ABSTRACT
Fusarium root rot is a major pea disease in Canada and only partial tolerance exists in germplasm. Transgenic technologies may hold promise but the economic benefits of genetically modified (GM) pea will need to surpass the regulatory costs, time and labor involved in bringing a GM crop to market. European pea (Pisum sativum L.) cultivars expressing four antifungal genes, 1-3 β glucanase (G), endochitinase (C) (belonging to PR proteins family), polygalacturonase inhibiting proteins (PGIPs) (P) and stilbene synthase (V) have been transformed for disease tolerance and showed disease tolerance under laboratory conditions. Transgenic lines with four antifungal genes inserted either individually or stacked through crossing were tested for their efficacy against Fusarium root rot (Fusarium avenaceum) in confined trials over three years (2013 to 2015) in comparison with two parental German lines and three Canadian lines. Superior emergence, higher fresh weight or lower disease ratings above and below ground, of transgenic lines in presence of disease inoculum were not observed consistently in the three years of field experiments when compared to the parental and Canadian lines in the presence of disease inoculum. No indication of an advantage of stacked genes over single genes was observed. Most transgenic lines had lower relative gene expression in the roots than in the leaves in greenhouse trials suggesting a possible explanation for poor tolerance to Fusarium root rot. Field trials are necessary to verify the agronomic performance and ecological relevance of the promising effects detected under laboratory conditions.
KEYWORDS: antifungal genes, genetically modified, disease tolerance, gene stacking, confined field trial, Pisum Sativum L
INTRODUCTION
Grain legumes are valuable worldwide, for their nutritional and health benefits and contribution towards agricultural sustainability. Among grain legumes, pea (Pisum sativum L.) is economically important next only to soybean and bean worldwide and is mostly grown in temperate regions. Pea is the largest pulse crop in the multi-billion dollar Canadian pulse industry, grown primarily in the prairie provinces of Saskatchewan, Alberta and Manitoba, with harvested area of 1.68 M ha in 2016.1 The ability of pea to fix nitrogen promotes environmental stewardship by decreasing fertilizer application, reducing greenhouse gas emissions while increasing nitrogen availability for subsequent crops. Development of field pea cultivars with improved yield, disease resistance, abiotic stress tolerance and seed quality have been identified as major research priorities by Canadian pulse growers.2 However, further improvement to yield and seed quality is limited by pea diseases, particularly the fungal endemic disease complex, Fusarium root rot (Fusarium spp.) which could be responsible for up to 60% yield losses in pea crops commercially in Canada.3
Recent disease surveys in Alberta have pinpointed Fusarium species associated with pea root rot complex,4 with F. avenaceum as the primary causal agent, whose increasing populations may have been benefitted by the crop rotations with canola.5 Fusarium avenaceum isolates show genetic and ecological plasticity, occupying various ecological niches such as root tissues of legumes, head and root tissues of cereals6 and hence, is also the causal agent for root rots associated with other common crop crops in the prairies including canola7 and wheat.8 Currently, no fungicides are effective against Fusarium root rot of pea and only partial disease resistance in pea has been identified.9 Management strategies are reliant on crop rotation, although species involved in this fungal complex can survive for several years in soil.5 Disease resistance would have positive economic impact on the pea industry in prairies. Genetic transformation could aid classical breeding techniques, by overcoming sexual incompatibility of related species and lack of natural sources of resistance in pea. Because of its self-pollinating nature, low degree of outcrossing and low allergenicity, pea is a good candidate for genetic modification.2,10 However, the economic benefits of genetically modified (GM) pea will need to surpass the regulatory costs, time and labor involved in bringing a GM crop to market11 and no GM pea has been commercialized to date for disease tolerance.
Despite the effort in crop improvement via transgenic technology, there are few examples of commercially successful transgenic legumes beyond soybean.12 Particularly in the case of pea, biotechnological approaches have been restricted to development of insect resistance traits13,14 or drought tolerance (Kahlon et al. 2017 unpublished). Successful examples of commercial release of GM disease resistant crops in general, are rare, currently limited to the example of ring spot virus resistant papaya genetically modified with the coat proteins from mild virus strains of the pathogen inserted.15 The lack of GM disease resistant crops could be attributed to lower level of disease resistance conferred (compared to other traits such as herbicide resistance), which is below economic threshold for producers or a high level of resistance but only to a very specific pathogen.16
Several approaches have been used to engineer plants for fungal resistance (for reviews, see ref16-18 such as introduction of resistance-genes (R-genes)(utilizing plants basal defense responses),19 detoxification of virulence factors,16 expression of antimicrobial secondary metabolites like phytoalexins and pathogenesis related (PR) proteins (inhibiting the pathogen's capacity to degrade polysaccharides within cell wall or RNA),16,20 and modification of plant signaling pathways including transcription factors genetically. It is notable that for achieving enhanced diseases tolerance, working on pathogens with a wide host range (particularly for seedling infecting pathogens), have been more successful.17 Fusarium spp. are often classified as hemibiotrophs because their infection pattern initially resembles that of a biotrophic pathogen (relying on living host) and gradually transitions into a necrotropic pathogen (consumer of host cells after killing them).21 Such pathogens invade living plant cells and subvert the metabolism in favor of their own growth, hence with such specialized plant-hemibiotrophic pathogen interactions, even minor changes in either host or pathogen can upset the balance.22 Achieving genetic resistance/tolerance becomes even more difficult when pathogens are genetic variable, as has been reported in cases of F. graminearum,23 F. avenaceum24 and other Fusarium species, because of a variation facilitated rapid evolution of resistance.5 Often, disease resistance is a highly complex multigenic trait and thus single gene transformations may be insufficient and/or offer limited spectrum disease resistance,25 or single gene resistance could be circumvented by a mutation reducing the function of the introduced gene.26 Hence, stacking/pyramiding more than one gene decreases the risk of resistance development.27 However, coordinated expression of several genes in one plant could pose an additional challenge.27,28
Recently, European pea (Pisum sativum L.) cultivars expressing four antifungal genes, 1-3 β glucanase (G), endochitinase (C) (belonging to PR proteins family), polygalacturonase inhibiting proteins (PGIPs) (P) and stilbene synthase (V) have been transformed for disease tolerance29-31 The PR proteins (1-3 β glucanase and endochitinase) degrade microbial cell wall components (i.e. glucan and chitin)32 and PGIPs can inhibit fungal endopolygalacturonases causing fungal wall degradation and plant tissue maceration33; both are important components of quantitative plant defense responses. Stilbene synthase belongs to the phytoalexins class of secondary metabolites that possess biological activity against a wide range of pathogens.34 Transgenic plants showed enhanced tolerance to fungi in in vitro testing (inhibition of fungal spore germination of Trichoderma harzianum (T12 strain).29 However, field trial is crucial to establish trait efficacy, especially with soil dwelling pathogens because of the complexity and high degree of temporal and spatial variation in soil based ecosystems.35 However, the field-testing of GM crops in Germany is complicated due to non-transparent legislation of GM crops.36 An experienced regulatory framework exists in Canada and Canada regulates products derived from biotechnology processes under “novel products”. Therefore GM plants are known as Plant with Novel Trait (PNT) and regulated under the auspices of Canadian Food Inspection Agency (CFIA), Health Canada and Environment Canada.37
We report here three years of investigation of confined field trial of transgenic disease tolerant pea stably expressing antifungal genes single and stacked against Fusarium avenaceum in comparison to parental lines and well adapted Canadian pea lines. Our hypothesis was that pea transformed with antifungal proteins would tolerate Fusarium root rot better than Canadian conventional pea lines and parental lines. We also hypothesize that transgenic lines with stacked genes will have an advantage over single gene insertions in response to Fusarium root rot in the field.
MATERIALS AND METHODS
Plant Material
Four antifungal genes V, P, G, C encoding for disease resistance were transformed into European pea cultivars “Baroness” and “Sponsor” at Institute of Plant Genetics, Department of Plant Biotechnology, Leibniz University Hannover, Germany. Embryo axis excised from mature seeds using the modified protocol by Schroeder et al.,38 of European pea cv. Baroness (P. sativum L. cv. Baroness) for transformation with V, P, G genes (V had inducible Vst promoter, P, G had constitutive D35S promoter), and similarly, European pea cv. Sponsor (P. sativum L. cv. Sponsor) was employed for transformation with C gene (promoter D35S), served as explants for Agrobacterium-mediated transformation using Agrobacterium strain EHA105.39 Herbicide resistant bar gene from Streptomyces hygroscopicus was present along with the genes as selectable marker40,41 (for functional map of each transgene, details of choice of promoters, sources of genes, please refer to.42 Two single Chitinase gene lines, have either D35S (double 35S promoter from CaMV {Cauliflower Mosaic Virus}) (line 18) or Vst (Stilbene synthase) promoter from grape (line 20). Conventional breeding was employed to incorporate genes into a single line (V:P43 x G:C29 = V:P:G:C.44
Gene Expression
To determine if gene expression in transgenic and non-transgenic lines used in the field experiment was maintained in subsequent generations, they were grown in a greenhouse in a separate experiment. Plants (10 plants line−1) were seeded in pre-autoclaved vermiculite-perlite mixture (Sunshine Mix®#4, Sun Gro Horticulture, Canada) and retained at 25 ± 2°C with a 16/8-h light/dark photoperiod for four weeks after which root and leaf samples were collected and cleaned thrice with RNAse free water. According to manufacturer (Qiagen, Canada)'s recommended protocol, total RNA was extracted from tissues using the Qiagen RNeasy Plant Mini Kit. Briefly, frozen tissues were ground into a fine powder with liquid nitrogen using baked (250°C, 3 hr) and chilled (−80°C) mortar and pestle. Then a 100 mg of the powder was taken and mixed with 450 µl of buffer RLT (containing β-ME) and vortexed to obtain slurry, incubated at 56°C for 3 min, passed through the QIAshredder column and centrifuged at 21,000xg for 2 min. In a collect tube (with flow through), 200 µl of absolute ethanol was added and the mix applied to RNeasy column and centrifuged at 10,000×g for 30 sec. The flow-through was dispensed off and column washed by adding 350 µl RW1 buffer, centrifuged again and subjected to on-column DNA digestion using RNase free DNase set (Qiagen) by adding 27.27 U DNase in 80 µl RDD buffer to the column and incubated at room temperature for 15 min. This was followed by additions of 700 µl RW1, 500 µl RPE and 50 µl RPE and centrifugation at 10,000xg and elimination of the flow-through to the column at each step. In a new 2 ml collection tube, the column was centrifuged at 12,000xg for 2 min and finally, transferred to a 1.5ml microfuge tube. Addition of 50 µl of nuclease free water to the centre of the column, and centrifugation of RNA eluted at 12,0000xg for 30 sec was next. A NanoDrop™ spectrophotometer (Thermo Fisher Scientific™) was used to quantify the extracted RNA and it was stored at −80°C until further analysis.
cDNA Synthesis
In a 20 µl volume, complimentary DNA (cDNA) was synthesized using 1 µg of total RNA using RevertAid RT kit (Thermo Fisher Scientific™) as per the recommended protocol. Briefly, 1µg of total RNA was used as the template in a 20 µl reaction containing 100 nmole of random hexamer primer, 20 U/µl of RiboLock RNase inhibitor, 10 nmole of dNTP and 200 U/µl of RevertAid reverse transcriptase in 1x reaction buffer. The contents were mixed, centrifuged and incubated at 25°C for 5 min, followed by 60 min at 42°C for cDNA synthesis followed by heating the tubes to 70°C for 5min to inactivate the enzyme. Products were stored at −80°C.
Quantitation of Gene Expression by Real-Time PCR
Gene expression levels (of V, P, G, and C) in transgenic pea line's tissues (from root and leaf), were quantified using SYBR Green based q-RT-PCR on a StepOnePlus™ instrument (Applied Biosystems®, Canada) with quantitation employing ΔΔCT method with melt curve. A 10 µl reaction contained 5 µl of 2 x KAPA SYBR® Fast Master Mix (Kapa Biosystems, Boston, MA, USA), 1 µl of 1:15 diluted cDNA, and 5 pmol of each (forward and reverse) gene specific primers which were designed using either Primer Express 3.0 (Applied biosystems) or PrimerQuest (Integrated DNA technologies, Coralville, Iowa) with Tm of 60°C and amplicon sizes between 100-140bp. Elongation factor 1a was used as endogenous control. Primers used were “P” forward: 5′-CTTCGAAATCAAGACAGCCTTCA-3′; reverse: 5′-GGGATCACACTCGACGCAGTA-3′; “V” forward: 5-AGAAATGCCCGGTGCAGAT-3′, reverse: 5′-TTCCACCTGCATAGCAACCTT-3′; “G” forward: 5′- AAC GCG CGG AAC TAC AA -3′, reverse: 5′- CTC GTT GAA CAT GGC GAA TAT G -3′; “C” forward: 5′- GAA CCG GAA CTC CTT CTA CAG -3′, reverse: 5′- TCC TGC TTC TTG GTG GTG -3′ and endogenous control forward: 5′-GATGGATGCTACCACCCCTAAG-3′, reverse: 5′-GAGATGGGAACGAAGGGAATT-3′). Every reaction was carried out in triplicate using 6 cDNA samples from individual plants from each line and the average CT values were used for calculating gene expression. The detection limit for the plasmid copies was obtained with a dilution series between 107 and 101 copies per reaction, a linear range of detection was established and were added to the German parental lines (Sponsor and Baroness)'s cDNA sample to serve as the baseline for estimating relative expression. These methods are previously published.42
Field Trials
A confined field trial was established at a secure field site located at the Crop Diversification Center (CDC) North, Alberta Agriculture and Forestry (AAF), north east of Edmonton, AB (lat. 53°38′N, long.113°22′W), on a black chernozemic sandy loam soil in spring of 2013, 2014 and 2015, following the regulatory guidelines for field testing of PNTs outlined by the CFIA. Seventeen treatments, comprised of nine transgenic lines (five lines with single gene insertions {5(G), 18(C), 20(C), 21(V) and 23(P)} one line with double gene insertion {4(V:P)}, one line with triple gene insertion {11(P:C:G)}, two lines with four gene insertions {8(V:P:G:C), 10(V:P:G:C)} and as comparator, four lines including two German parental lines, Sponsor and Baroness and three Canadian lines with/without disease inoculum : Agassiz ({resistant to Powdery mildew (Erysiphe pisi Syd.) and moderately susceptible to Mycosphaerella blight (Mycosphaerella pinodes) }45), AC Earlystar ({Resistant to powdery mildew moderately resistant to Mycosphaerella blight and Fusarium wilt (Fusarium oxysporum)}46) and AAC Royce ({resistant to powdery mildew, moderately susceptible to Mycosphaerella blight and Fusarium wilt47). Seeds were individually planted by hand at 30 seeds plot−1 (1 x 0.5m) at 5cm depth. Each plot was separated by seeded rows of conventional AC Ultima triticale to better delineate one genetic composition to another and limit tangling of pea lines between plots. All plots were inoculated with Rhizobium leguminosarum bv. Viciae (1.6 × 109 viable cells g−1) (Galloway seeds Ltd. Fort Saskatchewan, AB) at a rate of 291.58 g ha−1 @0.004g for 2.5g seeds for promoting root nodulation. All transgenic lines, German parental controls and three Canadian lines were also treated with Fusarium avenaceum inoculum @5g1m−1 row, ground into fine power from previous year's infected wheat plants and applied in contact with the seed at the time of seeding to promote disease establishment. After soil testing each year, appropriate fertilizers were added and plots were hand weeded throughout the growing season. The plots were arranged in randomized complete block design with pea lines as treatment randomly arranged in blocks, six replicates per treatment.
Confirmation of Pathogen Presence/ Disease Verification
Pea roots, the Fusarium avenaceum inoculum and random soil samples from the field site for 2013, 2014 and 2015 trials were used for characterization of the pathogen using plating and polymerase chain reaction (PCR).
Inoculum Plating
Approximately 1mg of ground Fusarium avenaceum inoculum used in the field experiment each year was plated onto potato dextrose agar (PDA) with antibiotics on petri dishes. Plates were incubated for 7-10 days at room temperature and resulting cultures were sub cultured and confirmed as F. avenaceum based on the colony morphology using culture identification techniques outlined in.48
Root Sample Plating
Five random roots plot−1 were selected in each replicate. Tap roots showing necrosis were cut into 1cm pieces and 3 pieces plot−1 were randomly selected each field season, transferred into 15 ml culture tube and surface sterilized using 70% ethanol for 30s followed by 0.5% NaOCl (10% bleach) for 2 min, rinsed 3X in distilled water and blotted dry on sterile filter paper. Three root pieces plot−1 were plated on acidified potato dextrose agar (APDA) in 90 mm Petri dish and incubated on the laboratory benchtop for 7 -10 days (8 h light, 16 h dark, 22°C). Colonies growing from the root pieces were transferred to a new APDA plate, using hyphal tip transfer under a dissecting microscope. Presumptive identification of F. avenaceum, F. solani, F. redolens and F. oxysporum were then made based on the distinct morphological characteristics of these species (pink/yellow/dark red/or purple pigmentation and shape of macroconidia under microscope) when possible, and in comparison to stock cultures as per,48 and confirmed with PCR. Number of roots plate−1 yielding a Fusarium culture was recorded. In 2014 and 2015, the symptomatic areas of roots were also used in a series of multiplex PCR reactions to assess for presence of 10 Fusarium species and Aphanomyces euteiches (another pathogen commonly associated with root rots in prairies). Briefly, root samples were lyophilized for 48 h (4.5 L FreeZone, LabConco, Kansas City, Missouri, USA) and then 30 mg of tissue transferred to collection microtubes in a 96-well plate format. Samples were then ground using a TissueLyzer II (Qiagen, Carlsbad, California, USA) and DNA extracted using the PlantDNABiosprint kit according to manufacturer's instructions (Qiagen, Carlsbad, California, USA). The multiplex reactions were performed using the Qiagen Multiplex PCR Kit according to manufacturer's directions with 2 µL of DNA and 0.2 µM of each primer (details of primers included in supplementary online material). Multiplex combinations were as follows: 1) F. graminearum, F. poae, F. oxysporum and A. euteiches; 2) F. acuminatum, F. solani and F. equisiti; 3) F. culmorum, F. redolens, F. sporotrichoides and A. euteiches; and 4) F. avenaceum (2 primer pairs). A positive DNA standard from stock cultures of all species was included with all multiplex reactions to ensure there was no cross-reactivity between primer pairs. There was cross-reactivity between the F. avenaceum primer pairs with F. acuminatum DNA. A combination of the two F. avenaceum and F. acuminatum primers could usually differentiate between these two species, but in some cases the reaction was scored as mixed for avenaceum/acuminatum when results were not clear.
Soil Sample Plating
Soil samples were collected from randomly chosen plots immediately prior to seeding of the trials each year. Soils were diluted 1:50 in sterile distilled water and 1ml was plated onto potato dextrose agar (PDA) with antibiotics on petri dishes. Plates were incubated for 7-10 days at room temperature and the resulting mixture of cultures were scraped from agar surface and subjected to DNA extractions and a multiplex PCR, as described above.
Plant Growth Assessment and Disease Ratings
Data was collected for seedling emergence (14 days after planting (DAP)), plant heights and root diameters (28, 42 DAP, 5 plants plot−1). The disease symptomology and severity for above49 and below ground plant parts50 (Table 1) were recorded from all plants in each plot after destructive sampling at 8 weeks after planting. Fresh weight (gm plot−1) was recorded as an indicator of the potential yield, because of the destructive nature of sampling for disease severity ratings.
TABLE 1.
| Tissue used | Observations recorded | Rating | ||
|---|---|---|---|---|
| Above ground | Healthy plant | 1 | ||
| Slight yellowing of lower leaves | 2 | |||
| Yellowing of lower leaves upto the 3rd or 4th node, some stunting | 3 | |||
| Necrosis of at least half or more of the plants with some stunting | 4 | |||
| All plants dead or nearly so | 5 | |||
| Below ground | Lesions | (%) Root discoloration | Root mass reduction | |
| 0 | 0 | 0 | 1 | |
| 0.1-0.2cm, small reddish brown at hypocotyl base | 0 | 0 | 2 | |
| Coalescing of localized root/hypocotyl lesions from 0.5-1cm, around the stem | 10-20% | 0 | 3 | |
| Lesions extending and completely encircling the stem | 95% | 5-10% | 4 | |
| Increasingly discolored and extended hypocotyl lesions | 100% | 20-50% | 5 | |
| Hypocotyl lesions encircling the stem extending up to 2 cm | 100% | 50-80% | 6 | |
| Pithy or hollow hypocotyl with very extended lesions | Dead | Dead | 7 | |
Statistical Analysis
Data generated from field experiment were analyzed using analysis of variance (ANOVA) using PROC MIXED in SAS 9.4,51 where lines and years were used as fixed effects and blocks as the random effect. LSmeans were compared using pre-planned orthogonal contrasts statements.
RESULTS
Gene Expression Analysis
The relative gene expression was higher in leaf tissue than in root tissue in all of the transgenic pea lines except C, where expression was similar (Table 2, previously published, in part.42 Gene expression between genes and lines was highly variable. Relative V expression was significantly higher (p < 0.05) in leaves compared to roots for line 21(V) (p = 0.0011) and line 10(V:P:G:C) (p = <0.0001). It was highest in line 10(V:P:G:C) followed by line 21(V) and line 4(V:P), and negligible in line 8(V:P:G:C). Interestingly, P had significantly lower relative root P expression than in leaf, for line 10(V:P:G:C) (p = <0.0001). Highest relative P expression was observed in leaf tissue of line 10(V:P:G:C) followed by line 4(V:P), 11(P:C:G) but lower in roots of these lines and negligible in root tissue of line 8(V:P:G:C). Relative G expression of root tissue of line 10(V:P:G:C) was significantly lower than leaf tissue (p = <0.0001) which is a four gene lines, but there was no significant difference among leaf and root tissue was recorded for the four genes stacked line 8(V:P:G:C) and three genes stacked line 11(P:C:G) as well as single gene line 5(G). All lines tested for relative expression levels of C were not significantly different between leaf and root tissues, however, line 18(C) had some low, relative gene expression (14.62 ± 4.01in leaf and 18.01 ± 2.56 in root) in comparison to negligible expression in stacked lines like 8(V:P:G:C), 10(V:P:G:C) and 11(P:C:G) and same gene but under a different promoter line 20(C). Variable relative gene expression among various genetic compositions of pea lines suggests gene silencing or possibly unequal efficacy of promoters. All transgenic lines were confirmed for homozygosity (data not shown).
TABLE 2.
Relative gene expression ± standard error (SE) of each gene in roots and leaves, for each transgenic line used in all field experiments.
| Gene | Relative gene expression |
||
|---|---|---|---|
| Line | Root ± SE | Leaf ± SE | |
| V | 4(V:P) | 16.87 ± 2.45 | 44.42 ± 4.40 |
| 21(V) | 32.85 ± 2.36 | 558.48 ± 84.99 | |
| 8(V:P:G:C) | 0.02 ± 0.00 | 1.17 ± 0.35 | |
| 10(V:P:G:C) | 40.47 ± 12.54 | 699.14 ± 220.76 | |
| P | 4(V:P) | 268.79 ± 159.53 | 32963 ± 14166.04 |
| 23(P) | |||
| 11(P:C:G) | 122.66 ± 69.40 | 11892.53 ± 6171.69 | |
| 8(V:P:G:C) | 3.58 ± 0.47 | 469.48 ± 93.35 | |
| 10(V:P:G:C) | 614.05 ± 72.30 | 111577.50 ± 27728.24 | |
| G | 5(G) | 127.21 ± 71.77 | 452.07 ± 214.25 |
| 11(P:C:G) | 1.53 ± 0.40 | 4.26 ± 1.03 | |
| 8(V:P:G:C) | 0.32 ± 0.20 | 1.77 ± 0.32 | |
| 10(V:P:G:C) | 94.94 ± 31.61 | 1566.15 ± 462.75 | |
| C | 18(C) | 18.01 ± 2.56 | 14.62 ± 4.01 |
| 20(C) | 0.10 ± 0.02 | 0.19 ± 0.03 | |
| 11(P:C:G) | 0.25 ± 0.04 | 0.08 ± 0.01 | |
| 8(V:P:G:C) | 0.24 ± 0.03 | 0.10 ± 0.01 | |
| 10(V:P:G:C) | 0.08 ± 0.01 | 0.09 ± 0.01 | |
data not available
Confirmation of Pathogen
In 2013, the majority of cultures resulting from the root samples were identified and confirmed by PCR as Fusarium spp. The most prevalent Fusarium species identified were Fusarium solani, F. avenaceum and F. redolens while other Fusarium species identified (for example, F. acuminatum) were relatively less abundant. Presence of other common soil inhabiting fungus like Rhizoctonia spp., Rhizopus spp., Trichoderma spp. and Clonostachys rosea were also recorded. The soil plating followed similar trends and F. redolens, F. solani and F. avenaceum were identified as the predominant Fusarium species, present in the soil. In 2014 and 2015, F. redolens and F. solani were the most abundant in soil samples with lesser F. avenaceum present than in 2013. The root plating and PCR confirmations for 2014 and 2015 resulted in fewer Fusarium spp identified from all samples than in 2013. For 2014 samples, the major Fusarium spp. identified and confirmed were F. solani, F. equisiti, F. oxysporum and F. avenaceum. For 2015, the pathogens present were characterized as Fusarium spp. and were not found in all the samples tested.
Growth Assessment
The precipitation accumulated during the growing period for three years, at the trial location was 58.43%, 75.59% and 60.17% of the long term average (LTA) for 2013, 2014 and 2015 respectively (Table 3). The percentage emergence of plants plot−1 14 DAP are summarized in Fig. 1. The line*year interaction was significant (p < 0.0001) at 0.05% level of significance and hence data is presented per year. Overall, all transgenic, parental and Canadian lines which received F. avenaceum inoculum showed reduced emergence (%) plot−1 than the Canadian lines that did not receive the inoculum which indicates that disease inoculum challenged pea seedlings.
TABLE 3.
Precipitation at field trial location conducted in three sequential years.
| ————–mm———— |
|||
|---|---|---|---|
| Location | Accumulated from May-October | Long term Average (LTA) | –%–% of LTA |
| 2013 | 201.3 | 344.5 | 58.43 |
| 2014 | 260.4 | 344.5 | 75.59 |
| 2015 | 207.3 | 344.5 | 60.17 |
FIGURE 1.
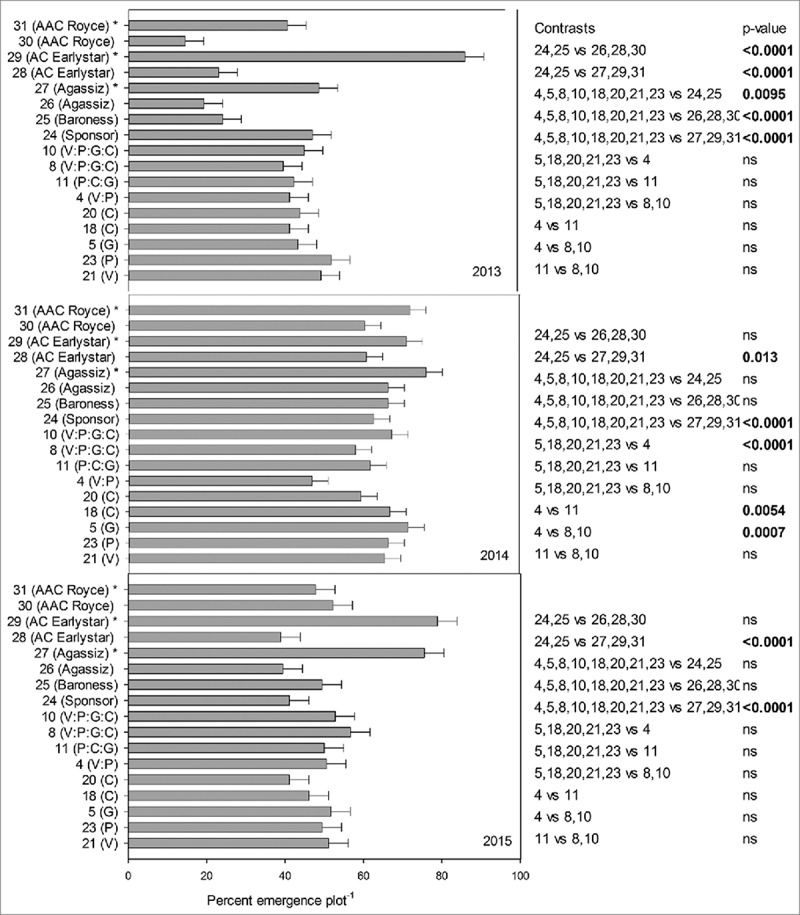
Emergence percent (plot−1) in presence or absence of disease inoculum, of transgenic lines and conventional pea lines (transgenic lines: 21(V), 23(P), 5(G), 18(C), 20(C), 4(V:P), 11(P:C:G), 8(V:P:G:C), 10(V:P:G:C), 24(Sponsor), 25(Baroness), 26(Agassiz), 27(Agassiz)*, 28(AC Earlystar), 29(AC Earlystar)*, 30(AAC Royce), 31(AAC Royce)* in three year confined field trial. * Disease inoculum was not provided.
In 2013, transgenic lines were significantly different from German parental lines 24 (Sponsor) and 25(Baroness) (p = 0.0095) and Canadian lines (with inoculum), 26(Agassiz), 28(AC Earlystar) and 30(AAC Royce) (p = <0.0001), the highest emergence (%) being for lines 23(P) & 21(V) of the transgenic lines. However, this trend did not continue in 2014 and 2015 where transgenic lines were not significantly different than the parental lines 24(Sponsor) and 25(Baroness) and Canadian lines (with inoculum), 26(Agassiz), 28(AC Earlystar) and 30(AAC Royce). We observed significant differences among some genes in 2014, for instance, two genes stacked line 4(V:P) had significantly lower percent emergence than three genes stacked line 11(P:C:G) (p = 0.0054) and four genes stacked lines 8 and 10(V:P:G:C) (p = 0.0007). In 2015, the highest emergence percent in presence of disease was recorded with transgenic line 8(V:P:G:C) but it was not significantly different from parental lines 24(Sponsor) and 25(Baroness) and Canadian lines (with inoculum), 26(Agassiz), 28(AC Earlystar) and 30(AAC Royce). In conclusion, we did not observe consistent pattern of superior emergence of transgenic line(s) in presence of disease in the three years of field experiments.
Pea plants were tallest (height cm) in 2013 followed by 2014 and 2015 but not statistically significant and no stunting or dwarfing was observed in plants that received inoculum compared to those that did not (data not shown). Because of the destructive nature of the experiment and as an indication of yield, fresh weight (gm plot−1) was recorded in three years and all lines, years and line*year interaction were significant (p = <0.001) at 0.05% level of significance, therefore, data is presented separately for each year. Fresh weight (gm plot−1) production was highest for lines 21(V) and 23(P), respectively, for 2013 and 2014 however, this pattern wasn't consistent for 2015 where line 5(G) had the highest fresh weight (Fig. 2). All transgenic lines had significantly higher fresh weight than their parents in 2013 (p = 0.007), but not in 2014 and 2015 and higher than Canadian lines that received disease inoculation in 2013 (p = <0.0001) and 2015 (p = 0.0396) but not in 2014. We observed that in presence of disease, single gene line 5(G), 18(C), 20(C), 21(V) and 23(P) had significantly higher fresh weight than four genes stacked lines 8, 10(V:P:G:C) in 2013 (p = 0.0222) and 2014 (p = 0.0304) but not in 2015. Transgenic lines were significantly different in all three years (2013 and 2015 (p = <0.0001), 2014 (p = 0.014)) from the Canadian lines that did not receive inoculum (line 27, 29 and 30) asserting that disease inoculum was effective. In general, fresh weight production among the three years did not identify any transgenic line producing significantly higher fresh weight among others or in comparison to parents and Canadian lines in presence of disease or indicating an advantage of stacked genes over single genes. Interestingly, having same chitinases gene with two different promoters (line 18(C)(D35S) and 20(C)(Vst), did not yield any significant differences among the two transgenic lines for emergence (%) and fresh weight (gm plot−1) throughout the three field seasons. Ample precipitation in 2014 possibly explains the higher emergence percent plot −1 and higher fresh weight production plot −1 in comparison to 2013 and 2015 which received lesser precipitation.
FIGURE 2.
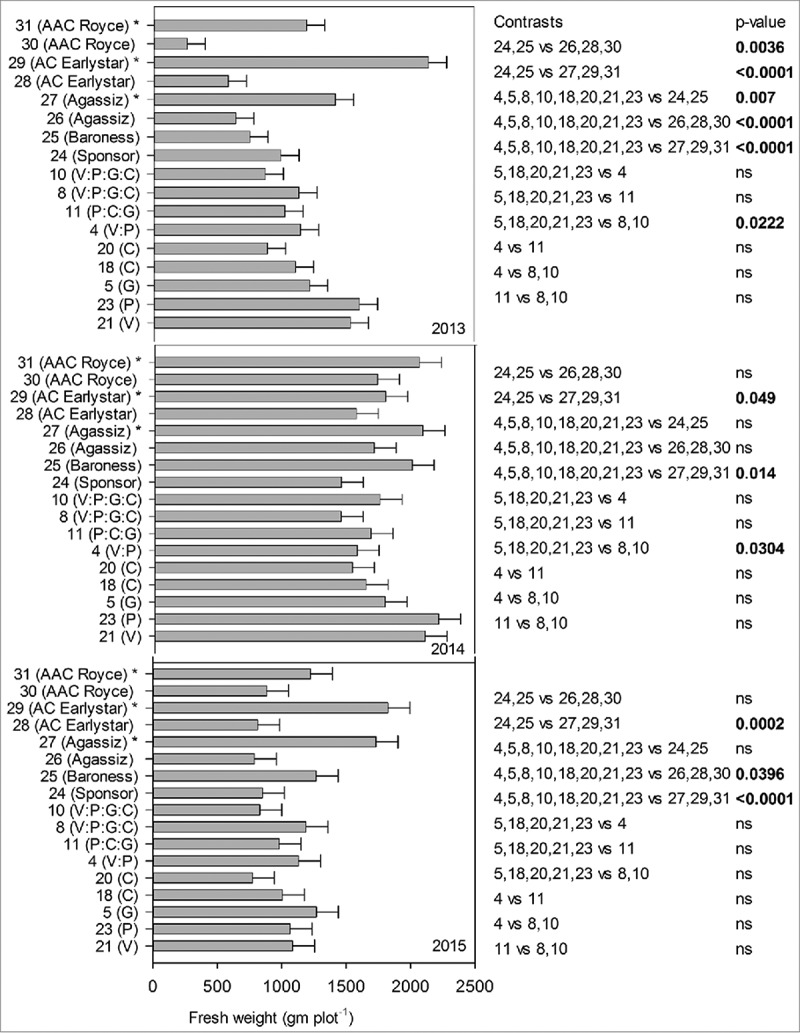
Fresh weight biomass plot−1 (gm) in presence or absence of disease inoculum, of transgenic lines and conventional pea lines (transgenic lines: 21(V), 23(P), 5(G), 18(C), 20(C), 4(V:P), 11(P:C:G), 8(V:P:G:C), 10(V:P:G:C), 24(Sponsor), 25(Baroness), 26(Agassiz), 27(Agassiz)*, 28(AC Earlystar), 29(AC Earlystar)*, 30(AAC Royce), 31(AAC Royce)* in three year confined field trial. * Disease inoculum was not provided.
Disease Severity Ratings
The line*year interaction for disease severity ratings was significant (p<0.0001) at 0.05% and hence data is presented for each year (Fig. 3 (above) and Fig. 4 (below)). Transgenic lines had significantly lower above ground disease ratings than German parental lines (24(Sponsor), 25(Baroness)) in 2013 (p = 0.0016) but not in 2014 or 2015 and Canadian lines that received disease inoculum (26(Agassiz), 28(AC Earlystar, 30(AAC Royce)) in 2013 and 2015 (p = <0.0001) but not in 2014. No significant differences were observed among transgenic lines having single or two, three and four genes stacked in 2013, but there were some significant differences between single or multiple gene lines in 2014 and 2015. For instance, single gene lines (5(G), 18(C), 20(C) and 21(V)) had significantly lower above ground disease rating vs two gene line 4(V:P) in 2014 (p = 0.0016) and two gene line 4(V:P) (p = 0.0308) and three gene line 11(P:C:G) (p = 0.0334) in 2015.
FIGURE 3.
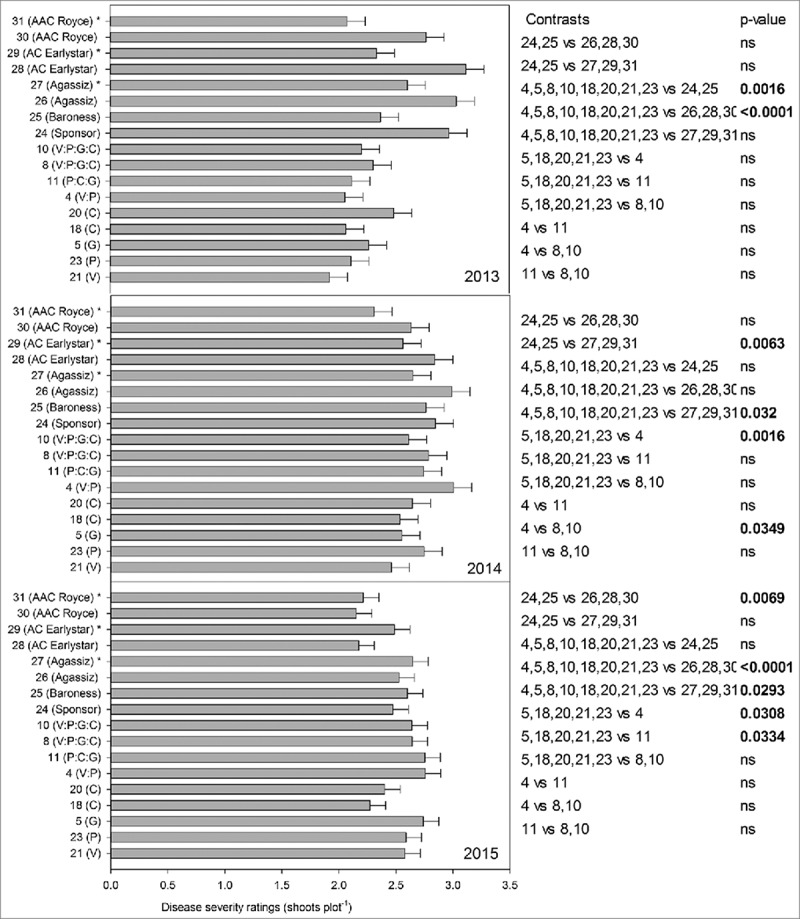
Disease severity ratings above ground (plot−1) in presence or absence of disease inoculum, of transgenic lines and conventional pea lines (transgenic lines: 21(V), 23(P), 5(G), 18(C), 20(C), 4(V:P), 11(P:C:G), 8(V:P:G:C), 10(V:P:G:C), 24(Sponsor), 25(Baroness), 26(Agassiz), 27(Agassiz)*, 28(AC Earlystar), 29(AC Earlystar)*, 30(AAC Royce), 31(AAC Royce)* in three year confined field trial. *Disease inoculum was not provided.
FIGURE 4.
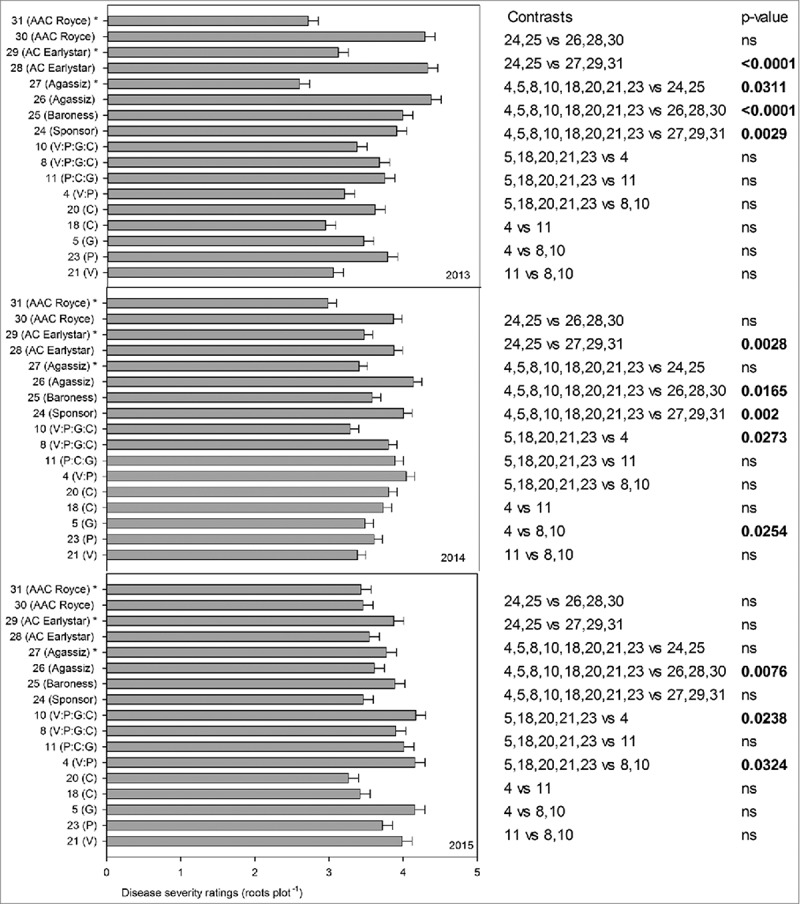
Disease severity ratings below ground (plot−1) in presence or absence of disease inoculum, of transgenic lines and conventional pea lines (transgenic lines: 21(V), 23(P), 5(G), 18(C), 20(C), 4(V:P), 11(P:C:G), 8(V:P:G:C), 10(V:P:G:C), 24(Sponsor), 25(Baroness), 26(Agassiz), 27(Agassiz)*, 28(AC Earlystar), 29(AC Earlystar)*, 30(AAC Royce), 31(AAC Royce)* in three year confined field trial. * Disease inoculum was not provided.
Similar results were recorded for disease ratings below ground. Transgenic lines had significantly lower below ground disease severity ratings than German parental lines (24(Sponsor), 25(Baroness)) in 2013 (p = 0.0311), but not in 2014 and 2015 and lower than Canadian lines that received disease inoculum (26(Agassiz), 28(AC Earlystar, 30(AAC Royce)) in 2013 (p = <0.0001), 2014 (p = 0.0165) and 2015 (p = 0.0076). In 2013, no significant advantage of single vs multiple genes in transgenic lines for disease severity ratings below ground was recorded however, differences were observed in 2014 and 2015. For example, in 2014 two gene line 4(V:P) had significantly higher below ground disease ratings than single gene (line 5(G), 18(C), 20(C), 21(V) and 23(P)) (p = 0.0273) and four gene lines 8, 10(V:P:G:C) (p = 0.0254) and single gene lines 5(G), 18(C), 20(C), 21(V) and 23(P) had significantly lower disease severity ratings than two gene line 4(V:P) (p = 0.0238) and four gene lines 8,10(V:P:G:C) (p = 0.0324) in 2015. Interestingly, throughout the three trial years (except above ground disease ratings in 2014), disease severity ratings above and below ground, were significantly lower in than the Canadian lines (26(Agassiz), 28(AC Earlystar, 30(AAC Royce)) that received disease inoculum and had some genetic advantage of partial disease resistance. It is notable that the Canadian lines have been selected in the presence (inadvertently) of similar races of these pathogens, whereas the German breeding efforts would have occurred under different selection pressure. As observed for growth parameters, lines with chitinases genes with two different promoters (line 18(C)(D35S) and 20(C) (Vst), did not yield significant differences for disease severity ratings (above and below) throughout the three field seasons. Overall, contrary to our expectation, we did not observe any transgenic line or gene combination that performed better than parental lines for disease tolerance performance in three consecutive years of field trials.
DISCUSSION
In this study, confined field trials were established for three consecutive years to test nine transgenic pea lines with four antifungal genes, present singly or stacked against Fusarium root rot and were compared to their parental lines as well as Canadian pea lines in presence/absence of disease inoculum. The variability found in transgenic lines throughout three years of field trial reveal the complexity of the disease tolerance traits and their interaction with variable environmental conditions over the different years of the study. However, we were not able to identify transgenic pea lines that outperformed parental lines or well adapted Canadian lines in presence of disease over the course of three consequent field seasons. Although some transgenes (for example, line 21(V) and 23(P) in 2013 and 2014) did indicate better performance (higher emergence, more biomass production and lower disease ratings) in presence of the pathogen than other transgenic lines, but that did not translate into consistent performance or differences in statistical significance vs comparators over the three trial years. No advantage of gene pyramiding over having individual genes was recorded, contrary to our initial hypothesis. Our results are consistent with other researchers who found that transgene insertions can have variable or no effect on disease tolerance or resistance. For example, high-level expression of tobacco chitinases gene in Nicotiana sylvestris did not increase the resistance to Cercospora nicotiana,52 bean (Phaseolus vulgaris) gene PGIP2 expressed in transgenic wheat did not reduce Claviceps purpurea symptoms53 and β-1,3 glucanase constitutively expressed in alfalfa did not decrease root severity of fungi containing chitin.54 However, contrary to what we observed, many successful transgenics, particularly in legumes, have been reported to achieving antifungal activity. For example, rice chitinases under control of CaMV35S promoter improved resistance against leaf spot (Cercospora arachidicola)55 and late leaf spot (Phaeoisariopsis personata), rust (Puccinia arachidis Speg.) and Aspergillus flavus56 in peanut (Arachis hypogaea L.). Similarly, barley chitinases (AAA56786) improved resistance to Corynespora leaf spot disease (Corynespora cassiicola) in Blackgram (Vigna mungo L. Hepper).57 Resveratrol synthase (a stilbene) from peanut decreased Black stem and leaf spot disease (Phoma medicaginis) in Alfalfa (Medicago sativa L.).58 Overexpression of tobacco β-1,3 glucanase in transgenic peanut lead to enhanced protection against Cercospora arachidicola and Aspergillus flavus59 and Cercospora personanta.60 All of these reports, however, were conducted in laboratory conditions and against single pathogen. Under laboratory conditions, differences between tested lines may be easier to differentiate as a single challenge could be imposed whereas in field trials, the number of pathogens, differences in soil moisture and weather variance more accurately represent the true agronomic effect of transgenes. Also, having multiple pathogens acting on the plant at the same time (as observed in this experiment) increase occurrence of disease symptoms.61
Gene pyramiding did not consistently have resistance advantages in trials conducted in the field as suggested by other researches. For instance,62 observed no resistance to Rhizoctonia solani in a hyphal extension assay using extracts from potato transgenics with chitinases and glucanase gene. Transgene combinations often result in successful inhibition of fungal growth in vitro but fail to translate the same success in greenhouse or field conditions, as was the case of our transgenes. Oilseed rape transformed with double gene construct of chitinases and β-1,3-glucanase genes from barley both driven by CaMV 35S promoter did not increase fungal resistance against Trichoderma sp., Alternaria brassicae, A. brassicola, Verticillium longisporum and L. maculans when assayed in greenhouse whereas purified chitinases and β-1,3-glucanase did reduce the fungal grown in vitro.63 This can be attributed to PR protein's differential activity against fungal cell wall targets and that they can be very specific for their resistance reaction against few pathogens yet be completely ineffective against some others in respect to same crop. This creates a challenge for continuous and sustainable resistance.20 For example, no known PGIP is able to inhibit polygalacturonases produced by the very harmful fungal pathogen F. verticillioides64 and PGIP1 which is unable to inhibit F. moniliforme, partially inhibits F. oxysporum f. sp. lycopersici polygalacturonases in bean (Phaseolus vulgaris)65 and chitinases obtained from Trichoderma sp are considered more effective in conferring disease resistance.66 The capacity of many fungal pathogens such as Fusarium spp, to change their genetic structure in the face of selection forces such as resistance genes and environmental factors,17 when they possess high inherent genetic variation,5 coupled with their hemibiotrophic nature,21 can also contribute to difficult to achieve disease resistance. Environmental variation has been attributed to be the most important factor contributing to disease progress and resistance responses in the case of Fusarium root rot, making this trait highly challenging to accommodate.67
Genes used in the experiment had variable relative gene expression in transgenic lines and a lower relative gene expression in the roots as compared to the shoots in general. In particular C had very low gene expression in both root and shoot tissues, which may have contributed to a lack of resistance. No significant differences amongst single or stacked genes, or choice of promoters for the same gene were recorded. The major constraint in co-expression of different transgenes is that the gene expression remains uncoordinated even with physically linked genes68 and transcriptional silencing of transgene may occur. Other factors, including choice of promoters, which can affect the strength, tissue specificity, timing69 or unexpected gene silencing70 or transgene copy number and epigenetic effects71-73 can contribute to varied gene expression. However, the presence of two different promoters driving the same chitinases gene (line 18 with Vst from Stilbene synthase and line 20 with D35SP from CamV) in the current experiment did not yield any different responses for antifungal activity.
Innumerable genes encoding for antifungal proteins have been isolated, cloned, sequenced and expressed in plants against multifarious phytopathogenic fungi with success (for a latest review, see20). Examples of a synergistic effect of pyramiding genes similar to what we had in this experiment on combating fungal diseases are numerous.25,29,31,74 However, most of these results were obtained via in vitro and/or greenhouse testing and few crops have been released employing this strategy. Long term field testing is required to test the agronomic performance and ecological relevance of the promising effects detected under laboratory conditions in the complex environment of field especially in the local environment.75 Field studies can help to evaluate any yield reduction which can occur due to introduction of new genes76 or to study the interactions and interplay between various biotic and abiotic stresses in their natural form,77 to identify and rectify issues with stability and resultant pleotropic effects78 and to satisfy regulatory agencies who rely on results from field trials conducted at several locations and are representative of the actual target area of crop cultivation.79 Since the goal of such research programs is improving grower yield and productivity,80 strategic field trial experiment allowing realistic evaluation of genotype x environment interaction become crucial.
Research efforts for legume crop improvement should be encouraged as a low level of public acceptance and high costs incurred in developing and deregulating transgenic crops11 makes it difficult for them to fit into a farmer's diversification strategy.10 Ideally, for addressing efficacy of transgenic disease resistant plants, multiyear, multi-location field trials are desired. However, the limited seed and cost of bringing a transgenic crop like peas is beyond the budgetary scope of public institutions,81 including ours. Often, research without significant differences between treatments are not published yet are very important for the scientific community,82 especially considering the resources required to conduct confined release trials.
Funding Statement
Alberta Pulse Growers (N/A) Alberta Crop Industry Development Fund (2012F068R; 2014F120R) The research fund was provided by the Alberta Crop Industry Development Fund Ltd. (ACIDF) and Alberta Pulse Growers (APG).
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
No potential conflicts of interest were disclosed.
ACKNOWLEDGEMENTS
We greatly acknowledge Lisa Raatz, Judy Irving, Keith Topinka, graduate students and summer staff at weed science and environmental biosafety lab at University of Alberta and plant pathology lab staff at Agriculture and Agri-food Canada (AAFC), Lethbridge, for assistance with the experiments. We are also thankful to Dr. Deng-Jin Bing, AAFC, Lacombe, for kindly supplying the seed of AC Early star, AAC Royce and Agassiz for the experiments, Dr. Muhammed Rahman, AFNS, University of Alberta, for conducting the gene expression analysis and Alberta Agriculture and Forestry for providing land for field experiments.
REFERENCES
- 1.Statistics Canada CANSIM Table 001–0010; 2016. Available at: http://www5.statcan.gc.ca/cansim/a47. Accessed December/7, 2016.
- 2.Warkentin TD, Smýkal P, Coyne CJ, Weeden N, Domoney C, Bing DJ, Leonforte A, Xuxiao Z, Dixit GP, Boros L, et al.. Pea. In: De Ron A, editor. Handbook of plant breeding: grain legumes. New York: Springer Science and Business Media; 2015. p. 37–83. [Google Scholar]
- 3.Basu PK, Brown NJ, Crête R, Gourley CO, Johnston HW, Pepin HS, Seaman WL.. Yield loss conversion factors for Fusarium root rot of pea. Can Plant Dis Surv (Canada). 1976;56:25–32. [Google Scholar]
- 4.Chang KF, Bowness R, Hwang SF, Turnbull GD, Bing DJ, DeMilliano E, Howard RJ.. Occurrence of pea diseases in central Alberta in 2006. Can Plant Dis Surv. 2007;87:122–23. [Google Scholar]
- 5.Feng J, Hwang R, Chang KF, Hwang SF, Strelkov SE, Gossen BD, Conner RL, Turnbull GD. Genetic variation in Fusarium avenaceum causing root rot on field pea. Plant Pathol. 2010;59(5):845–52. doi: 10.1111/j.1365-3059.2010.02313.x. [DOI] [Google Scholar]
- 6.Abdellatif L, Fernandez M, Bouzid S, Vujanovic V. Characterization of virulence and PCR-DGGE profiles of Fusarium avenaceum from western Canadian Prairie Ecozone of Saskatchewan. Canadian J Plant Pathol. 2010;32(4):468–80. doi: 10.1080/07060661.2010.510643. [DOI] [Google Scholar]
- 7.Chen Y, Zhou Q, Strelkov SE, Hwang S. Genetic diversity and aggressiveness of Fusarium spp. isolated from canola in Alberta, Canada. Plant Dis. 2014;98(6):727–38. doi: 10.1094/PDIS-01-13-0061-RE. [DOI] [PubMed] [Google Scholar]
- 8.Tyburski J, Kurowski T, Adamiak E. Root and foot rot diseases of winter wheat grown in conventional and organic systems. J Agric Chem Environ. 2014;3(03):1. [Google Scholar]
- 9.Bodah ET, Porter LD, Chaves B, Dhingra A. Evaluation of pea accessions and commercial cultivars for fusarium root rot resistance. Euphytica. 2016;208(1):63–72. doi: 10.1007/s10681-015-1545-6. [DOI] [Google Scholar]
- 10.Rubiales D, Fondevilla S, Chen W, Gentzbittel L, Higgins TJV, Castillejo MA, Singh KB, Rispail N.. Achievements and challenges in legume breeding for pest and disease resistance. Crit Rev Plant Sci. 2015;34(1–3):195–236. doi: 10.1080/07352689.2014.898445. [DOI] [Google Scholar]
- 11.Smyth SJ, Kerr WA, Phillips PW. Domestic regulatory approval costs. Biotechnology Regulation and Trade Cham (Switzerland): Springer; 2017. p. 33–53. [Google Scholar]
- 12.Eapen S. Advances in development of transgenic pulse crops. Biotechnol Adv. 2008. 0;26(2):162–8. doi: 10.1016/j.biotechadv.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Schroeder HE, Gollasch S, Moore A, Tabe LM, Craig S, Hardie DC, Chrispeels MJ, Spencer D, Higgins T.. Bean [alpha]-Amylase Inhibitor Confers Resistance to the Pea Weevil (Bruchus pisorum) in Transgenic Peas (Pisum sativum L.). Plant Physiol. 1995 Apr;107(4):1233–9. doi: 10.1104/pp.107.4.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Negawo AT, Aftabi M, Jacobsen H, Altosaar I, Hassan FS. Insect resistant transgenic pea expressing cry1Ac gene product from Bacillus thuringiensis. Biol Control. 2013;67(3):293–300. doi: 10.1016/j.biocontrol.2013.09.016. [DOI] [Google Scholar]
- 15.Gonsalves D, Ferreira S. Transgenic papaya: a case for managing risks of papaya ringspot virus in Hawaii. Plant Health Prog. 2003;10. doi: 10.1094/PHP-2003-1113-03-RV. [DOI] [Google Scholar]
- 16.Wally O, Punja ZK. Genetic engineering for increasing fungal and bacterial disease resistance in crop plants. GM Crops. 2010;1(4):199–206. doi: 10.4161/gmcr.1.4.13225. [DOI] [PubMed] [Google Scholar]
- 17.Punja ZK. Genetic engineering of plants to enhance resistance to fungal pathogens—a review of progress and future prospects. Can J Plant Pathol. 2001;23(3):216–35. doi: 10.1080/07060660109506935. [DOI] [Google Scholar]
- 18.Saharan V, Jain D, Pareek S, Pal A, Kumaraswamy R, Jakhar SK, Singh M. Viral, fungal and bacterial disease resistance in transgenic plants. In: Al-Khayri JM, Jain SM, Johnson DV, editors. Advances in plant breeding strategies: Agronomic, abiotic and biotic stress traits. Cham (Switzerland): Springer; 2016. p. 627–56. [Google Scholar]
- 19.Gururani MA, Venkatesh J, Upadhyaya CP, Nookaraju A, Pandey SK, Park SW. Plant disease resistance genes: current status and future directions. Physiol Mol Plant Pathol. 2012 4;78:51–65. doi: 10.1016/j.pmpp.2012.01.002. [DOI] [Google Scholar]
- 20.Moosa A, Farzand A, Sahi ST, Khan SA. Transgenic expression of antifungal pathogenesis-related proteins against phytopathogenic fungi–15 years of success. Isr J Plant Sci. 2017;32:1–17. doi: 10.1080/07929978.2017.1288407. [DOI] [Google Scholar]
- 21.Ma L, Geiser DM, Proctor RH, Rooney AP, O'Donnell K, Trail F, Gardiner DM, Manners JM, Kazan K. Fusarium pathogenomics. Annu Rev Microbiol. 2013;67:399–416. doi: 10.1146/annurev-micro-092412-155650. [DOI] [PubMed] [Google Scholar]
- 22.Hammond-Kosack KE, Jones JD. Plant disease resistance genes. Annu Rev Plant Biol. 1997;48(1):575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 23.O'Donnell K, Ward TJ, Geiser DM, Corby Kistler H, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet Biol. 2004 6;41(6):600–23. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Holtz M, Chang K, Hwang S, Gossen B, Strelkov S. Characterization of Fusarium avenaceum from lupin in central Alberta: genetic diversity, mating type and aggressiveness. Can J Plant Pathol. 2011;33(1):61–76. doi: 10.1080/07060661.2011.536651. [DOI] [Google Scholar]
- 25.Anand A, Zhou T, Trick HN, Gill BS, Bockus WW, Muthukrishnan S. Greenhouse and field testing of transgenic wheat plants stably expressing genes for thaumatin-like protein, chitinase and glucanase against Fusarium graminearum. J Exp Bot. 2003 Mar;54(384):1101–11. doi: 10.1093/jxb/erg110. [DOI] [PubMed] [Google Scholar]
- 26.Gurr SJ, Rushton PJ. Engineering plants with increased disease resistance: what are we going to express? Trends Biotechnol. 2005;23(6):275–82. doi: 10.1016/j.tibtech.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Halpin C. Gene stacking in transgenic plants–the challenge for 21st century plant biotechnology. Plant Biotechnol J. 2005;3(2):141–55. doi: 10.1111/j.1467-7652.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- 28.Que Q, Chilton MM, de Fontes CM, He C, Nuccio M, Zhu T, Wu Y, Chen JS, Shi L. Trait stacking in transgenic crops: challenges and opportunities. GM Crops. 2010;1(4):220–9. doi: 10.4161/gmcr.1.4.13439. [DOI] [PubMed] [Google Scholar]
- 29.Amian A, Papenbrock J, Jacobsen H, Hassan F. Enhancing transgenic pea (Pisum sativum L.) resistance against fungal diseases through stacking of two antifungal genes (chitinase and glucanase). GM Crops. 2011 04;2(2):1–6. doi: 10.4161/gmcr.2.2.16125. [DOI] [PubMed] [Google Scholar]
- 30.Hassan F, Kiesecker H, Jacobsen H, Meens J. A family 19 chitinase (Chit30) from Streptomyces olivaceoviridis ATCC 11238 expressed in transgenic pea affects the development of T. harzianum in vitro [electronic resource]. J Biotechnol. 2009 09/25;143(4):302–8. doi: 10.1016/j.jbiotec.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 31.Richter A, Jacobsen H, De Kathen A, De Lorenzo G, Briviba K, Hain R, Ramsay G, Kiesecker H. Transgenic peas (Pisum sativum) expressing polygalacturonase inhibiting protein from raspberry (Rubus idaeus) and stilbene synthase from grape (Vitis vinifera). Plant Cell Rep. 2006;25(11):1166–73. doi: 10.1007/s00299-006-0172-z. [DOI] [PubMed] [Google Scholar]
- 32.Van Loon LC, Rep M, Pieterse CMJ. Significance of inducible defense-related proteins in infected plants. Annu Rev Phytopathol. 2006;44:135–62. doi: 10.1146/annurev.phyto.44.070505.143425. [DOI] [PubMed] [Google Scholar]
- 33.De Lorenzo G, D'Ovidio R, Cervone F. The role of polygalacturonase-inhibiting proteins (PGIPs) in defense against pathogenic fungi. Annu Rev Phytopathol. 2001;39(1):313–35. doi: 10.1146/annurev.phyto.39.1.313. [DOI] [PubMed] [Google Scholar]
- 34.Jeandet P, Douillet-Breuil A, Bessis R, Debord S, Sbaghi M, Adrian M. Phytoalexins from the Vitaceae: biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J Agric Food Chem. 2002;50(10):2731–41. doi: 10.1021/jf011429s. [DOI] [PubMed] [Google Scholar]
- 35.Birch ANE, Griffiths BS, Caul S, Thompson J, Heckmann LH, Krogh PH, Cortet J. The role of laboratory, glasshouse and field scale experiments in understanding the interactions between genetically modified crops and soil ecosystems: A review of the ECOGEN project. Pedobiologia. 2007 8/15;51(3):251–60. doi: 10.1016/j.pedobi.2007.04.008. [DOI] [Google Scholar]
- 36.Nelissen H, Moloney M, Inzé D. Translational research: from pot to plot. Plant Biotechnol J. 2014;12(3):277–85. doi: 10.1111/pbi.12176. [DOI] [PubMed] [Google Scholar]
- 37.CFIA Novelty and plants with novel traits; 2017. Accessed December 15, 2017. [Google Scholar]
- 38.Schroeder HE, Schotz AH, Wardley-Richardson T, Spencer D, Higgins T. Transformation and regeneration of two cultivars of Pea (Pisum sativum L.). Plant Physiol. 1993 Mar;101(3):751–7. doi: 10.1104/pp.101.3.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hood EE, Gelvin SB, Melchers LS, Hoekema A. NewAgrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 1993;2(4):208–18. doi: 10.1007/BF01977351. [DOI] [Google Scholar]
- 40.Murakami T, Anzai H, Imai S, Satoh A, Nagaoka K, Thompson CJ. The bialaphos biosynthetic genes of Streptomyces hygroscopicus: Molecular cloning and characterization of the gene cluster. Mol Gen Genet MGG. 1986;205(1):42–53. doi: 10.1007/BF02428031. [DOI] [Google Scholar]
- 41.Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J. Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J. 1987 Sep;6(9):2519–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kahlon JG, Jacobsen H, Cahill JF, Hall LM. Antifungal genes expressed in transgenic pea (Pisum sativum L.) do not affect root colonization of arbuscular mycorrhizae fungi. Mycorrhiza. 2017;27:683–94. doi: 10.1007/s00572-017-0781-0. [DOI] [PubMed] [Google Scholar]
- 43.Richter A, Jacobsen HJ, Kathen A, de Lorenzo GD, Briviba K, Hain R, Ramsay G, Kiesecker H.. Transgenic peas (Pisum sativum) expressing polygalacturonase inhibiting protein from raspberry (Rubus idaeus) and stilbene synthase from grape (Vitis vinifera) [electronic resource]. Plant Cell Rep. 2006 11;25(11):1166–73. doi: 10.1007/s00299-006-0172-z. [DOI] [PubMed] [Google Scholar]
- 44.Hassan F, Muller A, Jacobsen H. Gene stacking of antifungal genes in Transgenic Pea to enhance the level of resistance. In Cell Dev Biol Animal. 2010;46(Suppl 1):1. doi: 10.1007/s11626-010-9338-7. [DOI] [Google Scholar]
- 45.CFIA (Canadian Food Inspection Agency) Agassiz; 2017. Available at: http://www.inspection.gc.ca/english/plaveg/pbrpov/cropreport/pea/app00006069e.shtml.
- 46.CFIA (Canadian Food Inspection Agency) Earlystar; 2017. Available at: http://www.inspection.gc.ca/english/plaveg/pbrpov/cropreport/pea/app00008219e.shtml.
- 47.Bing D, Beauchesne D, McLaren D, Vera CL. AAC Royce field pea. Can J Plant Sci. 2016;97(2):365–7 [Google Scholar]
- 48.Taheri AE, Chatterton S, Foroud N, Gossen B, McLaren D. Identification and community dynamics of fungi associated with root, crown, and foot rot of field pea in western Canada. Eur J Plant Pathol. 2017;147(3):489–500. doi: 10.1007/s10658-016-1017-4. [DOI] [Google Scholar]
- 49.Infantino A, McPhee KE, Grunwald NJ, Coyne CJ, Kharrat M, Riccioni L. Screening techniques and sources of resistance to root diseases in cool season food legumes [electronic resource]. Euphytica. 2006 01;147(1–2):201–21. doi: 10.1007/s10681-006-6963-z. [DOI] [Google Scholar]
- 50.Bilgi VN, Grafton KF, Rasmussen JB, Bradley CA, Khot SD. Response of Dry Bean Genotypes to Fusarium Root Rot, Caused by Fusarium solani f. sp. phaseoli, Under Field and Controlled Conditions [electronic resource]. Plant Dis. 2008 08;92(8):1197–200. doi: 10.1094/PDIS-92-8-1197. [DOI] [PubMed] [Google Scholar]
- 51.SAS Institute Inc SAS OnlineDoc 9.4. Cary, NC, USA: SAS Institute Inc.; 2014. [Google Scholar]
- 52.Neuhaus J, Ahl-Goy P, Hinz U, Flores S, Meins Jr F. High-level expression of a tobacco chitinase gene in Nicotiana sylvestris. Susceptibility of transgenic plants to Cercospora nicotianae infection. Plant Mol Biol. 1991;16(1):141–51. doi: 10.1007/BF00017924. [DOI] [PubMed] [Google Scholar]
- 53.Volpi C, Raiola A, Janni M, Gordon A, O'Sullivan DM, Favaron F, D'Ovidio R.. Claviceps purpurea expressing polygalacturonases escaping PGIP inhibition fully infects PvPGIP2 wheat transgenic plants but its infection is delayed in wheat transgenic plants with increased level of pectin methyl esterification. Plant Physiol Biochem. 2013 12;73:294–301. doi: 10.1016/j.plaphy.2013.10.011. [DOI] [PubMed] [Google Scholar]
- 54.Masoud SA, Zhu Q, Lamb C, Dixon RA. Constitutive expression of an inducible β-1, 3-glucanase in alfalfa reduces disease severity caused by the oomycete pathogen Phytophthora megasperma f. spmedicaginis, but does not reduce disease severity of chitin-containing fungi. Transgenic Res. 1996;5(5):313–23. doi: 10.1007/BF01968941. [DOI] [Google Scholar]
- 55.Iqbal MM, Nazir F, Ali S, Asif MA, Zafar Y, Iqbal J, Ali GM. Over expression of rice chitinase gene in transgenic peanut (Arachis hypogaea L.) improves resistance against leaf spot. Mol Biotechnol. 2012;50(2):129–36. doi: 10.1007/s12033-011-9426-2. [DOI] [PubMed] [Google Scholar]
- 56.Prasad K, Bhatnagar-Mathur P, Waliyar F, Sharma KK. Overexpression of a chitinase gene in transgenic peanut confers enhanced resistance to major soil borne and foliar fungal pathogens. J Plant Biochem Biotechnol 2013;22(2):222–33. doi: 10.1007/s13562-012-0155-9. [DOI] [Google Scholar]
- 57.Chopra R, Saini R. Transformation of blackgram (Vigna mungo (L.) Hepper) by barley chitinase and ribosome-inactivating protein genes towards improving resistance to Corynespora leaf spot fungal disease. Appl Biochem Biotechnol. 2014;174(8):2791–800. doi: 10.1007/s12010-014-1226-2. [DOI] [PubMed] [Google Scholar]
- 58.Hipskind JD, Paiva NL. Constitutive accumulation of a resveratrol-glucoside in transgenic alfalfa increases resistance to Phoma medicaginis. Mol Plant-Microbe Interact. 2000;13(5):551–62. doi: 10.1094/MPMI.2000.13.5.551. [DOI] [PubMed] [Google Scholar]
- 59.Sundaresha S, Kumar AM, Rohini S, Math S, Keshamma E, Chandrashekar S, Udayakumar M. Enhanced protection against two major fungal pathogens of groundnut, Cercospora arachidicola and Aspergillus flavus in transgenic groundnut over-expressing a tobacco β 1–3 glucanase. Eur J Plant Pathol. 2010;126(4):497–508. doi: 10.1007/s10658-009-9556-6. [DOI] [Google Scholar]
- 60.Qiao L, Ding X, Wang H, Sui J, Wang J. Characterization of the β-1, 3-glucanase gene in peanut (Arachis hypogaea L.) by cloning and genetic transformation. Genet Mol Res. 2014;13:1893–904. doi: 10.4238/2014.March.17.17. [DOI] [PubMed] [Google Scholar]
- 61.Willsey T, Thomas J, Chatterton S. Intra-host interactions of the pea root rot pathogens Aphanomyces euteiches and Fusarium spp; 2016. Available at: http://phytopath.ca/wp-content/uploads/2016/06/2016-CPS-Annual-meeting-Program_Full_PDF.pdf. Accessed 09/15, 2017.
- 62.Moravčíková J, Matušíková I, Libantová J, Bauer M, Mlynárová L. Expression of a cucumber class III chitinase and Nicotiana plumbaginifolia class I glucanase genes in transgenic potato plants. Plant Cell, Tissue Organ Cult. 2004;79(2):161–8. doi: 10.1007/s11240-004-0656-x. [DOI] [Google Scholar]
- 63.Melander M, Kamnert I, Happstadius I, Liljeroth E, Bryngelsson T. Stability of transgene integration and expression in subsequent generations of doubled haploid oilseed rape transformed with chitinase and β-1, 3-glucanase genes in a double-gene construct. Plant Cell Rep. 2006;25(9):942–52. doi: 10.1007/s00299-006-0153-2. [DOI] [PubMed] [Google Scholar]
- 64.Kalunke RM, Tundo S, Benedetti M, Cervone F, De Lorenzo G, D'Ovidio R. An update on polygalacturonase-inhibiting protein (PGIP), a leucine-rich repeat protein that protects crop plants against pathogens. Front Plant Sci. 2015;6:146. doi: 10.3389/fpls.2015.00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Desiderio A, Aracri B, Leckie F, Mattei B, Salvi G, Tigelaar H, Van Roekel JS, Baulcombe DC, Melchers LS, De Lorenzo G, et al.. Polygalacturonase-inhibiting proteins (PGIPs) with different specificities are expressed in Phaseolus vulgaris. Mol Plant-Microbe Interact. 1997;10(7):852–60. doi: 10.1094/MPMI.1997.10.7.852. [DOI] [PubMed] [Google Scholar]
- 66.Sandhu JS, Sidhu MK, Yadav IS. Control of fungal diseases in agricultural crops by chitinase and glucanase transgenes. In: Lichtfouse E, editor. Sustainable agriculture reviews. Vol. 22. Cham (Switzerland): Springer; 2017. p. 163–212. [Google Scholar]
- 67.Foroud NA, Chatterton S, Reid LM, Turkington TK, Tittlemier SA, Gräfenhan T. Fusarium diseases of Canadian grain crops: impact and disease management strategies. In: Goyal A, Manoharachary C, editors. Future challenges in crop protection against fungal pathogens. Fungal Biology. New York (NY): Springer; 2014. p. 267–316. [Google Scholar]
- 68.Maqbool SB, Christou P. Multiple traits of agronomic importance in transgenic indica rice plants: analysis of transgene integration patterns, expression levels and stability. Mol Breed. 1999;5(5):471–80. doi: 10.1023/A:1009634226797. [DOI] [Google Scholar]
- 69.Qu LQ, Takaiwa F. Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J. 2004;2(2):113–25. doi: 10.1111/j.1467-7652.2004.00055.x. [DOI] [PubMed] [Google Scholar]
- 70.Daxinger L, Hunter B, Sheikh M, Jauvion V, Gasciolli V, Vaucheret H, Matzke M, Furner I. Unexpected silencing effects from T-DNA tags in Arabidopsis. Update. 2008;13(1):4–6. [DOI] [PubMed] [Google Scholar]
- 71.Dietz-Pfeilstetter A. Stability of transgene expression as a challenge for genetic engineering. Plant Sci. 2010 9;179(3):164–7. doi: 10.1016/j.plantsci.2010.04.015. [DOI] [Google Scholar]
- 72.Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33:245–54. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- 73.Finnegan J, McElroy D. Transgene inactivation: plants fight back. Bio Technol. 1994;12:883–88. doi: 10.1038/nbt0994-883. [DOI] [Google Scholar]
- 74.Ziaei M, Motallebi M, Zamani MR, Panjeh NZ. Co-expression of chimeric chitinase and a polygalacturonase-inhibiting protein in transgenic canola (Brassica napus) confers enhanced resistance to Sclerotinia sclerotiorum. Biotechnol Lett. 2016;38(6):1021–32. doi: 10.1007/s10529-016-2058-7. [DOI] [PubMed] [Google Scholar]
- 75.Wozniak CA, McHughen A. Regulation of agricultural biotechnology: the United States and Canada. (Netherlands): Springer Science & Business Media; 2012. [Google Scholar]
- 76.Cohen JI. Poorer nations turn to publicly developed GM crops. Nat Biotechnol. 2005;23(1):27. doi: 10.1038/nbt0105-27. [DOI] [PubMed] [Google Scholar]
- 77.Bostock RM, Pye MF, Roubtsova TV. Predisposition in plant disease: exploiting the nexus in abiotic and biotic stress perception and response. Annu Rev Phytopathol. 2014;52:517–49. doi: 10.1146/annurev-phyto-081211-172902. [DOI] [PubMed] [Google Scholar]
- 78.Pons E, Peris JE, Peña L. Field performance of transgenic citrus trees: assessment of the long-term expression of uidA and nptII transgenes and its impact on relevant agronomic and phenotypic characteristics. BMC Biotechnol. 2012;12(1):41. doi: 10.1186/1472-6750-12-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Romeis J, Bartsch D, Bigler F, Candolfi MP, Gielkens MM, Hartley SE, Hellmich RL, Huesing JE, Jepson PC, Layton R, et al.. Assessment of risk of insect-resistant transgenic crops to nontarget arthropods. Nat Biotechnol 2008;26(2):203–8. doi: 10.1038/nbt1381. [DOI] [PubMed] [Google Scholar]
- 80.Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, Pretty J, Robinson S, Thomas SM, Toulmin C.. Food security: the challenge of feeding 9 billion people. Science. 2010 Feb 12;327(5967):812–8. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- 81.Parisi C, Tillie P, Rodríguez-Cerezo E. The global pipeline of GM crops out to 2020. Nat Biotechnol. 2016;34(1):31–36. doi: 10.1038/nbt.3449. [DOI] [PubMed] [Google Scholar]
- 82.Knight J. Negative results: Null and void. Nature. 2003;422(6932):554–5. doi: 10.1038/422554a. [DOI] [PubMed] [Google Scholar]


