Abstract
Human immunodeficiency virus type 1 (HIV-1) hijacks the host Endosomal Sorting Complex Required for Transport (ESCRT) proteins in order to release infectious viral particles from the cell. ESCRT recruitment is virtually essential for the production of infectious virus, despite that the main structural protein of HIV-1, Gag, is capable of self-assembling and eventually budding from membranes on its own. Recent data have reinforced the paradigm of ESCRT-dependent particle release while clarifying why this rapid release is so critical. The ESCRTs were originally discovered as integral players in endosome maturation and are now implicated in many important cellular processes beyond viral and endosomal budding. Nearly all of these roles have in common that membrane scission occurs from the inward face of the membrane neck, which we refer to as “reverse topology” scission. A satisfactory mechanistic description of reverse-topology membrane scission by ESCRTs remains a major challenge both in general and in the context of HIV-1 release. New observations concerning the fundamental scission mechanism for ESCRTs in general, and the process of HIV-1 release specifically, have generated new insights in both directions, bringing us closer to a mechanistic understanding.
Introduction and history
The human immunodeficiency virus type 1 (HIV-1), like other enveloped viruses that emerge from the plasma membrane (PM), must bud away from infected cells in order to complete its replication cycle (1, 2). The budding process, as opposed to the subsequent release of the virion, is driven entirely by the viral Gag protein. Gag consists of four domains connected by linker peptides: matrix (MA), capsid (CA), nucleocapsid (NC), and p6 (Fig. 1A). MA is myristoylated, binds to the PM lipid phosphatidylinositol (4,5)-bisphosphate (PI(4,5)P2), and is the main driver of Gag membrane localization. CA assembles into a quasi-spherical lattice consisting of an incomplete array of CA hexamers. CA is responsible for scaffolding the shape of the membrane bud (3–5). NC packages the RNA genome (6). Crosslinking of Gag mediated by the large genomic RNA is important for the assembly of a tight immature lattice (7), and there is interplay between CA assembly and the specificity of RNA packaging (8). Truncated Gag constructs consisting of CA and NC are able to form spherical shells of appropriate dimensions in the presence of RNA and inositol hexakisphosphate, but in the absence of membranes or host proteins (7). Thus, there is abundant evidence that HIV-1 encodes its own ability to self-assemble and deform membranes into attached buds.
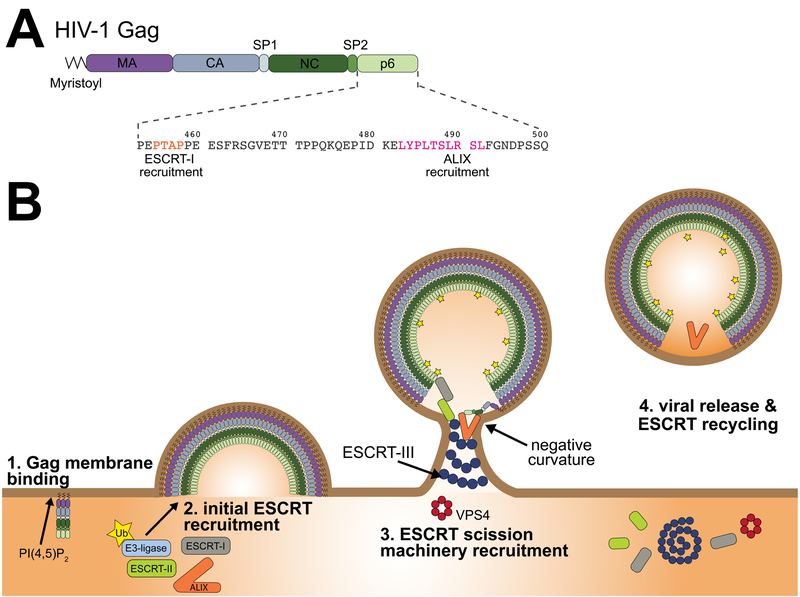
Figure 1. How ESCRTs and Gag collaborate in HIV-1 viral particle formation and release.
(A) The major domains of the HIV-1 Gag polyprotein are shown arranged from N- to C-terminus (left to right) and the N-terminal glycine is post-translationally modified by a myristoyl moiety. The late domain, p6, harbors parallel recruitment motifs for the early acting ESCRT proteins: ESCRT-I and ALIX. (B) The MA domain targets Gag to the plasma membrane (PM) by binding PI(4,5)P2. Additionally, the N-terminal myristoylation of Gag promotes its insertion into the inner leaflet of the PM. As Gag further accumulates at the PM it gets ubiquitinated and recruits the ESCRT complex via PTAP-TSG101 or LYPXnL-ALIX binding. The ESCRT-III and VPS4 scission machinery is eventually recruited to the growing virion and catalyze the viral particle release of HIV-1 Gag from the PM.
HIV-1 does not, in practice, release from the host cell PM by itself. The first clue that host factors were involved came from the isolation of mutants within the last domain of Gag, p6, which potently blocked virion release (9). These regions are termed late domains because their function is required late in the replication cycle (10), and they are present in many retroviruses and other viruses (11). In HIV-1, the two key sequences have the motifs PTAP and LYPXnL. The search for late domain interactors led to the 2001 discovery that PTAP targets the protein TSG101 (tumor susceptibility gene 101) (12–15). The other motif, LYPXnL, was shown two years later to target ALIX (apoptosis-linked gene 2-interacting protein X) (16–18). TSG101 is the mammalian ortholog of the yeast protein Vps23. Vps23 is one of four subunits of the ESCRT-I complex, which was also discovered in 2001 as an essential component of multivesicular body biogenesis, an endosomal membrane budding pathway that mediates lysosomal transport of integral membrane proteins (19). ALIX is the mammalian ortholog of Bro1, which was shown to be an ESCRT-associated protein (20, 21) in the same year the ALIX-viral late domain connection was made. These and related principles also apply to the release of many other, though not all, membrane enveloped viruses (22–24). These seminal discoveries set the stage for the current paradigm and questions in the field of HIV-1 release.
Here we review our current understanding of ESCRT-mediated HIV-1 release. We begin by revisiting the basic question why the ESCRTs are needed at all, and whether in fact the underlying premise of the field as is as solid as commonly presented. We then consider the circuitry of Gag-ESCRT and intra-ESCRT linkages and some recently discovered nuances. We address the very basic yet still unresolved question of the sidedness of ESCRT activity at the bud neck. ESCRTs direct membrane scission in a vast array of cell biological processes in addition to enveloped virus release, including membrane abscission in cytokinesis, extracellular vesicle biogenesis, membrane wound repair, nuclear envelope reformation and quality control, and many others (25–27). We end by addressing the prospects for resolving the larger question as to how ESCRTs cut membranes in general, and consider how the fields of HIV release and membrane scission biophysics might synergize to find the answer.
Why bother with ESCRTs?
The observation of HIV Gag virus-like particle (VLP) budding from cells (28–32) in the absence of ESCRTs begged the question as to why ESCRTs are needed, given that at least slow release is possible in their absence. VLPs released in the absence of ESCRTs have sharply reduced infectivity, and the basis for this defect was recently elucidated. The Gag-Pol polyprotein, which includes the essential viral enzymes reverse-transcriptase (RT), viral protease (PR), and integrase (IN), is packaged together with Gag during the assembly of the immature viral lattice. HIV-1 maturation entails the proteolytic cleavage by PR of Gag and Gag-Pol. The formation of non-infectious ESCRT-independent VLPs is attributed to premature cleavage of Gag-Pol caused by the delay in the kinetics of their release (33). The delay induced by deletion of PTAP as compared to LYPXnL is 10-fold longer, explaining the difference in these phenotypes. Hence, proper ESCRT recruitment accelerates the release of VLPs far above that of the basal ESCRT-independent process such that virions are severed prior to PR activation.
Circuitry of ESCRT recruitment
Based on the strengths of phenotypes observed upon mutation or deletion of the motif, or depletion of the host factors, PTAP binding to the ubiquitin-conjugating enzyme E2 variant (UEV) domain of ESCRT-I has the dominant role in HIV-1 release (12–15). LYPXnL binding to the Bro1 domain of ALIX has a milder phenotype when perturbed, but still plays an important role (16–18). The structures of the molecular complexes have been determined (34–36), and the recruitment cascade has been reconstituted from start to finish (37). Artificial protein nanocages designed from first principles were fused to HIV-1 Gag p6 and found to cluster and release in an ESCRT-dependent manner (38), fulfilling a strong prediction of the current paradigm. Thus our overall understanding of the recruitment pathways is on an exceptionally solid footing.
One of the few incompletely resolved points is the extent to which ESCRT-II plays a role during HIV-1 VLP release. ESCRT-II bridges ESCRT-I to -III by binding to the ESCRT-III associated upstream subunit CHMP6 (charged multivesicular body protein, CHMP) in an in vivo minimal system (37). However, depletion of ESCRT-II subunits and CHMP6 was found to have no HIV-1 release phenotype (39). A more recent report suggested that ESCRT-II is in fact involved in viral egress (40), and ESCRT-II has also been suggested to have a role in gRNA trafficking (41).
In the normal course of events, both the ESCRT-I and ALIX-based mechanisms seem to operate simultaneously (42). The partial redundancy of these branches of the ESCRT recruitment pathway presumably is adaptive for the virus by conferring additional robustness on the critical step. If two mechanisms are better than one, it should not be surprising that additional mechanisms seem to be operative beyond just PTAP and LYPXnL recognition. ESCRTs can be recruited by the Gag NC domain (43, 44). This interaction appears to involve basic residues on both the NC domain and the Bro1 domain of ALIX, with the membrane acting as a bridge (45). This model is appealing because NC and p6 are next to one another in Gag, and NC binding to the membrane helps position the p6 of just those Gag molecules at the edge of the lattice so that they can recruit ESCRTs. This suggests an elegant mechanism to potentially inactivate bulk Gag distal to the membrane neck. In regards to neck localization, it has been found that negative membrane curvature potently promotes ESCRT-III nucleation (46), another factor that might explain the restriction of ESCRT-III to the membrane neck seen in most studies (Fig. 1B).
Gag ubiquitination also plays a role in ESCRT recruitment. ESCRTs contain several ubiquitin binding domains (UBDs) whose normal function is to recognize ubiquitinated cargo proteins. In particular, TSG101 contains the UEV domain. ALIX (47, 48) and ESCRT-II (49–51) also contain ubiquitin binding domains (UBDs). HIV-1 Gag ubiquitination is increased when Gag assembles on membranes (52). Gag ubiquitination is mediated by Nedd4 family ubiquitin ligases (53, 54), which are also responsible for ubiquitination of endogenous ESCRT substrates. Fusion of a deubiquitinating enzyme (DUB) to ESCRT-I suppresses HIV-1 release (55), a strong indicator that ubiquitination cooperates with other Gag elements in ESCRT targeting.
The topology of ESCRTs in HIV budding
One of the most basic questions about the HIV-1 release mechanism is whether the ESCRTs sever the neck in cis or in trans with respect to the contents of the virion (Fig. 2). Most models of ESCRT action in general and ESCRT release of HIV-1 in particular are based on the idea that ESCRTs localize and act at the membrane neck (23, 56, 57). In endosomal sorting pathways, ESCRTs are thought to act in trans with respect to the contents of the budded vesicles, such that most or all of the ESCRTs are retained in the cytosol (58–60). The analogy to the endosomal budding has led many to expect the ESCRTs to sever HIV-1 by a similar trans mechanism.
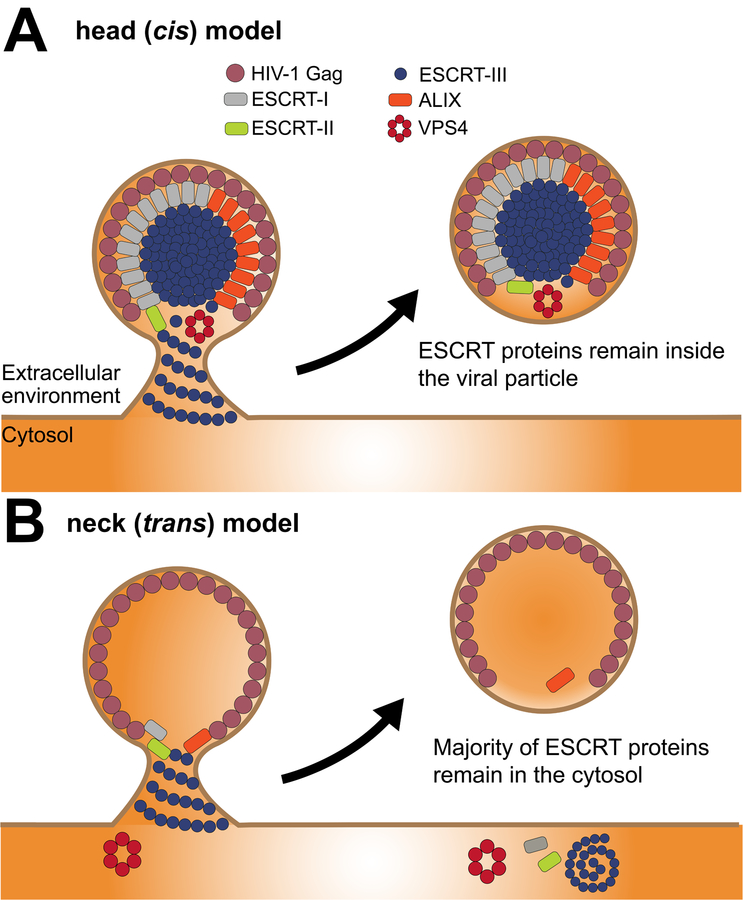
Figure 2. ESCRT topology in HIV-1 viral particle release.
An overview of the proposed HIV-1 Gag and ESCRT interactions during the viral release mechanism. (A) The cis model (cis with respect to the contents of the viral bud) postulates that ESCRTs form assemblies and sever the neck from within. This could be driven very locally from the vicinity of the neck (not shown), or in the model shown here, from the head of the virion. The cis mechanism must result in at least some ESCRT proteins departing with the viral particle when it liberated from the PM, but the number of molecules need could be small enough to be difficult to detect. The head-driven version shown here would lead to large amounts of ESCRTs incorporated into viral particles, which does not fit most of the data in the field, but does have its advocates (65). (B) In comparison with the cis model, the trans model or neck model shows that ESCRT subunits polymerize and recruit VPS4 at the neck where membrane abscission is achieved. Finally, ESCRTs are recycled back to the cytosolic pool although substantial amounts of ALIX and trace amounts of other ESCRTs may be found present inside the virion.
This question has been studied most directly by super-resolution imaging, but conclusions are still ambiguous. One report is consistent with the neck model (61) and the other, in contrast, found that ESCRTs localize predominantly in the head (62). The observation of substantial viral head localization in one report seems consistent with a cis mechanism. Indeed, it might seem more intuitive to sever the membrane in cis since ESCRTs are recruited by Gag. Seemingly, consistent with this idea, ALIX has been reproducibly seen in proteomic studies of HIV-1 virions (63–65). However, other ESCRTs, including the critical ESCRT-III CHMP subunits of membrane scission generally have not been reported in released virions, with one major exception (62). Possible explanations for these differences were recently reviewed in depth (66). While large-scale virion head localization of ESCRT-III is difficult to reconcile with much of the available data, the larger question of cis vs. trans mechanisms of release is still, in our view, open. Further improvements in high-speed high-resolution imaging of reconstituted and cellular systems will likely be needed to resolve this question.
How do ESCRTs cut membrane necks?
There is a consensus by now that the ESCRTs themselves comprise the machinery that cuts membrane necks in HIV-1 release, endosome budding, and most of the other areas of biology where they function. More specifically, the filamentous ESCRT-III subunits CHMP2A/B, CHMP3, and CHMP4A-C, and the hexameric AAA+ ATPase VPS4A/B are thought to comprise the main scission engine in mammalian cells. The structures, kinetic behavior, and mechanistic models were recently reviewed in much greater depth than is possible here (67). Here we recap only the most pressing questions, and provide a few updates since ref (67) was published.
Beyond the question of cis vs. trans action described above for HIV-1 release, there are several other pressing mechanistic questions that concern ESCRT membrane scission across diverse areas of biology. In all models, scission is thought to be driven by a conical or funnel shaped helical assembly. It is uncertain, however, whether scission is driven by the narrowing of an inward-growing ESCRT funnel to a fine tip (68), buckling of an outward-growing funnel to a disc (69), or some other type of transition. The role of the ATPase VPS4 has also been uncertain. At a minimum, VPS4 is a recycling factor to replenish the pool of soluble ESCRTs after scission. However, VPS4 arrives at HIV-1 budding sites prior to scission (70, 71), suggesting a direct role in the scission itself, as opposed to merely recycling. In fact, one recent report found that VPS4 not only arrives, but also departs, prior to scission (72). Our lab recently reconstituted membrane scission in a synthetic nanotube system, and found that ATP and enzymatically active VPS4 were required for scission (73). In this study, a pulling force was observed, which could be related to a VPS4-driven contraction of the membrane tube corresponding to buckling, but this remains to be further explored. VPS4 acts on CHMPs by unfolding them and threading them through the central pore of the hexamer (74, 75), but how molecular-level protein unfolding is related to membrane scission is still uncertain.
Concluding remarks
In this short minireview, we hope to have given readers new to the field of HIV-1 release an express tour of its origins, pillars of established knowledge, and current open questions and controversies. For a more complete account we refer the reader to several full-length reviews on the topic (2, 57, 66, 67). For our colleagues in the field, we hope you found our perspective on the open questions interesting, and apologize for omitting to cite many important contributions due to the limited space of the minireview format.
Acknowledgements
We would like to thank T. Flower, M.R. Pavlin, J. Schöneberg, the rest of the Hurley laboratory, and colleagues who attended the recent Biochemical Society symposium on ESCRTs, for stimulating discussions.
Funding information
Research on HIV-1 release and ESCRTs in the Hurley laboratory is supported by the US National Institutes of Health grant R01 AI112442.
Abbreviations:
- AAA+
ATPases associated with various cellular activities
- ALIX
apoptosis-linked gene 2-interacting protein X
- CA
capsid protein p24
- CHMP
charged multivesicular body protein
- DUB
deubiquitinating enzyme
- ESCRT
endosomal sorting complex required for transport
- IN
integrase
- MA
matrix protein p17
- NC
nucleocapsid protein p7
- Nedd4
neural-precursor-cell-expressed developmentally down-regulated 4-like ubiquitin ligase
- PM
plasma membrane
- PR
viral protease
- RT
reverse-transcriptase
- TSG101
tumor susceptibility gene 101
- UBD
ubiquitin binding domain
- UEV
ubiquitin-conjugating enzyme E2 variant
- VLP
virus-like particle
- VPS
vacuolar protein sorting-associated protein
Footnotes
Competing interests
The authors declare no competing interests
References
- 1.Sundquist WI, Krausslich HG. HIV-1 assembly, budding, and maturation. Cold Spring Harbor Perspectives on Medicine. 2012;2:a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed EO. HIV-1 assembly, release and maturation. Nature reviews Microbiology. 2015. August;13(8):484–96. PubMed PMID: Epub 2015/06/30. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schur FKM, Obr M, Hagen WJH, Wan W, Jakobi AJ, Kirkpatrick JM, et al. An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science. 2016. July;353(6298):506–8. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 4.Mattei S, Schur FK, Briggs JA. Retrovirus maturation-an extraordinary structural transformation. Curr Opin Virol. 2016 Jun;18:27–35. PubMed PMID: Epub 2016/03/25. eng. [DOI] [PubMed] [Google Scholar]
- 5.Perilla JR, Gronenborn AM. Molecular Architecture of the Retroviral Capsid. Trends Biochem Sci. 2016. May;41(5):410–20. PubMed PMID: Pubmed Central PMCID: PMC4879823. Epub 2016/04/04. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comas-Garcia M, Davis SR, Rein A. On the Selective Packaging of Genomic RNA by HIV-1. Viruses. 2016. September 12;8(9). PubMed PMID: Pubmed Central PMCID: PMC5035960. Epub 2016/09/15. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, et al. Modulation of HIV-like particle assembly in vitro by inositol phosphates. Proc Natl Acad Sci U S A. 2001. September;98(19):10875–9. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carlson LA, Bai Y, Keane SC, Doudna JA, Hurley JH. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. Elife. 2016. June;5:pii: e14663. doi: 10.7554/eLife. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gottlinger HG, Dorfman T, Sodroski JG, Haseltine WA. Effect of Mutations Affecting the P6 Gag Protein on Human-Immunodeficiency-Virus Particle Release. Proc Natl Acad Sci U S A. 1991. April;88(8):3195–9. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang MJ, Orenstein JM, Martin MA, Freed EO. P6(Gag) Is Required for Particle-Production from Full-Length Human-Immunodeficiency-Virus Type-1 Molecular Clones Expressing Protease. J Virol. 1995. November;69(11):6810–8. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed EO. Viral late domains. J Virol. 2002. May;76(10):4679–87. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.VerPlank L, Bouamr F, LaGrassa TJ, Agresta B, Kikonyogo A, Leis J, et al. Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc Natl Acad Sci U S A. 2001. July 3;98(14):7724–9. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, et al. Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell. 2001. October 5;107(1):55–65. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 14.Martin-Serrano J, Zang T, Bieniasz PD. HIV-I and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med. 2001. December;7(12):1313–9. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 15.Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci U S A. 2002. January 22;99(2):955–60. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strack B, Calistri A, Craig S, Popova E, Gottlinger HG. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell. 2003. September 19;114(6):689–99. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 17.von Schwedler UK, Stuchell M, Muller B, Ward DM, Chung HY, Morita E, et al. The protein network of HIV budding. Cell. 2003. September 19;114(6):701–13. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 18.Martin-Serrano J, Yaravoy A, Perez-Caballero D, Bieniasz PD. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci U S A. 2003. October 14;100(21):12414–9. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katzmann DJ, Babst M, Emr SD. Ubiquitin-Dependent Sorting into the Multivesicular Body Pathway Requires the Function of a Conserved Endosomal Protein Sorting Complex, ESCRT-I. Cell 2001;106(2):145–55. [DOI] [PubMed] [Google Scholar]
- 20.Nikko E, Marini AM, Andre B. Permease recycling and ubiquitination status reveal a particular role for Bro1 in the multivesicular body pathway. J Biol Chem. 2003. December 12;278(50):50732–43. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 21.Odorizzi G, Katzmann DJ, Babst M, Audhya A, Emr SD. Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci. 2003;116(Pt 10):1893–903. [DOI] [PubMed] [Google Scholar]
- 22.Ren X, Hurley JH. Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic. 2011;12:1282–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weissenhorn W, Poudevigne E, Effantin G, Bassereau P. How to get out: ssRNA enveloped viruses and membrane fission. Curr Opin Virol. 2013. April;3(2):159–67. PubMed PMID: Epub 2013/04/16. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rossman JS, Lamb RA. Viral membrane scission. Annu Rev Cell Dev Biol. 2013;29:551–69. PubMed PMID: Pubmed Central PMCID: PMC4286373. Epub 2013/10/09. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hurley JH. ESCRTs are everywhere. EMBO J. 2015;34:2398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campsteijn C, Vietri M, Stenmark H. Novel ESCRT functions in cell biology: spiraling out of control? Curr Opin Cell Biol. 2016;41:1–8. [DOI] [PubMed] [Google Scholar]
- 27.Olmos Y, Carlton JG. The ESCRT machinery: new roles at new holes. Curr Opin Cell Biol. 2016;38:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilk T, Gross I, Gowen BE, Rutten T, de Haas F, Welker R, et al. Organization of immature human immunodeficiency virus type 1. J Virol. 2001. January;75(2):759–71. PubMed PMID: Pubmed Central PMCID: PMC113972. Epub 2001/01/03. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ako-Adjei D, Johnson MC, Vogt VM. The retroviral capsid domain dictates virion size, morphology, and coassembly of gag into virus-like particles. J Virol. 2005. November;79(21):13463–72. PubMed PMID: Pubmed Central PMCID: PMC1262573. Epub 2005/10/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fang Y, Wu N, Gan X, Yan WH, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007. June;5(6):1267–83. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gan X, Gould SJ. HIV Pol inhibits HIV budding and mediates the severe budding defect of Gag-Pol. PLoS One. 2012;7(1):e29421 PubMed PMID: Pubmed Central PMCID: PMC3250436. Epub 2012/01/12. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Norgan AP, Lee JRE, Oestreich AJ, Payne JA, Krueger EW, Katzmann DJ. ESCRT-Independent Budding of HIV-1 Gag Virus-Like Particles from Saccharomyces cerevisiae Spheroplasts. PLoS One. 2012. December;7(12):e52603 PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bendjennat M, Saffarian S. The Race against Protease Activation Defines the Role of ESCRTs in HIV Budding. PLoS Path. 2016. June;12(6):e1005657 PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pornillos O, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9:812–7. [DOI] [PubMed] [Google Scholar]
- 35.Im YJ, Kuo L, Ren X, Burgos PV, Zhao XZ, Lui F, et al. Crystallographic and functional analysis of the ESCRT-1/HIV-1 Gag PTAP interaction. Structure. 2010. 18:1536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhai Q, Fisher RD, Chung HY, Myszka DG, Sundquist WI, Hill CP. Structural and functional studies of ALIX interactions with YPXnL late domains of HIV-1 and EIAV. Nat Struct Mol Biol. 2008. January;15(1):43–9. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 37.Carlson L-A, Hurley JH. In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters. Proceedings of the National Academy of Sciences. 2012. October 16, 2012;109(42):16928–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Votteler J, Ogohara C, Yi S, Hsia Y, Nattermann U, Belnap DM, et al. Designed proteins induce the formation of nanocage-containing extracellular vesicles. Nature. 2016. December;540(7632):292–5. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morita E, Sandrin V, McCullough J, Katsuyama A, Hamilton IB, Sundquist WI. ESCRT-III Protein Requirements for HIV-1 Budding. Cell Host Microbe. 2011. March;9(3):235–42. PubMed PMID: English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meng B, Ip NCY, Prestwood LJ, Abbink TEM, Lever AML. Evidence that the endosomal sorting complex required for transport-II (ESCRT-II) is required for efficient human immunodeficiency virus-1 (HIV-1) production. Retrovirology. 2015;12:72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ghoujal B, Milev MP, Ajamian L, Abel K, Mouland AJ. ESCRT-II’s involvement in HIV-1 genomic RNA trafficking and assembly. Biol Cell. 2012. December;104(12):706–21. PubMed PMID: Epub 2012/09/18. eng. [DOI] [PubMed] [Google Scholar]
- 42.Christ L, Wenzel EM, Liestol K, Raiborg C, Campsteijn C, Stenmark H. ALIX and ESCRT-I/II function as parallel ESCRT-III recruiters in cytokinetic abscission. J Cell Biol. 2016;212:499–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dussupt V, Javid MP, Abou-Jaoude G, Jadwin JA, de La Cruz J, Nagashima K, et al. The Nucleocapsid Region of HIV-1 Gag Cooperates with the PTAP and LYPXnL Late Domains to Recruit the Cellular Machinery Necessary for Viral Budding. PLoS Path. 2009. March;5(3). PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Popov S, Popova E, Inoue M, Gottlinger HG. Divergent Bro1 Domains Share the Capacity To Bind Human Immunodeficiency Virus Type 1 Nucleocapsid and To Enhance Virus-Like Particle Production. J Virol. 2009. July 15;83(14):7185–93. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sette P, O’Connor SK, Yerramilli VS, Dussupt V, Nagashima K, Chutiraka K, et al. HIV-1 Nucleocapsid Mimics the Membrane Adaptor Syntenin PDZ to Gain Access to ESCRTs and Promote Virus Budding. Cell Host Microbe. 2016. March;19(3):336–48. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee IH, Kai H, Carlson LA, Groves JT, Hurley JH. Negative membrane curvature catalyzes nucleation of endosomal sorting complex required for transport (ESCRT)-III assembly. Proc Natl Acad Sci U S A. 2015. December;112(52):15892–7. PubMed PMID: English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Keren-Kaplan T, Attali I, Estrin M, Kuo LS, Farkash E, Jerabek-Willemsen M, et al. Structure-based in silico identification of ubiquitin-binding domains provides insights into the ALIXV: ubiquitin complex and retrovirus budding. EMBO J. 2013. February 20;32(4):538–51. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowlatshahi DP, Sandrin V, Vivona S, Shaler TA, Kaiser SE, Melandri F, et al. ALIX Is a Lys63-Specific Polyubiquitin Binding Protein that Functions in Retrovirus Budding. Dev Cell. 2012. December 11;23(6):1247–54. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hirano S, Suzuki N, Slagsvold T, Kawasaki M, Trambaiolo D, Kato R, et al. Structural basis of ubiquitin recognition by mammalian Eap45 GLUE domain. Nat Struct Mol Biol. 2006. November;13(11):1031–2. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 50.Alam SL, Langelier C, Whitby FG, Koirala S, Robinson H, Hill CP, et al. Structural basis for ubiquitin recognition by the human ESCRT-II EAP45 GLUE domain. Nat Struct Mol Biol. 2006. November;13(11):1029–30. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 51.Slagsvold T, Aasland R, Hirano S, Bache KG, Raiborg C, Trambaiolo D, et al. Eap45 in mammalian ESCRT-II binds ubiquitin via a phosphoinositide-interacting GLUE domain. J Biol Chem. 2005;280(20):19600–6. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 52.Jager S, Gottwein E, Krausslich HG. Ubiquitination of human immunodeficiency virus type 1 Gag is highly dependent on Gag membrane association. J Virol. 2007. September;81(17):9193–201. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss ER, Popova E, Yamanaka H, Kim HC, Huibregtse JM, Gottlinger H. Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog. 2010. September 16;6(9):e1001107 PubMed PMID: Pubmed Central PMCID: PMC2940739. Epub 2010/09/24. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sette P, Jadwin JA, Dussupt V, Bello NF, Bouamr F. The ESCRT-associated protein Alix recruits the ubiquitin ligase Nedd4–1 to facilitate HIV-1 release through the LYPXnL L domain motif. J Virol. 2010. August;84(16):8181–92. PubMed PMID: Pubmed Central PMCID: PMC2916511. Epub 2010/06/04. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sette P, Nagashima K, Piper RC, Bouamr F. Ubiquitin conjugation to Gag is essential for ESCRT-mediated HIV-1 budding. Retrovirology. 2013:doi:10.1186/742-4690-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol. 2010;11(8):556–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Votteler J, Sundquist WI. Virus Budding and the ESCRT Pathway. Cell Host Microbe. 2013. September 11;14(3):232–41. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hurley JH. ESCRT Complexes and the Biogenesis of Multivesicular Bodies. Curr Opin Cell Biol. 2008;20:4–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. [DOI] [PubMed] [Google Scholar]
- 60.Hanson PI, Cashikar A. Multivesicular Body Morphogenesis. Annu Rev Cell Dev Biol. 2012;28(1):337–62. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 61.Prescher J, Baumgartel V, Ivanchenko S, Torrano AA, Brauchle C, Muller B, et al. Super-resolution imaging of ESCRT-proteins at HIV-1 assembly sites. PLoS Path. 2015 2015;11(2):doi: 10.1371/journal.ppat.1004677. PubMed PMID: English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Engelenburg SB, Shtengel G, Sengupta P, Waki K, Jarnik M, Ablan SD, et al. Distribution of ESCRT Machinery at HIV Assembly Sites Reveals Virus Scaffolding of ESCRT Subunits. Science. 2014. February;343(6171):653–6. PubMed PMID: English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bregnard C, Zamborlini A, Leduc M, Chafey P, Camoin L, Saib A, et al. Comparative proteomic analysis of HIV-1 particles reveals a role for Ezrin and EHD4 in the Nef-dependent increase of virus infectivity. J Virol. 2013. April;87(7):3729–40. PubMed PMID: Pubmed Central PMCID: PMC3624205. Epub 2013/01/18. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Denard J, Rundwasser S, Laroudie N, Gonnet F, Naldini L, Radrizzani M, et al. Quantitative proteomic analysis of lentiviral vectors using 2- DE. Proteomics 2009. July;9(14):3666–76. PubMed PMID: Epub 2009/07/30. eng. [DOI] [PubMed] [Google Scholar]
- 65.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Jr., et al. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol. 2006. September;80(18):9039–52. PubMed PMID: Pubmed Central PMCID: PMC1563931. Epub 2006/08/31. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lippincott-Schwartz J, Freed EO, van Engelenburg SB. A Consensus View of ESCRT-Mediated Human Immunodeficiency Virus Type 1 Abscission. Annu Rev Virol. 2017. September 29;4(1):309–25. PubMed PMID: Epub 2017/07/19. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Schöneberg J, Lee I-H, Iwasa JH, Hurley JH. Reverse-topology membrane scission by the ESCRT proteins. Nature Reviews Molecular Cell Biology. 2017;18:5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fabrikant G, Lata S, Riches JD, Briggs JAG, Weissenhorn W, Kozlov MM. Computational Model of Membrane Fission Catalyzed by ESCRT-III. PLoS Comput Biol. 2009;5(11):e1000575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carlson L-A, Shen Q-T, Pavlin MR, Hurley JH. ESCRT Filaments as Spiral Springs. Dev Cell. 2015. November 23;35(4):397–8. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 70.Jouvenet NJN, Zhadina M, Bieniasz PD, Simon SM. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol. 2011. April;13(4):394–401. PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baumgartel VBV, Ivanchenko S, Dupont A, Sergeev M, Wiseman PW, Krausslich HG, et al. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol. 2011. April;13(4):469–74. PubMed PMID: . [DOI] [PubMed] [Google Scholar]
- 72.Johnson DS, Bleck M, Simon SM. Timing of ESCRT-III protein recruitment and membrane scission during HIV-1 assembly. bioRxiv. 2018;10.1101/281774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schoeneberg J, Yan S, Righini M, Remec Pavlin M, Lee IH, Carlson LA, et al. ATP-dependent force generation and membrane scission by ESCRT-III and Vps4. bioRxiv. 2018;262170; doi: https://doi.org/10.1101/262170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang B, Stjepanovic G, Shen QT, Martin A, Hurley JH. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat Struct Mol Biol. 2015. June;22(6):492–8. PubMed PMID: English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Han H, Monroe N, Sundquist WI, Shen PS, Hill CP. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. Elife. 2017. November;6:pii: e31324 PubMed PMID: . [DOI] [PMC free article] [PubMed] [Google Scholar]