of true negatives were constructed as reported in Zheng et al 9,17.
Application of CellDMC to a buccal smoking-EWAS and validation in lung squamous cell carcinoma
The smoking buccal Illumina 450k dataset was processed as described in Teschendorff et al 30. Using EpiDISH and EpiFibIC DNAm reference, we estimated cell-type fractions of total epithelial cells, total fibroblasts, and total immune cells. Since the estimated fractions of fibroblasts in all samples were quite small with mean less than 0.05, we only included epithelial cells and immune cells when running CellDMC, which was run with smoking pack years as the phenotype. Among all 790 samples in the dataset, we only used 647 samples for which smoking pack year information was available.
DMCTs predicted to occur specifically in the epithelial compartment of buccal swabs were validated in the Illumina 450k lung squamous cell carcinoma (LSCC) data, generated as part of The Cancer Genome Atlas (TCGA) 32. We only validated DMCTs which exhibited similar DNAm levels in epithelial and blood cell-subtypes in samples not exposed to smoking, since only for these we would expect to see an association between their DNAm levels and the estimated epithelial fraction in the tumors. To this end, we collected Illumina 450k data of 11 different epithelial cell-lines (Hipe, Saec, Hre, Hae, Hrpe, Prec, Hee, Hcpe, Hnpce, Hmec, Hrce) from ENCODE (GSE40699) and a total of 42 purified samples representing all 7 major immune cell types (neutrophils, eosinophils, monocytes, CD4+ and CD8+ T-cells, B-cells and NK-cells) from Reinius et al 43 (GSE35069) to identify CpGs with the same ground state of DNAm level in epithelial and immune cells: in detail, we used limma (an empirical Bayes framework) 44, to select 277,801 CpGs with P-values greater than 0.5 or absolute DNAm change between epithelial and immune cells less than 0.05. Of 17,285 and 6658 DMCTs predicted by CellDMC in buccal swabs to be hypermethylated and hypomethylated in epithelial cells, respectively, 3629 and 816 of them overlapped with the 277,801 non-DMCs. We used EpiDISH with EpiFibIC reference to estimate cell-type fractions in the TCGA LSCC DNAm dataset (275 cancer samples). We computed the Pearson’s correlation coefficient between the DNAm beta values of these predicted epithelial DMCTs and the estimated fraction of epithelial cells across the 275 cancer samples. To test whether there is an association between DNAm variance and epithelial cell-type fraction, we used the Breusch-Pagan (BP) test with beta values as response and cell-type fraction as predictor. We used the signed BP-statistic to test for positive and negative associations between DNAm variance with epithelial and immune cell-type fractions, respectively.
We confirm that all work described herein complies with the Life Sciences Reporting Summary, published alongside this article.
Data availability
All data analyzed in this manuscript is already publicly available from Gene Expression Omnibus - GEO (www.ncbi.nlm.nih.gov/geo/), the TCGA data portal (https://gdc.cancer.gov), International Human Epigenome Consortium - IHEC (http://epigenomesportal.ca/ihec), or from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/). Accession codes for data from GEO include GSE31848, GSE59250, GSE71955, GSE71244, GSE43976, GSE50222, GSE56047, GSE42861, GSE69914, GSE40699, and GSE35069. Accession code for data from ArrayExpress is E-MTAB-2145. All DNA methylation reference matrices used to estimate cell-type fractions are available on Github (https://github.com/sjczheng/EpiDISH)
The DNAm dataset in buccal cells is available by submitting data requests to mrclha.swiftinfo@ucl.ac.uk; see full policy at http://www.nshd.mrc.ac.uk/data.aspx. Managed access is in place for this 69 year old NSHD study to ensure that use of the data are within the bounds of consent given previously by participants, and to safeguard any potential threat to anonymity since the participants are all born in the same week.
Code availability
CellDMC is freely available as a user-friendly R-script within the EpiDISH Bioconductor package, freely available from github (https://github.com/sjczheng/EpiDISH), and in due course also from Bioconductor (https://www.bioconductor.org).
Supplementary Material
Acknowledgements
This work was supported by NSFC (National Science Foundation of China) grants 31571359, 31771464 and 31401120, a Wellcome Trust grant (99148) and by a Royal Society Newton Advanced Fellowship (NAF project number: 522438, NAF award number: 164914).
Footnotes
Author Contributions: Study was conceived by AET. SCZ and AET contributed jointly to methods development. SCZ performed the statistical analyses. Manuscript was written by both SCZ and AET. SB and CEB contributed valuable feedback.
Competing Financial Interests Statement: The authors declare that they have no competing interests.
References
- 1.Petronis A. Epigenetics as a unifying principle in the aetiology of complex traits and diseases. Nature. 2010;465:721–7. doi: 10.1038/nature09230. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 3.Lappalainen T, Greally JM. Associating cellular epigenetic models with human phenotypes. Nat Rev Genet. 2017;18:441–451. doi: 10.1038/nrg.2017.32. [DOI] [PubMed] [Google Scholar]
- 4.Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011;12:529–41. doi: 10.1038/nrg3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu Y, et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat Biotechnol. 2013;31:142–7. doi: 10.1038/nbt.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Houseman EA, Molitor J, Marsit CJ. Reference-free cell mixture adjustments in analysis of DNA methylation data. Bioinformatics. 2014;30:1431–9. doi: 10.1093/bioinformatics/btu029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zheng SC, et al. Correcting for cell-type heterogeneity in epigenome-wide association studies: revisiting previous analyses. Nat Methods. 2017;14:216–217. doi: 10.1038/nmeth.4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Teschendorff AE, Relton CL. Statistical and integrative system-level analysis of DNA methylation data. Nat Rev Genet. 2018;19:129–147. doi: 10.1038/nrg.2017.86. [DOI] [PubMed] [Google Scholar]
- 11.Zou J, Lippert C, Heckerman D, Aryee M, Listgarten J. Epigenome-wide association studies without the need for cell-type composition. Nat Methods. 2014;11:309–11. doi: 10.1038/nmeth.2815. [DOI] [PubMed] [Google Scholar]
- 12.Rahmani E, et al. Sparse PCA corrects for cell type heterogeneity in epigenome-wide association studies. Nat Methods. 2016;13:443–5. doi: 10.1038/nmeth.3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahmani E, et al. Correcting for cell-type heterogeneity in DNA methylation: a comprehensive evaluation. Nat Methods. 2017;14:218–219. doi: 10.1038/nmeth.4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lutsik P, et al. MeDeCom: discovery and quantification of latent components of heterogeneous methylomes. Genome Biol. 2017;18:55. doi: 10.1186/s13059-017-1182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breeze CE, et al. eFORGE: A Tool for Identifying Cell Type-Specific Signal in Epigenomic Data. Cell Rep. 2016;17:2137–2150. doi: 10.1016/j.celrep.2016.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teschendorff AE, Breeze CE, Zheng SC, Beck S. A comparison of reference-based algorithms for correcting cell-type heterogeneity in Epigenome-Wide Association Studies. BMC Bioinformatics. 2017;18:105. doi: 10.1186/s12859-017-1511-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng SC, et al. A novel cell-type deconvolution algorithm reveals substantial contamination by immune cells in saliva, buccal and cervix. Epigenomics. 2018;10:925–940. doi: 10.2217/epi-2018-0037. [DOI] [PubMed] [Google Scholar]
- 18.Teschendorff AE, et al. DNA methylation outliers in normal breast tissue identify field defects that are enriched in cancer. Nat Commun. 2016;7:10478. doi: 10.1038/ncomms10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–35. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McGregor K, et al. An evaluation of methods correcting for cell-type heterogeneity in DNA methylation studies. Genome Biol. 2016;17:84. doi: 10.1186/s13059-016-0935-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandoval J, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6:692–702. doi: 10.4161/epi.6.6.16196. [DOI] [PubMed] [Google Scholar]
- 22.Moran S, Arribas C, Esteller M. Validation of a DNA methylation microarray for 850,000 CpG sites of the human genome enriched in enhancer sequences. Epigenomics. 2016;8:389–99. doi: 10.2217/epi.15.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stunnenberg HG, International Human Epigenome, C. Hirst M. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell. 2016;167:1145–1149. doi: 10.1016/j.cell.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Julia A, et al. Epigenome-wide association study of rheumatoid arthritis identifies differentially methylated loci in B cells. Hum Mol Genet. 2017;26:2803–2811. doi: 10.1093/hmg/ddx177. [DOI] [PubMed] [Google Scholar]
- 25.Koboldt DC, et al. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones A, et al. Role of DNA Methylation and Epigenetic Silencing of HAND2 in Endometrial Cancer Development. PLoS Med. 2013;10:e1001551. doi: 10.1371/journal.pmed.1001551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jiao Y, Widschwendter M, Teschendorff AE. A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics. 2014;30:2360–6. doi: 10.1093/bioinformatics/btu316. [DOI] [PubMed] [Google Scholar]
- 28.Kandoth C, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan T, et al. An integrative multi-scale analysis of the dynamic DNA methylation landscape in aging. PLoS Genet. 2015;11:e1004996. doi: 10.1371/journal.pgen.1004996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teschendorff AE, et al. Correlation of Smoking-Associated DNA Methylation Changes in Buccal Cells With DNA Methylation Changes in Epithelial Cancer. JAMA Oncol. 2015;1:476–85. doi: 10.1001/jamaoncol.2015.1053. [DOI] [PubMed] [Google Scholar]
- 31.Gao X, Jia M, Zhang Y, Breitling LP, Brenner H. DNA methylation changes of whole blood cells in response to active smoking exposure in adults: a systematic review of DNA methylation studies. Clin Epigenetics. 2015;7:113. doi: 10.1186/s13148-015-0148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cancer Genome Atlas Research N. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Y, Widschwendter M, Teschendorff AE. Systems-epigenomics inference of transcription factor activity implicates aryl-hydrocarbon-receptor inactivation as a key event in lung cancer development. Genome Biol. 2017;18:236. doi: 10.1186/s13059-017-1366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Venables WN, Ripley BD. Modern Applied Statistics with S. Springer; New York: 2002. [Google Scholar]
- 35.Newman AM, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12:453–7. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nazor KL, et al. Recurrent variations in DNA methylation in human pluripotent stem cells and their differentiated derivatives. Cell Stem Cell. 2012;10:620–34. doi: 10.1016/j.stem.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Absher DM, et al. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Limbach M, et al. Epigenetic profiling in CD4+ and CD8+ T cells from Graves' disease patients reveals changes in genes associated with T cell receptor signaling. J Autoimmun. 2016;67:46–56. doi: 10.1016/j.jaut.2015.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Marabita F, et al. An evaluation of analysis pipelines for DNA methylation profiling using the Illumina HumanMethylation450 BeadChip platform. Epigenetics. 2013;8:333–46. doi: 10.4161/epi.24008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nestor CE, et al. DNA methylation changes separate allergic patients from healthy controls and may reflect altered CD4+ T-cell population structure. PLoS Genet. 2014;10:e1004059. doi: 10.1371/journal.pgen.1004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reynolds LM, et al. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat Commun. 2014;5:5366. doi: 10.1038/ncomms6366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zilbauer M, et al. Genome-wide methylation analyses of primary human leukocyte subsets identifies functionally important cell-type-specific hypomethylated regions. Blood. 2013;122:e52–60. doi: 10.1182/blood-2013-05-503201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reinius LE, et al. Differential DNA methylation in purified human blood cells: implications for cell lineage and studies on disease susceptibility. PLoS One. 2012;7:e41361. doi: 10.1371/journal.pone.0041361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analyzed in this manuscript is already publicly available from Gene Expression Omnibus - GEO (www.ncbi.nlm.nih.gov/geo/), the TCGA data portal (https://gdc.cancer.gov), International Human Epigenome Consortium - IHEC (http://epigenomesportal.ca/ihec), or from ArrayExpress (https://www.ebi.ac.uk/arrayexpress/). Accession codes for data from GEO include GSE31848, GSE59250, GSE71955, GSE71244, GSE43976, GSE50222, GSE56047, GSE42861, GSE69914, GSE40699, and GSE35069. Accession code for data from ArrayExpress is E-MTAB-2145. All DNA methylation reference matrices used to estimate cell-type fractions are available on Github (https://github.com/sjczheng/EpiDISH)
The DNAm dataset in buccal cells is available by submitting data requests to mrclha.swiftinfo@ucl.ac.uk; see full policy at http://www.nshd.mrc.ac.uk/data.aspx. Managed access is in place for this 69 year old NSHD study to ensure that use of the data are within the bounds of consent given previously by participants, and to safeguard any potential threat to anonymity since the participants are all born in the same week.