Abstract
In breast cancer, 17β-estradiol (E2) plays critical roles mainly by binding to its canonical receptor, estrogen receptor (ER) α66, and eliciting genomic effects. E2 also triggers rapid, nongenomic responses. E2 activates sphingosine kinase 1 (SphK1), increasing sphingosine-1-phosphate (S1P) that binds to its receptors, leading to important breast cancer signaling. However, the E2 receptor responsible for SphK1 activation has not yet been identified. Here, we demonstrate in triple-negative breast cancer cells, which lack the canonical ERα66 but express the novel splice variant ERα36, that ERα36 is the receptor responsible for E2-induced activation of SphK1 and formation and secretion of S1P and dihydro-S1P, the ligands for S1PRs. Tamoxifen, the first-line endocrine therapy for breast cancer, is an antagonist of ERα66, but an agonist of ERα36, and, like E2, activates SphK1 and markedly increases secretion of S1P. A major problem with tamoxifen therapy is development of acquired resistance. We found that tamoxifen resistance correlated with increased SphK1 and ERα36 expression in tamoxifen-resistant breast cancer cells, in patient-derived xenografts, and in endocrine-resistant breast cancer patients. Our data also indicate that targeting this ERα36 and SphK1 axis may be a therapeutic option to circumvent endocrine resistance and improve patient outcome.
Keywords: estradiol, estrogen receptor-α, splice variant, sphingosine-1-phosphate
Breast cancer is the most prevalent type of cancer in women, and 1 in 8 women in the United States will develop breast cancer over their lifetime. The estrogen receptor-α (ERα) and its ligand 17β-estradiol (E2) play important roles in cancer pathogenesis, progression, and metastasis. Patients with tumors that express the full-length ERα66 are termed ERα-positive and those lacking it as ERα-negative. Endocrine therapy, such as tamoxifen, is a first-line treatment of ERα-positive breast cancer (1, 2). Unfortunately, more than 50% of these patients will ultimately fail therapy due to acquired resistance. Moreover, triple-negative breast cancers (TNBCs) lacking ERα66, progesterone receptor (PR), and human epidermal growth factor receptor (EGFR) are aggressive cancers with high recurrence, metastatic, and mortality rates and limited treatment options, and they do not respond to hormonal therapy (3, 4). Understanding the mechanisms responsible for de novo and acquired hormonal therapy resistance may provide clues to better treatments.
E2 elicits most of its cellular effects by binding to ERα66 in the cytosol, followed by receptor dimerization and translocation to the nucleus, where it regulates expression of genes that are important for tumor growth and survival by binding to estrogen response elements on target genes (5, 6). These genomic responses are slow and take hours to days to induce effects. However, it has become apparent that E2 also exerts rapid, within minutes, nongenomic effects through membrane-associated receptors (7, 8). Most of the nongenomic responses of E2 have been linked to ERα36, a 36-kDa splice variant of ERα66 (9–15) that is mainly expressed on the plasma membranes of breast cancer cells, particularly in TNBC (15, 16). ERα36 has a novel noncoding exon as its first exon, which is spliced into exons 2–6 of the ERα66 gene, and also has a unique 27 amino acid domain (exon 9) that replaces the last 138 amino acids encoded by exons 7 and 8 of the ERα66 gene. Although tamoxifen is an antagonist of ERα66, it activates ERα36, which lacks both transcriptional activation domains of ERα66 (AF-1 and AF-2) and a large portion of the ligand-binding domain (16–18). It has been suggested that tamoxifen resistance is due to upregulation of ERα36 (18, 19). Moreover, recent studies correlated increased expression of ERα36 with advanced severity of breast cancer and endocrine therapy resistance, as well as increased distant metastasis and poor prognosis (17, 19, 20).
Abundant evidence indicates that sphingosine-1-phosphate (S1P), a pleiotropic bioactive sphingolipid metabolite formed by sphingosine kinase 1 (SphK1), is involved in breast cancer growth, progression, transformation, and metastasis (21–23). Overexpression of SphK1 in ERα-positive breast cancer cells promotes tumorigenesis and angiogenesis of xenografts (24). SphK1 is commonly upregulated in breast tumors and has been linked to worse prognosis and progression, possibly leading to resistance to certain anticancer therapies (25–29). There is an emerging idea that S1P may play an important role in the nongenomic responses mediated by E2 (30, 31). Several studies have shown that E2 treatment of MCF-7 breast cancer cells that express all ERα splice variants (ERα66, ERα46, and ERα36) rapidly activates SphK1, leading to formation and secretion of S1P (32–34). S1P in turn binds to S1P receptors (S1PRs) present on breast cancer cells, leading to downstream signaling pathways, including ERK1/2, Akt, protein kinase C, and even transactivation of EGFR, events important for breast cancer progression and metastasis (30, 31, 35–37). Therefore, it has been suggested that some of the nongenomic effects of E2 are mediated via the SphK1/S1P/S1PR axis. However, the E2 receptor responsible for SphK1 activation has not yet been identified. In this work, we have demonstrated that the novel ERα splice variant ERα36 is the major membrane surface receptor for E2 that rapidly activates SphK1 and subsequent S1P signaling and that hormone therapy resistance occurs through upregulation of ERα36 and SphK1.
MATERIALS AND METHODS
Materials
E2 (catalog no. 50282), E2-BSA (catalog no. E5630), tamoxifen (catalog no. T5648), and FA-free BSA (catalog no. A8806) were from MilliporeSigma (St. Louis, MO). ERα36-neutralizing Ab was from Alpha Diagnostic (San Diego, CA). HALT protease and phosphatase inhibitor (catalog no. 78440) was from ThermoFisher (Waltham, MA). The SphK1-specific inhibitor SK1-I (catalog no. BML-EI411) was from Enzo Life Sciences (Farmingdale, NY). RPMI 1640 phenol red-free medium and DMEM phenol red-free medium were from Gibco, and FBS (catalog no. ES-009-B) was from MilliporeSigma.
Cell culture
HCC38 and MDA-MB-231 human breast cancer cells were from American Type Culture Collection (Manassas, VA). HCC38 cells were cultured in phenol red-free RPMI 1640 containing 10% FBS and penicillin/streptomycin. MDA-MB-231 cells were cultured in phenol red-free DMEM supplemented with 10% FBS, 1% l-glutamine, and antibiotics. MCF-7/S0.5 (catalog no. SCC100) and MCF-7/TAMR-7 (catalog no. SCC101) cells were from MilliporeSigma. MCF-7/S0.5 and MCF-7/TAMR were grown in DMEM/F12 medium without phenol red, containing 10% FBS, 2.5 mM l-glutamine, and 6 ng/ml insulin. MCF-7/TAMR cell medium also contained 1 nM tamoxifen.
In some experiments, cells were transfected with ON-TARGET plus siRNAs specific for the unique 27 amino acid sequence at the N terminus of ERα36 (#1 UCUCACAUGUAGAAGCAAAUU; #3 GCAAAGAAGAGAAUCCUGAUU) or control siRNA (Dharmacon, Lafayette, CO), according to the manufacturer’s protocol.
Patient-derived xenografts
Patient-derived xenograft (PDX) lines were obtained from Washington University in St. Louis (WHIM2 and WHIM30) (38), the University of Utah (HCI lines) (39), and the University of Colorado (UCD18 and UCD52) (40). They were implanted in the fourth mammary fat pads of nonobese diabetic severe combined immunodeficient γ (NSG) mice until tumors reached a size of ∼10 × 10 mm. Tumor pieces were flash frozen, and protein extracts were prepared in RIPA buffer.
SphK1 translocation and activity assays
To determine translocation of SphK1 to membranes, cells were lysed, and membrane fractions were prepared by 100,000 g centrifugation as described (41). SphK1 activity was determined with sphingosine (50 μM) and ATP (1 mM) containing MgCl2 (10 mM) in the presence of 0.25% Triton X-100, which inhibits SphK2, as described previously (42). SphK2 activity was determined similarly when sphingosine was added as a complex with 4 mg/ml BSA in the presence of 1 M KCl, which inhibits SphK1 activity (42). S1P formation was measured by LC-ESI-MS/MS. Activity is expressed as pmol of S1P formed per min/mg protein.
Treatment with E2 and measurement of phosphorylated sphingoid bases by LC/ESI/MS/MS
Cells cultured in 6-well tissue culture plates (105/well) were washed twice with 1 ml of PBS and starved in 0.5 ml of serum-free medium containing 1% FA-free BSA for 2 h. Cells were then treated with vehicle or E2 (100 nM) in 0.5 ml of the same serum-free medium containing 1% FA-free BSA for the indicated times. After treatments, plates were placed on ice, and the medium was removed and added to prechilled 13 × 100 mm borosilicate tubes containing 1 ml of ice-cold LC/MS grade methanol. Cells were washed two times with prechilled PBS and 300 µl of ice cold-PBS containing 1:100 HALT protease, and phosphatase inhibitor was added. Cells were scraped, and suspensions (200 µl) were added to 13 × 100 mm borosilicate tube containing 0.5 ml of ice-cold LC/MS grade methanol. An aliquot of the remaining 100 µl cell suspension was used for protein quantification with the Bio-Rad Protein Assay. Sphingolipids were measured by LC/ESI/MS/MS (Sciex 5500 QTRAP; ABSciex, Farmingham, MA). Cellular sphingolipid levels were expressed as pmol per milligram of protein and secreted as pmol per milliliter of medium.
Confocal microscopy
Immunofluorescent localization was performed essentially as described previously (43). Briefly, MDA-MB-231 cells were seeded onto glass coverslips in 6-well dishes and transfected with V5-SphK1 using Lipofectamine Plus. After treatment with E2, cells were washed twice with room temperature PBS and then fixed with 3.7% paraformaldehyde with 0.1% Triton X-100 for 10 min. Fixative was quenched by extensive washing with 10 mM glycine-PBS, and cells permeabilized for 3 min with 0.5% Triton X-100. After washing, coverslips were incubated for 20 min with mouse anti-V5 Ab (1:100; ThermoFisher) in 1% IgG-free BSA in glycine-PBS, washed three times, and then incubated with Alexa 488-labeled anti-mouse Ab (1:400; ThermoFisher). Coverslips were washed and mounted with 10 mM n-propylgallate in 100% glycerol and visualized with a Zeiss LSM 700 confocal microscope (Carl Zeiss, Oberkochen, Germany).
Western blotting
Cells were washed with ice-cold PBS and scraped into lysis buffer containing 20 mM Tris-HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 1% Triton X-100, 1 mM β-mercaptoethanol, 1 mM Na3VO4, and 1:100 HALT protease and phosphatase inhibitor. Lysates were sonicated three times for 10 s each and centrifuged at 10,000 g for 10 min. Protein was measured by the Bio-Rad protein assay, and equal amounts were separated by SDS-PAGE and blotted onto nitrocellulose. Membranes were incubated overnight at 4°C with the following specific primary Abs: p-SphK1 (Ser225) (1:1,000; ECM Bioscience, catalog no. SP1641), SphK1 (1:1,000; Sigma, catalog no. HPA0229829), ERα36 (1:1,000; Cell Application, catalog no. CY1109), GAPDH (1:3,000; Cell Signaling, catalog no. 2218L), tubulin (1:3,000, Cell Signaling, catalog no. 2146S), p-Akt (S473) (1:1,000; Cell Signaling, catalog no. 9271S), Akt (1:1,000; Cell Signaling, catalog no. 9272S), ERα (H222) (1:500; Santa Cruz, catalog no. sc-53492), transferrin receptor (1:1,000; Cell Signaling, catalog no. 3113S), ABCC1 (1:2,000; Cell Signaling, catalog no. 72202S), ABCG2 (1:3,000; Cell Signaling, catalog no. 42078), and Spns2 (1:3,000; Sigma-Aldrich, catalog no. SAB2104271). Immunopositive bands were visualized by ECL after 2 h incubations at room temperature with secondary Abs conjugated with HRP (1:10,000) and Super-Signal West Pico chemiluminescent substrate (ThermoFisher). Blots were stripped and reprobed with anti-tubulin or anti-GAPDH as loading controls. Optical densities of bands associated with proteins of interest were quantified with NIH ImageJ and normalized to the optical densities of their respective loading control bands.
Cell proliferation
Cell proliferation was determined with WST-8 [2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium and monosodium salt] or with Alamar blue as previously described (44).
Statistical analysis
Statistical analyses were performed using an unpaired two-tailed Student’s t-test for comparison of two groups or ANOVA with Bonferroni post hoc analyses for multiple groups (GraphPad Prism; GraphPad, San Diego, CA). All experiments were repeated independently at least three times, and representative data are shown. A value of P < 0.05 was considered significant.
RESULTS
SphK1 is activated by E2 to produce S1P in cells only expressing ERα splice variant ERα36
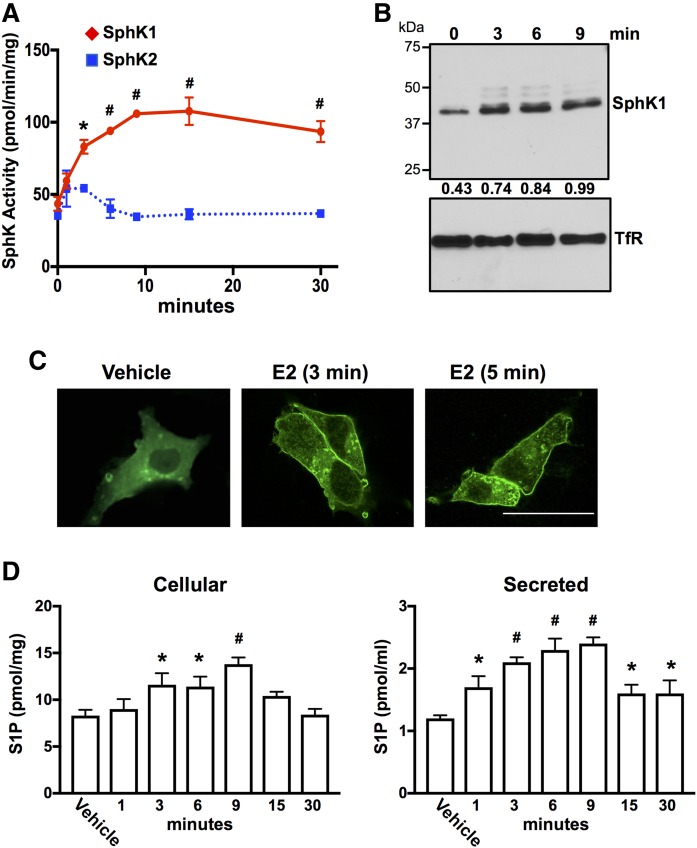
Previous studies have shown that E2 stimulates formation and secretion of S1P in a SphK1-dependent manner in MCF-7 breast cancer cells (32–34). However, the E2 receptor involved in the activation of SphK1 has not been identified. Because MCF-7 cells express ERα66, ERα46, ERα36, a classical G protein-coupled E2 receptor GPER1, and a low level of ERβ, we sought to examine the effects of E2 on SphK1 activation and formation of S1P in TNBC cells that lack both ERα66 and ERα46 and only express ERα36 and GPER1, such as MDA-MB-231 cells (45). Similar to previous studies with MCF-7 cells (32–34), SphK1 was rapidly activated by E2 in MDA-MB-231 cells, although little to no increased SphK2 activity was detected (Fig. 1A). Multiple studies have demonstrated that SphK1 is translocated to the plasma membrane upon its activation (46–48). Consistent with these studies, Western blotting with anti-SphK1 confirmed that SphK1 is translocated to MDA-MB-231 cell membranes within 3 min after E2 treatment (Fig. 1B). Moreover, confocal immunofluorescence microscopy revealed that E2 also induced rapid translocation of epitope-tagged SphK1 from the cytosol to the plasma membrane in MDA-MB-231 cells within 3–5 min (Fig. 1C). There was also a significant increase in cellular and secreted S1P within 3 min that remained elevated for at least 9 min and declined thereafter (Fig. 1D).
Fig. 1.
E2 activates SphK1 and increases production and secretion of S1P in triple-negative MDA-MB-231 breast cancer cells. A, B, D: Serum-starved MDA-MB-231 cells were treated without (vehicle) or with E2 (100 nM) for the indicated time. A: SphK1 and SphK2 enzymatic activities in lysates were determined with isoenzyme-specific assays. * P ≤ 0.05; # P < 0.001 (compared with 0 time). B: Equal amounts of membrane proteins were separated by SDS-PAGE and immunoblotted with anti-SphK1 Ab. Blots were stripped and reprobed with antitransferrin receptor (TfR) Ab as loading control. Proteins were quantified by densitometry, and numbers indicate relative optical density of SphK1 normalized to TfR. Similar results were obtained in two additional experiments. C: MDA-MB-231 cells were transfected with V5-SphK1, serum-starved, and stimulated with vehicle or with E2 for 3 or 5 min. Cells were stained with anti-V5 Ab (green) and visualized by confocal microscopy. Scale bar, 25 µm. D: Cellular S1P and S1P released into the medium during 30 min secretion assay were measured by LC/ESI/MS/MS. Data are mean ± SD. * P ≤ 0.05; # P < 0.001 (compared with vehicle).
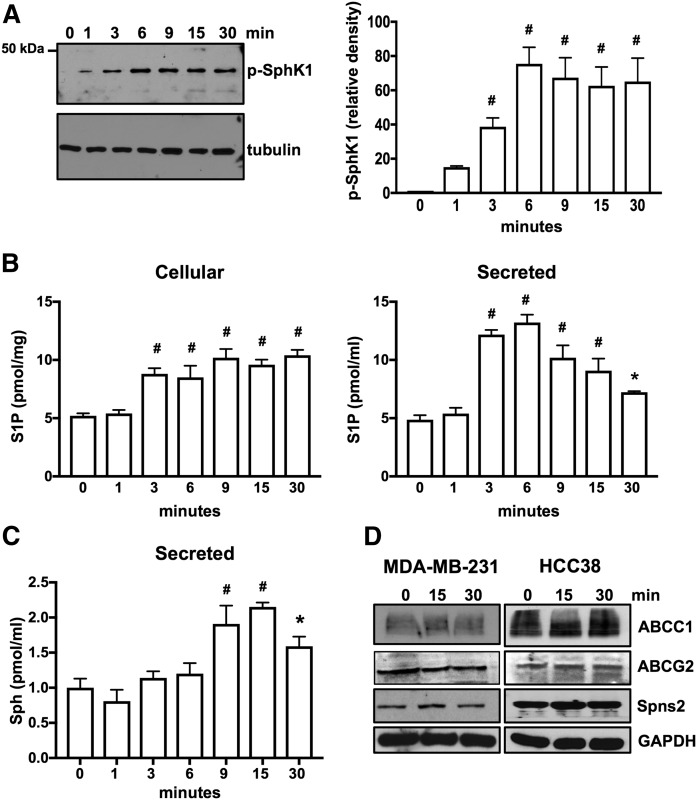
Similar results were observed in ERα-negative HCC38 breast cancer cells that express only ERα36 and GPER1 (Fig. 2). Moreover, E2 also increased phosphorylation of SphK1 on serine 225 (Fig. 2A), which is known to enhance its enzymatic activity (47). This rapid increase in phosphorylated SphK1 was evident within 1 min and remained elevated for at least 30 min (Fig. 2A). In agreement, E2 induced rapid increases of cellular S1P and secreted S1P, reaching a maximum at 6 min and declining thereafter (Fig. 2B). This decline is partly due to rapid degradation of S1P to sphingosine (Fig. 2C) by lipid phosphate phosphatases known to be present on the outer leaflet of the plasma membrane (49). Moreover, there were no changes in expression of known S1P transporters (31, 50), including ABCC1, ABCG2, and Spns2 (Fig. 2D).
Fig. 2.
E2 induces phosphorylation of SphK1 for production and secretion of S1P in HCC38 breast cancer cells that express only ERα36. A, B: Serum-starved HCC38 cells were treated with E2 (100 nM) for the indicated time. A: Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with anti-phospho-SphK1 Ab. Blots were stripped and reprobed with antitubulin to show equal transfer and loading. Phospho-SphK1 was quantified by densitometry and data expressed as relative density of p-SphK1 normalized to tubulin. B, C: Cellular S1P and S1P released into the medium during 30 min secretion assays as well as sphingosine (Sph) were measured by LC/ESI/MS/MS. Data are mean ± SD. * P ≤ 0.05; # P < 0.001 (compared with 0 time). D: Serum-starved MDA-MB-231 and HCC38 cells were treated with E2 (100 nM) for the indicated time, and cell lysate proteins were immunoblotted with the indicated Abs. Blots were stripped and reprobed with anti-GAPDH to show equal transfer and loading.
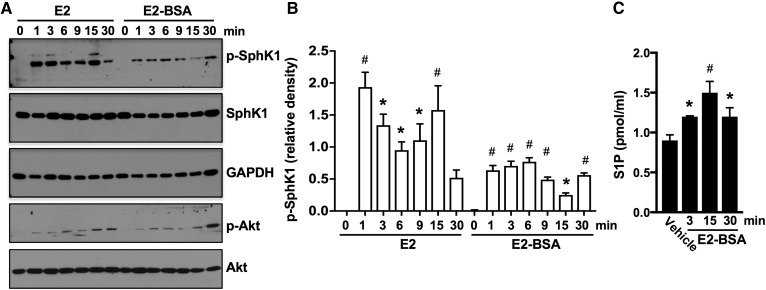
To further substantiate that these effects were mediated through a plasma membrane receptor, cells were treated with E2 conjugated to BSA (E2-BSA) that does not cross the plasma membrane and reach intracellular receptors, yet elicits many of the same effects as E2 (16). E2-BSA not only stimulated nongenomic pathways, such as Akt activation, in agreement with previous reports (13, 16, 51), but it also stimulated SphK1, as shown by immunoblotting with a phospho-specific Ab that recognizes activated SphK1, albeit to a lesser extent than E2 (Fig. 3A, B). Moreover, like E2, E2-BSA also induced secretion of S1P (Fig. 3C). Taken together, these results indicate that E2 activates SphK1 by a plasma membrane receptor in TNBC cells.
Fig. 3.
Membrane-impermeable E2-BSA activates SphK1 and enhances secretion of S1P. A, B: MDA-MB-231 cells treated with E2 (100 nM) or with E2-BSA (100 nM) for the indicated times. A: Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with the indicated Abs. B: Phospho-SphK1 was quantified by densitometry, and data are expressed as relative density of p-SphK1 normalized to SphK1. C: Cells from duplicate MDA-MB-231 cultures were treated without or with E2-BSA (100 nM) for the indicated times. S1P released into the medium during 30 min secretion assays was measured by LC/ESI/MS/MS. Data are mean ± SD. * P ≤ 0.05; # P < 0.001 (compared with control).
Role of ERα36 in E2-mediated formation and secretion of S1P
Because previous studies have implicated ERα36, which is present on the plasma membrane, in nongenomic effects of E2, we focused our attention on its involvement (10–14). As a first approach, cells were treated with a rabbit Ab for human ERα36 raised against the unique peptide sequence of ERα36 that blocks ERα36 downstream signaling (16). Neutralizing ERα36 with this Ab that does not cross-react with other ERα family members markedly blocked activation of SphK1 by E2 (Fig. 4A). This Ab also greatly reduced activation of Akt induced by E2 (Fig. 4A), consistent with previous reports (13, 16, 51–53). Treatment with anti-ERα36 also almost completely suppressed E2-induced formation and secretion of S1P (Fig. 4B) and dihydro-S1P (Fig. 4C).
Fig. 4.
ERα36 neutralizing Ab attenuates E2-induced SphK1 activation and S1P formation and secretion. MDA-MB-231 cells were pretreated without or with anti-ERα36 neutralizing Ab and then stimulated with vehicle or with E2 (100 nM) for the indicated times. A: Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with the indicated Abs. p-SphK1 and p-Akt were quantified by densitometry, and data are expressed as relative densities normalized to GAPDH. Cellular S1P and S1P released into the medium (B) and cellular S1P and dihydro-S1P (DHS1P) (C) released into the medium during 30 min secretion assays were measured by LC/ESI/MS/MS. Data are mean ± SD. * P ≤ 0.05; # P < 0.001 (compared with vehicle).
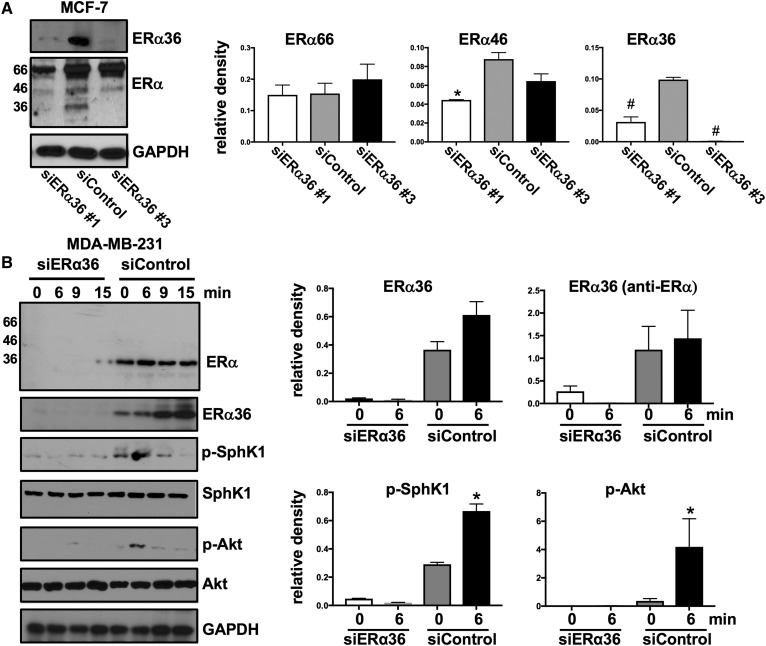
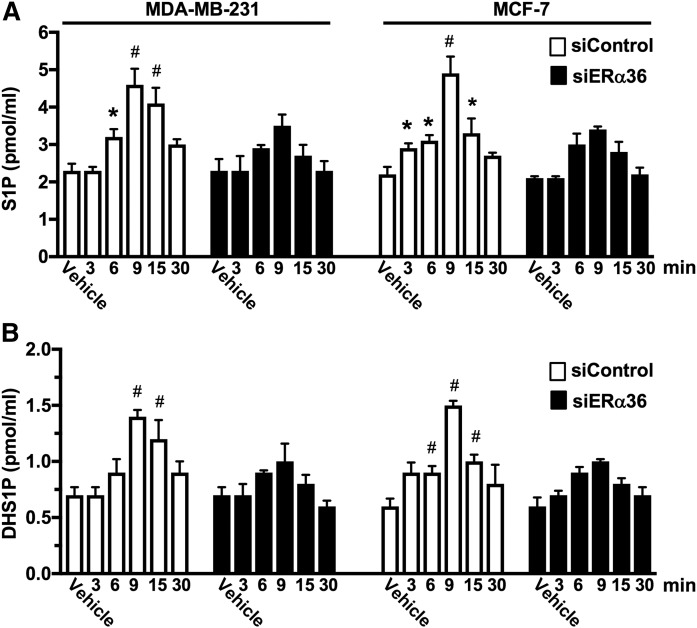
To confirm the role of ERα36 in activation of the SphK1 by E2, ERα36 was downregulated with two siRNAs specific for the unique 27 amino acid sequence at the N terminus of ERα36. siERα36 #3 that markedly reduced ERα36 levels in MCF-7 cells without reducing levels of ERα66 and ERα46 present in this breast cancer cell line that expresses all three ERα splice variants was selected for further studies (Fig. 5A). This siRNA significantly reduced ERα36 expression in MDA-MB-231 cells detected with either an anti-ERα Ab that recognizes all three splice variants or with anti-ERα36-specific Ab (Fig. 5B). Downregulation of ERα36 in MDA-MB-231 cells abrogated E2-mediated SphK1 activation and stimulation of Akt, a downstream signaling pathway (Fig. 5B). In both MDA-MB-231 and MCF-7 cells, knockdown of ERα36 significantly reduced secretion of S1P and dihydro-S1P (Fig. 6A, B). These results suggest that ERα36, a plasma membrane E2 receptor, plays a role in the rapid, nongenomic E2 activation of SphK1 and subsequent S1P signaling.
Fig. 5.
Downregulation of ERα36 decreases E2-mediated SphK1 activation. A: MCF-7 cells were transfected with control siRNA or with the indicated siRNA targeted to ERα36. Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with the indicated Abs. Protein bands of ERα66, ERα46, and ERα36 were quantified by densitometry and data are expressed as relative densities normalized to GAPDH. * P ≤ 0.05; # P < 0.001 (compared with siControl). B: MDA-MB-231 cells transfected with control siRNA or with siRNA targeted to ERα36 (#3) were stimulated with vehicle or with E2 (100 nM) for the indicated times. Proteins in cell lysates were separated by SDS-PAGE and immunoblotted with the indicated Abs. ERα36, p-SphK1, and p-Akt were quantified by densitometry, and data are expressed as relative densities normalized to GAPDH, SphK1, and Akt, respectively. * P ≤ 0.05; # P < 0.001 (compared with time 0).
Fig. 6.
Downregulation of ERα36 reduces S1P and dihydro-S1P secretion stimulated by E2. MDA-MB-231 cells and MCF-7 cells transfected with control siRNA or with siRNA targeted to ERα36 (#3) were stimulated with vehicle or with E2 (100 nM) for the indicated times. S1P (A) and dihydro-S1P (DHS1P) (B) released into the medium during 30 min secretion assays were measured by LC/ESI/MS/MS. Data are mean ± SD. * P ≤ 0.05; # P < 0.001 (compared with vehicle).
Role of ERα36 and SphK1 in acquired tamoxifen resistance
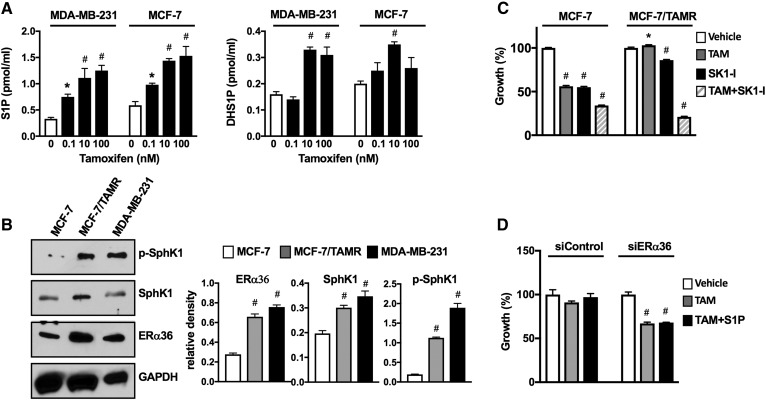
Tamoxifen, an antagonist of ERα66 and one of the first-line endocrine therapies for treatment of ERα-positive breast cancer (1, 2), was shown to be an agonist of ERα36 (9, 17). Therefore, we next examined the ability of tamoxifen to stimulate SphK1 and S1P secretion from ERα-negative MDA-MB-231 cells. Like E2, tamoxifen activated SphK1 and markedly increased secretion of S1P in a dose-dependent manner (Fig. 7A). A concentration of tamoxifen as low as 0.1 nM significantly increased secretion of S1P from MCF-7 cells expressing all three splice variants and from MDA-MB-231 cells. Maximum secretion of S1P and dihydro-S1P from both cell lines was observed at tamoxifen concentrations of 10–100 nM (Fig. 7A).
Fig. 7.
Tamoxifen stimulates S1P production, and tamoxifen resistance is associated with increased ERα36 and SphK1 activation. A: MDA-MB-231 and MCF-7 cells were treated without or with the indicated concentrations of tamoxifen for 30 min. S1P and DHS1P released into the medium were measured by LC/ESI/MS/MS. Data are mean ± SD. * P ≤ 0.05. B: Expression levels of SphK1, p-SphK1, and ERα36 were determined by immunoblot analysis in MDA-MB-231, MCF-7/TAMR-1, and parental MCF-7/S0.5 cells. Blots were stripped and reprobed with anti-GAPDH Ab to show equal loading and transfer. Indicated proteins were quantified by densitometry, and data are expressed as relative densities normalized to GAPDH. A, B: * P ≤ 0.05, #P < 0.001 (compared to vehicle). C: MCF-7/TAMR-1 and MCF-7/S0.5 cells were treated with vehicle, tamoxifen (10 µM), SK1-I (15 µM), or both for 2 days, and cell growth was determined. D: MCF-7/TAMR-1 cells transfected with control siRNA or with siRNA targeted to ERα36 (#3) were treated with vehicle or tamoxifen (1 µM) in the absence or presence of S1P (100 nM) for 2 days, and cell growth was determined. C, D: Data are expressed as percent of vehicle-treated control and are mean ± SEM. * P ≤ 0.05; # P < 0.001 (compared with vehicle).
A major problem with tamoxifen therapy in breast cancer is development of acquired resistance. It has been suggested that tamoxifen resistance correlates not only with decreased expression of ERα66 but also with upregulation or activation of ERα36, leading to increased nongenomic signaling induced by tamoxifen (17, 54). To further substantiate that tamoxifen resistance may be due to increased expression of ERα36 and SphK1, we utilized tamoxifen-resistant MCF-7 cells (MCF-7/TAMR-1), derived from the parental MCF-7/S0.5 cell line, which acquired resistance after long-term culturing in the presence of 1 µM tamoxifen. In both de novo tamoxifen-resistant MDA-MB-231 TNBC cells and acquired tamoxifen-resistant MCF-7/TAMR-1 cells, ERα36 was significantly upregulated compared with MCF-7 parental cells (Fig. 7B). Levels of SphK1 and particularly activated SphK1, determined with a phospho-specific Ab, were greatly elevated in tamoxifen-resistant cells (Fig. 7B). Growth of MCF-7 cells was significantly reduced by treatment with tamoxifen or SK1-I, a specific SphK1 inhibitor that does not inhibit SphK2 (55), and was further decreased by their combination (Fig. 7C). As expected, tamoxifen was not toxic to MCF-7/TAMR-1 cells. Although treatment with SK1-I had a marginal effect on MCF-7/TAMR-1 cells, it markedly sensitized them to tamoxifen, and combination treatment significantly decreased viability by almost 80% (Fig. 7C). Similar to previous reports (19, 56), knockdown of ERα36 restored the sensitivity of MCF-7/TAMR-1 cells to the growth-inhibitory effects of tamoxifen (Fig. 7D). However, addition of exogenous S1P (100 nM) did not reverse it (Fig. 7D).
Expression of SphK1 and ERα36 is increased in endocrine-resistant breast cancer patients
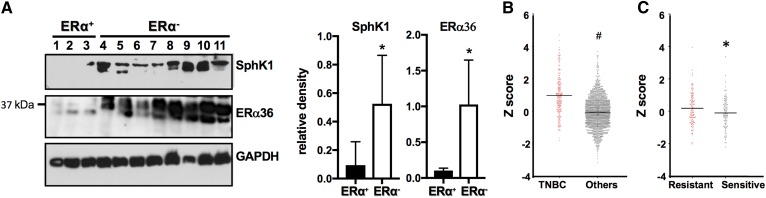
It has recently been noted that increased expression of ERα36 correlates with severity of breast cancer, metastasis and recurrence, and tamoxifen therapy resistance (17, 20). In addition, SphK1 is upregulated in drug- and endocrine-therapy-resistant breast cancers and correlates with poor prognosis (25, 26, 37). Consistent with these reports, immunoblotting of PDXs showed that ERα-positive tumors have low expression of ERα36 compared with ERα-negative tumors that have significantly higher ERα36 expression (Fig. 8A). Similarly, ERα-negative xenografts have significantly higher levels of SphK1 (Fig. 8A). Moreover, mining of The Cancer Genome Atlas (TCGA) breast tumor expression database indicated that TNBC patients have greater SphK1 expression compared with all other breast cancer patients (Fig. 8B). Furthermore, tumors from hormone therapy-resistant patients also have significantly higher SphK1 levels than patients that respond to hormone therapies, such as tamoxifen (Fig. 8C).
Fig. 8.
Tamoxifen resistance correlates with increased SphK1 and ERα36 expression in breast cancer patients. A: Expression levels of SphK1 and ERα36 in the indicated PDXs derived from ERα-positive (1–3) and -negative (4–11) breast cancer patients were determined by immunoblot analysis and quantified by densitometry. Data are expressed as relative densities normalized to GAPDH. * P ≤ 0.05. 1, HCI-03; 2, HCI-13; 3, HCI-11; 4, W2; 5, W30; 6, HCI-16; 7, HCI-10; 8, HCI-9; 9, HCI-1; 10, HCI-2; 11, UCD18. B, C: Breast cancer patient subtypes were from clinical expression information contained within TCGA datasets. B: SphK1 expression in TNBC versus other ERα-positive breast tumors. # P = 0.0001. C: Comparison of SphK1 expression in hormone therapy-sensitive and hormone therapy-resistant tumors. * P = 0.04.
DISCUSSION
E2 is traditionally considered to regulate complex functions by binding to its canonical receptor ERα66 and directing it to the nucleus, where it modulates gene expression (57). Although it has long been known that E2 also can elicit rapid, nongenomic signaling (58, 59), the identity of the receptor and the mechanisms involved has been a matter of great debate. Several receptor candidates have been proposed to mediate these rapid effects of E2, including the canonical ERα receptors, ERα66 and ERα46, and the noncanonical ERα36, as well as GPER1 (7, 8). However, it is still controversial whether E2 is a physiological agonist of GPER1 and whether this receptor is even plasma membrane-associated, as some groups have shown that it is endoplasmic reticulum-associated (60, 61). Moreover, several studies have implicated the involvement of ERα36, but not GPER1, in nongenomic signaling of E2 (10–14, 16, 62).
Ample studies in TNBC cells have shown that this novel splice variant ERα36 enhances cell growth and survival in response to E2 (9, 11–15). Binding of E2 to ERα36 initiates diverse downstream signaling, including activation of phospholipase C, leading to production of diacylglycerol and inositol trisphosphate, calcium signaling, and protein kinase C activation, as well as activation of ERK1/2 and PI3K/Akt, all important survival pathways for breast cancer cells (10–15). However, the mechanism by which activation of ERα36 by E2 leads to these downstream signaling pathways is still unclear. In this work, we have uncovered this missing link. We have shown that nongenomic effects of E2 occur through binding of E2 to ERα36 and subsequent stimulation of SphK1, resulting in the formation and secretion of S1P and dihydro-S1P. Binding of these ligands to S1PRs leads to activation of downstream signaling pathways important for breast cancer progression, metastasis, and hormone therapy resistance. In this regard, ligation of S1PR3 by S1P has been shown to lead to activation of Src and matrix metalloproteases, resulting in heparin-binding-EGF shedding and release that is necessary for transactivation of EGFR (22, 33, 35, 63). These signaling events explain how rapid increases in extracellular S1P and binding to S1PR3 after treatment with E2 can lead to long-lasting effects in cancer resistance to tamoxifen treatment.
We have now provided several lines of evidence that ERα36 is the E2 receptor responsible for activation of SphK1 by E2. First, in triple-negative MDA-MB-231 and ERα-negative HCC38 breast cancer cells that express only ERα36 and lack expression of ERα66 and ERα46, E2 activates SphK1, enhances its phosphorylation and translocation to the plasma membrane, and increases production and secretion of S1P. Second, in these cells, membrane-impermeable E2-BSA also activates SphK1 and enhances secretion of S1P. Third, ERα36-neutralizing Ab attenuates SphK1 activation and S1P formation and secretion following E2 stimulation. Finally, downregulation of ERα36 suppresses E2-mediated SphK1 activation and S1P and dihydro-S1P secretion. Interestingly, in MCF-7 breast cancer cells that are ERα-positive and express all three ERα splice variants, specific downregulation of ERα36 also significantly reduced the rapid secretion of S1P and dihydro-S1P induced by E2. This result suggests that, even in these cells, which have high expression levels of ERα66 and ERα46, low expression of ERα36 mediates the rapid, nongenomic activation of SphK1 and production of S1P and dihydro-S1P.
Despite extensive studies, endocrine resistance is still a major problem for adequate treatment of breast cancer, and 50% of patients that initially respond to tamoxifen treatment eventually acquire hormone-therapy resistance. Activation of ERα36 has also been associated with EGFR activation, and downregulation of ERα66 switches growth from E2-dependent to growth factor-dependent, suggesting that ERα36 is involved in hormone-therapy resistance (17, 54). Consistent with previous studies, we have shown that hormone-therapy resistance correlates with increased expression of ERα36 and SphK1 in tamoxifen-resistant breast cancer cell lines, as well as in PDXs from ERα-negative breast cancer patients. Similarly, data from TCGA show that tumors from ERα-negative as well as those from hormonal-resistant breast cancer patients have significantly higher expression of SphK1 compared with all other breast cancer patients. Our results support the notion that hormone-therapy resistance occurs through activation of ERα36, which in turn activates the SphK1/S1P axis important for growth, survival, switching breast cancer from E2-dependent to E2-independent progression, and resistance to hormonal therapies, such as tamoxifen. It should be noted that, although as was reported previously (19, 56), knockdown of ERα36 restored the sensitivity of tamoxifen-resistant cells to tamoxifen, addition of exogenous S1P did not rescue them. This could be due to degradation of exogenous S1P. Alternatively, intracellularly generated S1P rather than inside-out signaling via S1PR3 could contribute to cell growth and tamoxifen resistance, as both intracellular and secreted S1P are increased in response to activation of ERα36 by E2. These results are consistent with a previous study (64) and the notion that intracellular S1P might also contribute to cell growth and drug resistance (65–68). Moreover, in agreement with previous studies demonstrating that downregulation of SphK1 sensitized tamoxifen-resistant breast cancer cell lines to tamoxifen (64), we demonstrated that a specific SphK1 inhibitor also greatly restored sensitivity to tamoxifen in resistant cells. Taken together, our findings indicate that ERα36 and the SphK1 axis may play an important role in nongenomic effects of E2 and development of de novo and acquired resistance to hormone therapy of breast cancer. Therefore, targeting this axis should be explored as a therapeutic option to circumvent endocrine resistance with potential improvement of clinical outcome.
Acknowledgments
The authors thank Dr. Jeremy Allegood for skillful sphingolipid analyses. The authors acknowledge the Virginia Commonwealth University Lipidomics and Microscopy Cores, which are supported in part by funding from the National Institutes of Health-National Cancer Institute Cancer Center Support Grant P30 CA016059.
Footnotes
Abbreviations:
- E2
- 17β-estradiol
- E2-BSA
- 17β-estradiol conjugated to BSA
- EGFR
- epidermal growth factor receptor
- ERα
- estrogen receptor α
- PDX
- patient-derived xenograft
- PR
- progesterone receptor
- S1P
- sphingosine-1-phosphate
- S1PR
- sphingosine-1-phosphate receptor
- SphK
- sphingosine kinase
- TCGA
- The Cancer Genome Atlas
- TNBC
- triple-negative breast cancer
This work was supported by National Institutes of Health Grants F31 CA220798 (M.A.M.) and R01GM043880 (S.S.); and U.S. Department of Defense Breast Cancer Research Program Award W81XWH-14-1-0086 (S.S). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare no conflicts of interest.
REFERENCES
- 1.Clarke R., Leonessa F., Welch J. N., and Skaar T. C.. 2001. Cellular and molecular pharmacology of antiestrogen action and resistance. Pharmacol. Rev. 53: 25–71. [PubMed] [Google Scholar]
- 2.McDonnell D. P., and Norris J. D.. 2002. Connections and regulation of the human estrogen receptor. Science. 296: 1642–1644. [DOI] [PubMed] [Google Scholar]
- 3.Madaio R. A., Spalletta G., Cravello L., Ceci M., Repetto L., and Naso G.. 2010. Overcoming endocrine resistance in breast cancer. Curr. Cancer Drug Targets. 10: 519–528. [DOI] [PubMed] [Google Scholar]
- 4.Bayraktar S., and Gluck S.. 2013. Molecularly targeted therapies for metastatic triple-negative breast cancer. Breast Cancer Res. Treat. 138: 21–35. [DOI] [PubMed] [Google Scholar]
- 5.Klinge C. M. 2001. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 29: 2905–2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schutz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., et al. 1995. The nuclear receptor superfamily: the second decade. Cell. 83: 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poulard C., Treilleux I., Lavergne E., Bouchekioua-Bouzaghou K., Goddard-Leon S., Chabaud S., Tredan O., Corbo L., and Le Romancer M.. 2012. Activation of rapid oestrogen signalling in aggressive human breast cancers. EMBO Mol. Med. 4: 1200–1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levin E. R., and Hammes S. R.. 2016. Nuclear receptors outside the nucleus: extranuclear signalling by steroid receptors. Nat. Rev. Mol. Cell Biol. 17: 783–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Z., Zhang X., Shen P., Loggie B. W., Chang Y., and Deuel T. F.. 2006. A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc. Natl. Acad. Sci. USA. 103: 9063–9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L., Wang Q., Lv X., Sha L., Qin H., Wang L., and Li L.. 2015. Expression and localization of estrogen receptor in human breast cancer and its clinical significance. Cell Biochem. Biophys. 71: 63–68. [DOI] [PubMed] [Google Scholar]
- 11.Chamard-Jovenin C., Jung A. C., Chesnel A., Abecassis J., Flament S., Ledrappier S., Macabre C., Boukhobza T., and Dumond H.. 2015. From ERalpha66 to ERalpha36: a generic method for validating a prognosis marker of breast tumor progression. BMC Syst. Biol. 9: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pelekanou V., Notas G., Kampa M., Tsentelierou E., Radojicic J., Leclercq G., Castanas E., and Stathopoulos E. N.. 2012. ERalpha36, a new variant of the ERalpha is expressed in triple negative breast carcinomas and has a specific transcriptomic signature in breast cancer cell lines. Steroids. 77: 928–934. [DOI] [PubMed] [Google Scholar]
- 13.Chaudhri R. A., Olivares-Navarrete R., Cuenca N., Hadadi A., Boyan B. D., and Schwartz Z.. 2012. Membrane estrogen signaling enhances tumorigenesis and metastatic potential of breast cancer cells via estrogen receptor-alpha36 (ERalpha36). J. Biol. Chem. 287: 7169–7181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gu Y., Chen T., Lopez E., Wu W., Wang X., Cao J., and Teng L.. 2014. The therapeutic target of estrogen receptor-alpha36 in estrogen-dependent tumors. J. Transl. Med. 12: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omarjee S., Jacquemetton J., Poulard C., Rochel N., Dejaegere A., Chebaro Y., Treilleux I., Marangoni E., Corbo L., and Romancer M. L.. 2017. The molecular mechanisms underlying the ERalpha-36-mediated signaling in breast cancer. Oncogene. 36: 2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhri R. A., Hadadi A., Lobachev K. S., Schwartz Z., and Boyan B. D.. 2014. Estrogen receptor-alpha 36 mediates the anti-apoptotic effect of estradiol in triple negative breast cancer cells via a membrane-associated mechanism. Biochim. Biophys. Acta. 1843: 2796–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Su X., Xu X., Li G., Lin B., Cao J., and Teng L.. 2014. ER-alpha36: a novel biomarker and potential therapeutic target in breast cancer. Onco Targets Ther. 7: 1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Z. Y., and Yin L.. 2015. Estrogen receptor alpha-36 (ER-alpha36): a new player in human breast cancer. Mol. Cell. Endocrinol. 418: 193–206. [DOI] [PubMed] [Google Scholar]
- 19.Gu W., Dong N., Wang P., Shi C., Yang J., and Wang J.. 2017. Tamoxifen resistance and metastasis of human breast cancer cells were mediated by the membrane-associated estrogen receptor ER-alpha36 signaling in vitro. Cell Biol. Toxicol. 33: 183–195. [DOI] [PubMed] [Google Scholar]
- 20.Fahlén M., Zhang H., Löfgren L., Masironi B., Von Schoultz E., Von Schoultz B. O., and Sahlin L.. 2016. Expression of estrogen receptors in relation to hormone levels and the Nottingham prognostic index. Anticancer Res. 36: 2839–2847. [PubMed] [Google Scholar]
- 21.Pyne N. J., and Pyne S.. 2010. Sphingosine 1-phosphate and cancer. Nat. Rev. Cancer. 10: 489–503. [DOI] [PubMed] [Google Scholar]
- 22.Maczis M., Milstien S., and Spiegel S.. 2016. Sphingosine-1-phosphate and estrogen signaling in breast cancer. Adv. Biol. Regul. 60: 160–165. [DOI] [PubMed] [Google Scholar]
- 23.Geffken K., and Spiegel S.. 2018. Sphingosine kinase 1 in breast cancer. Adv. Biol. Regul. 67: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nava V. E., Hobson J. P., Murthy S., Milstien S., and Spiegel S.. 2002. Sphingosine kinase type 1 promotes estrogen-dependent tumorigenesis of breast cancer MCF-7 cells. Exp. Cell Res. 281: 115–127. [DOI] [PubMed] [Google Scholar]
- 25.Ruckhäberle E., Rody A., Engels K., Gaetje R., von Minckwitz G., Schiffmann S., Grösch S., Geisslinger G., Holtrich U., Karn T., et al. 2008. Microarray analysis of altered sphingolipid metabolism reveals prognostic significance of sphingosine kinase 1 in breast cancer. Breast Cancer Res. Treat. 112: 41–52. [DOI] [PubMed] [Google Scholar]
- 26.Watson C., Long J. S., Orange C., Tannahill C. L., Mallon E., McGlynn L. M., Pyne S., Pyne N. J., and Edwards J.. 2010. High expression of sphingosine 1-phosphate receptors, S1P1 and S1P3, sphingosine kinase 1, and extracellular signal-regulated kinase-1/2 is associated with development of tamoxifen resistance in estrogen receptor-positive breast cancer patients. Am. J. Pathol. 177: 2205–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ohotski J., Edwards J., Elsberger B., Watson C., Orange C., Mallon E., Pyne S., and Pyne N. J.. 2013. Identification of novel functional and spatial associations between sphingosine kinase 1, sphingosine 1-phosphate receptors and other signaling proteins that affect prognostic outcome in estrogen receptor-positive breast cancer. Int. J. Cancer. 132: 605–616. [DOI] [PubMed] [Google Scholar]
- 28.Gao Y., Gao F., Chen K., Tian M. L., and Zhao D. L.. 2015. Sphingosine kinase 1 as an anticancer therapeutic target. Drug Des. Devel. Ther. 9: 3239–3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta A., Loo S. Y., Huang B., Wong L., Tan S. S., Tan T. Z., Lee S. C., Thiery J. P., Lim Y. C., Yong W. P., et al. 2014. SPHK1 regulates proliferation and survival responses in triple-negative breast cancer. Oncotarget. 5: 5920–5933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sukocheva O., and Wadham C.. 2014. Role of sphingolipids in oestrogen signalling in breast cancer cells: an update. J. Endocrinol. 220: R25–R35. [DOI] [PubMed] [Google Scholar]
- 31.Takabe K., and Spiegel S.. 2014. Export of sphingosine-1-phosphate and cancer progression. J. Lipid Res. 55: 1839–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sukocheva O. A., Wang L., Albanese N., Pitson S. M., Vadas M. A., and Xia P.. 2003. Sphingosine kinase transmits estrogen signaling in human breast cancer cells. Mol. Endocrinol. 17: 2002–2012. [DOI] [PubMed] [Google Scholar]
- 33.Sukocheva O., Wadham C., Holmes A., Albanese N., Verrier E., Feng F., Bernal A., Derian C. K., Ullrich A., Vadas M. A., et al. 2006. Estrogen transactivates EGFR via the sphingosine 1-phosphate receptor Edg-3: the role of sphingosine kinase-1. J. Cell Biol. 173: 301–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takabe K., Kim R. H., Allegood J. C., Mitra P., Ramachandran S., Nagahashi M., Harikumar K. B., Hait N. C., Milstien S., and Spiegel S.. 2010. Estradiol induces export of sphingosine 1-phosphate from breast cancer cells via ABCC1 and ABCG2. J. Biol. Chem. 285: 10477–10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sukocheva O., Wadham C., and Xia P.. 2013. Estrogen defines the dynamics and destination of transactivated EGF receptor in breast cancer cells: role of S1P(3) receptor and Cdc42. Exp. Cell Res. 319: 455–465. [DOI] [PubMed] [Google Scholar]
- 36.Truman J. P., Garcia-Barros M., Obeid L. M., and Hannun Y. A.. 2014. Evolving concepts in cancer therapy through targeting sphingolipid metabolism. Biochim. Biophys. Acta. 1841: 1174–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pyne N. J., Ohotski J., Bittman R., and Pyne S.. 2014. The role of sphingosine 1-phosphate in inflammation and cancer. Adv. Biol. Regul. 54: 121–129. [DOI] [PubMed] [Google Scholar]
- 38.Huang K. L., Li S., Mertins P., Cao S., Gunawardena H. P., Ruggles K. V., Mani D. R., Clauser K. R., Tanioka M., Usary J., et al. 2017. Proteogenomic integration reveals therapeutic targets in breast cancer xenografts. Nat. Commun. 8: 14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeRose Y. S., Wang G., Lin Y. C., Bernard P. S., Buys S. S., Ebbert M. T., Factor R., Matsen C., Milash B. A., Nelson E., et al. 2011. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 17: 1514–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kabos P., Finlay-Schultz J., Li C., Kline E., Finlayson C., Wisell J., Manuel C. A., Edgerton S. M., Harrell J. C., Elias A., et al. 2012. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res. Treat. 135: 415–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sarkar S., Maceyka M., Hait N. C., Paugh S. W., Sankala H., Milstien S., and Spiegel S.. 2005. Sphingosine kinase 1 is required for migration, proliferation and survival of MCF-7 human breast cancer cells. FEBS Lett. 579: 5313–5317. [DOI] [PubMed] [Google Scholar]
- 42.Liu H., Sugiura M., Nava V. E., Edsall L. C., Kono K., Poulton S., Milstien S., Kohama T., and Spiegel S.. 2000. Molecular cloning and functional characterization of a novel mammalian sphingosine kinase type 2 isoform. J. Biol. Chem. 275: 19513–19520. [DOI] [PubMed] [Google Scholar]
- 43.Kim E. Y., Sturgill J. L., Hait N. C., Avni D., Valencia E. C., Maceyka M., Lima S., Allegood J., Huang W. C., Zhang S., et al. 2014. Role of sphingosine kinase 1 and sphingosine-1-phosphate in CD40 signaling and IgE class switching. FASEB J. 28: 4347–4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hait N. C., Avni D., Yamada A., Nagahashi M., Aoyagi T., Aoki H., Dumur C. I., Zelenko Z., Gallagher E. J., Leroith D., et al. 2015. The phosphorylated prodrug FTY720 is a histone deacetylase inhibitor that reactivates ERalpha expression and enhances hormonal therapy for breast cancer. Oncogenesis. 4: e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chaudhri R. A., Schwartz N., Elbaradie K., Schwartz Z., and Boyan B. D.. 2014. Role of ERalpha36 in membrane-associated signaling by estrogen. Steroids. 81: 74–80. [DOI] [PubMed] [Google Scholar]
- 46.Johnson K. R., Becker K. P., Facchinetti M. M., Hannun Y. A., and Obeid L. M.. 2002. PKC-dependent activation of sphingosine kinase 1 and translocation to the plasma membrane. Extracellular release of sphingosine-1-phosphate induced by phorbol 12-myristate 13-acetate. J. Biol. Chem. 277: 35257–35262. [DOI] [PubMed] [Google Scholar]
- 47.Pitson S. M., Moretti P. A., Zebol J. R., Lynn H. E., Xia P., Vadas M. A., and Wattenberg B. W.. 2003. Activation of sphingosine kinase 1 by ERK1/2-mediated phosphorylation. EMBO J. 22: 5491–5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jarman K. E., Moretti P. A., Zebol J. R., and Pitson S. M.. 2010. Translocation of sphingosine kinase 1 to the plasma membrane is mediated by calcium- and integrin-binding protein 1. J. Biol. Chem. 285: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brindley D. N., and Pilquil C.. 2009. Lipid phosphate phosphatases and signaling. J. Lipid Res. 50(Suppl): S225–S230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishi T., Kobayashi N., Hisano Y., Kawahara A., and Yamaguchi A.. 2014. Molecular and physiological functions of sphingosine 1-phosphate transporters. Biochim. Biophys. Acta. 1841: 759–765. [DOI] [PubMed] [Google Scholar]
- 51.Tong J. S., Zhang Q. H., Wang Z. B., Li S., Yang C. R., Fu X. Q., Hou Y., Wang Z. Y., Sheng J., and Sun Q. Y.. 2010. ER-alpha36, a novel variant of ER-alpha, mediates estrogen-stimulated proliferation of endometrial carcinoma cells via the PKCdelta/ERK pathway. PLoS One. 5: e15408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin S. L., Yan L. Y., Zhang X. T., Yuan J., Li M., Qiao J., Wang Z. Y., and Sun Q. Y.. 2010. ER-alpha36, a variant of ER-alpha, promotes tamoxifen agonist action in endometrial cancer cells via the MAPK/ERK and PI3K/Akt pathways. PLoS One. 5: e9013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H. B., Li T., Ma D. Z., and Zhi H.. 2018. ERalpha36 gene silencing promotes tau protein phosphorylation, inhibits cell proliferation, and induces apoptosis in human neuroblastoma SH-SY5Y cells. FASEB J. 22: fj201701386. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X. T., Kang L. G., Ding L., Vranic S., Gatalica Z., and Wang Z. Y.. 2011. A positive feedback loop of ER-alpha36/EGFR promotes malignant growth of ER-negative breast cancer cells. Oncogene. 30: 770–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Paugh S. W., Paugh B. S., Rahmani M., Kapitonov D., Almenara J. A., Kordula T., Milstien S., Adams J. K., Zipkin R. E., Grant S., et al. 2008. A selective sphingosine kinase 1 inhibitor integrates multiple molecular therapeutic targets in human leukemia. Blood. 112: 1382–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang X., and Wang Z. Y.. 2013. Estrogen receptor-alpha variant, ER-alpha36, is involved in tamoxifen resistance and estrogen hypersensitivity. Endocrinology. 154: 1990–1998. [DOI] [PubMed] [Google Scholar]
- 57.Jensen E. V., Desombre E. R., Kawashima T., Suzuki T., Kyser K., and Jungblut P. W.. 1967. Estrogen-binding substances of target tissues. Science. 158: 529–530. [DOI] [PubMed] [Google Scholar]
- 58.Szego C. M., and Davis J. S.. 1967. Adenosine 3′,5′-monophosphate in rat uterus: acute elevation by estrogen. Proc. Natl. Acad. Sci. USA. 58: 1711–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Björnström L., and Sjöberg M.. 2005. Mechanisms of estrogen receptor signaling: convergence of genomic and nongenomic actions on target genes. Mol. Endocrinol. 19: 833–842. [DOI] [PubMed] [Google Scholar]
- 60.Otto C., Rohde-Schulz B., Schwarz G., Fuchs I., Klewer M., Brittain D., Langer G., Bader B., Prelle K., Nubbemeyer R., et al. 2008. G protein-coupled receptor 30 localizes to the endoplasmic reticulum and is not activated by estradiol. Endocrinology. 149: 4846–4856. [DOI] [PubMed] [Google Scholar]
- 61.Revankar C. M., Cimino D. F., Sklar L. A., Arterburn J. B., and Prossnitz E. R.. 2005. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 307: 1625–1630. [DOI] [PubMed] [Google Scholar]
- 62.Kang L., Zhang X., Xie Y., Tu Y., Wang D., Liu Z., and Wang Z. Y.. 2010. Involvement of estrogen receptor variant ER-alpha36, not GPR30, in nongenomic estrogen signaling. Mol. Endocrinol. 24: 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sukocheva O., Wadham C., and Xia P.. 2009. Role of sphingolipids in the cytoplasmic signaling of estrogens. Steroids. 74: 562–567. [DOI] [PubMed] [Google Scholar]
- 64.Sukocheva O., Wang L., Verrier E., Vadas M. A., and Xia P.. 2009. Restoring endocrine response in breast cancer cells by inhibition of the sphingosine kinase-1 signaling pathway. Endocrinology. 150: 4484–4492. [DOI] [PubMed] [Google Scholar]
- 65.Olivera A., Rosenfeldt H. M., Bektas M., Wang F., Ishii I., Chun J., Milstien S., and Spiegel S.. 2003. Sphingosine kinase type 1 Induces G12/13-mediated stress fiber formation yet promotes growth and survival independent of G protein coupled receptors. J. Biol. Chem. 278: 46452–46460. [DOI] [PubMed] [Google Scholar]
- 66.Usatyuk P. V., He D., Bindokas V., Gorshkova I. A., Berdyshev E. V., Garcia J. G., and Natarajan V.. 2011. Photolysis of caged sphingosine-1-phosphate induces barrier enhancement and intracellular activation of lung endothelial cell signaling pathways. Am. J. Physiol. Lung Cell. Mol. Physiol. 300: L840–L850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., et al. 2010. Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature. 465: 1084–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rashad S., Niizuma K., Saigusa D., Han X., Sato-Maeda M., Saito R., Uruno A., Fujimura M., Ikawa S., Yamamoto M., et al. 2018. Intracellular S1P levels dictate fate of different regions of the hippocampus following transient global cerebral ischemia. Neuroscience. 384: 188–202. [DOI] [PubMed] [Google Scholar]