Abstract
Mass spectrometry imaging (MSI) is a tool to rapidly map the spatial location of analytes without the need for tagging or a reporter system. Niemann-Pick disease type C1 (NPC1) is a neurodegenerative, lysosomal storage disorder characterized by accumulation of unesterified cholesterol and sphingolipids in the endo-lysosomal system. Here, we use MSI to visualize lipids including cholesterol in cerebellar brain tissue from the NPC1 symptomatic mouse model and unaffected controls. To complement the imaging studies, a data-processing pipeline was developed to generate consensus mass spectra, thereby using both technical and biological image replicates to assess differences. The consensus spectra are used to determine true differences in lipid relative abundance; lipid distributions can be determined in an unbiased fashion without prior knowledge of location. We show the cerebellar distribution of gangliosides GM1, GM2, and GM3, including variants of lipid chain length. We also performed MALDI-MSI of cholesterol. Further analysis of lobules IV/V and X of the cerebellum gangliosides indicates regional differences. The specificity achieved highlights the power of MSI, and this new workflow demonstrates a universal approach for addressing reproducibility in imaging experiments applied to NPC1.
Keywords: gangliosides, lipidomics, statistics, cholesterol
MALDI-MS imaging (MALDI-MSI) enables label-free in situ analysis of molecules such as lipids (1–3), proteins (4, 5), or metabolites (6, 7) directly within tissue slices as demonstrated by Caprioli et al. (8). Each irradiated tissue area generates a mass spectrum from which ions of interest can be viewed relative to their location and intensity. As recently reviewed (9), MALDI-MSI of lipids has been quite successful, given that lipids are abundant in biological tissues. Additionally, with the exception of cardiolipins and gangliosides, lipids have molecular masses between 200 and 1,000 Da, an optimal operating mass range of MALDI-TOF instruments to achieve high mass resolving power and maintain accurate mass measurements.
Niemann-Pick disease type C (NPC) is a fatal, neurodegenerative, lysosomal storage disorder caused by mutations of the encoding regions of genes either for the lysosomal transmembrane protein NPC1 or the cholesterol-binding glycoprotein NPC2 (10), two proteins that work in tandem to mobilize cholesterol through the late endosomal/lysosomal system (11–14). As a result of the primary genetic defect, unesterified cholesterol and glycosphingolipids accumulate in late endosomes and lysosomes. Specifically, dysfunction of these proteins causes the inability to metabolize and traffic contents, and eventually cellular death. Cerebellar Purkinje neurons appear to be most susceptible though the cerebral cortex; thalamus and hippocampus also demonstrate neuronal loss (15–19). Systemic downstream effects include oxidative stress (20), defective calcium signaling (21), and neuroinflammation (22). The clinical phenotype of NPC is broad and includes vertical supranuclear gaze palsy, tremors, ataxia, and early dementia, and ultimately is fatal (23). While several studies have looked at transcript and protein-level alterations in NPC1 (22, 24–27), less is known about alterations in the lipid landscape in the disease. Early work on lipid accumulation demonstrated that several tissues are affected (28–30). A targeted MS-based profiling study was reported in which different sphingolipids species were observed to be differential in the null NPC1 mouse model (31) as well as in the recently generated I1061T point mutant model (32). Additionally, lyso-sphingolipids in plasma and amniotic fluid have been quantified using targeted MS methods (33). Although whole tissue lysate lipidomics studies are invaluable, as they are sensitive, specific and can be used to obtain precise quantities, any information regarding spatial distribution is lost.
In the context of lysosomal storage disorders, few studies have reported lipid imaging using MS. Lipid mapping has been performed in a mouse model of Hunter syndrome, a carbohydrate metabolism disorder in which spatial distributions of the gangliosides GM2 and GM3 were obtained alongside immunohistochemistry (34). A method for enhancing neutral lipid mapping was shown in a mouse model of Fabry disease (35). Other lysosomal disorders with imaging studies include Farber (36), Gaucher (37, 38), and Sandhoff disease (39, 40). Recently, the I1061T NPC1 point mutant model was used to develop an infrared spectroscopy-MALDI imaging workflow. In this approach initial Fourier-transform infrared imaging data is used to guide MALDI imaging to allow for tissue annotation. The statistical component, including a t-test based feature and peak-picking extraction, was performed; however, this approach required specialized tools to obtain segmentation information (41). It is also important to note that while an NPC1 model was used, this study did not look at cerebellar changes given the coronal sectioning performed nor did the authors provide insight into lipid mapping related to NPC1 disease. These reports reveal the importance of spatially localizing lipids, particularly in NPC1, which has been far more troublesome for antibody-based approaches and provide an insight into disease pathology.
Given the complexity and size of MSI experiments, reproducibility is a paramount consideration in experimental design. For example, the goal of many MSI tissue profiling studies is to identify disease markers relative to their location in the sample. In order to have significance of a specific mass-to-charge ratio, any finding must be spatially, analytically, and biologically reproducible. To avoid the pursuit of “beautiful noise,” various techniques in preprocessing MSI data have been implemented (42, 43) and reviewed in greater detail (44). Pre-processing strategies apply mathematical and logical operations to raw spectral data such that replicates can be combined, compared, and analyzed ostensibly with minimized contributions from biological, instrumental, and random variance. The most common of these approaches is to normalize the total ion current to generate the image. The difficulty with such an approach is that it only averages signal variance and does not take into account the overall spectral integrity. It thereby assumes that each spectrum collected over a given area has a comparable number of features. This assumption may be acceptable when dealing with homogenous results of different chemical samples. However, imaging of biological replicates must be considered as a set of separate experiments, and thus even multiple acquisitions of serial sections can lead to great mass spectral variability. Eijkel et al. (45) developed an algorithm to correlate secondary ion MS and MALDI imaging data from the same tissue. The authors attempted to address the data reduction of imaging experiments by conducting spectral binning to unit resolution, which is inappropriate to do when using high-resolution mass analyzers. They performed principal component analysis and canonical correlation analysis to determine unique features in their datasets, but replicate analysis was not addressed in the workflow, thus potentially leading to an underestimation of features (45). We argue that simply publishing MSI data as representative images rather than constructs of replicate spectra underestimates the true variance between analyses, thereby potentially misleading the reader as to the significance of features. Such an approach furthermore excludes rather than incorporates the majority of data generated in the investigation.
In the present study, we sought to develop an algorithm to evaluate both biological and technical replicate imaging experiments and then identify altered lipids and map their spatial distribution using a mouse model of NPC1. We imaged the cerebellum of 7-week-old null BALB/c-Npcnih, hereafter Npc1−/− (46), and control littermates, using MALDI-MSI. Relative to the disease progression, the 7-week time point displays classical phenotypes of ataxia and tremors that recapitulate the human disease. The newly developed algorithm was used to evaluate Npc1+/+ and Npc1−/− MSI spectra datasets of the cerebellum to validate reproducibility between replicates. This method provides an unbiased process to screen for unique and biologically interesting features that would not be detected from a single tissue image alone and also provides a way to form representative mass spectra for a dataset of replicates.
MATERIALS AND METHODS
All reagents were used as supplied unless otherwise noted. Purified water was obtained via a Barnstead GenPure (Thermo Fisher Scientific). All reagents were obtained from Sigma-Aldrich unless noted. Hematoxylin was purchased from Ricca Chemical Co.
Experimental model
All experiments were performed in accordance with University of Illinois at Chicago IACUC-approved protocols. Balb/c npcnih (Npc1+/−) mice were obtained from Jackson Laboratories (RRID:IMSR JAX:003092), and a breeding colony was maintained in our laboratory. Genotyping was performed using polymerase chain reaction as previously reported (17). The primer sequences used for genotyping were: (FWD8F) 5′-GGTGCTGGACAGCCAAGTA-3′ and (REVINTR3) 5′-GATGGTCTGTTCTCCCATG-3′. At 7 weeks of age, control (Npc1+/+) and null mutant (Npc1−/−) mice were euthanized via CO2 asphyxiation followed by decapitation. Whole brain tissue was dissected and immediately frozen in dry ice to maintain spatial integrity and stored at −80°C.
H&E staining
Tissue staining was performed using rehydration and dehydration steps. Briefly, tissue sections on microscope slides were immersed in 95% ethanol solution for 30 s, 70% ethanol solution for 30 s, purified deionized water for 30 s, hematoxylin solution for 30 s, 100 mM ammonium carbonate solution for 20 s, 70% ethanol solution for 30 s, 95% ethanol solution for 30 s, 30 s in eosin solutions, 30 s in 100% ethanol, and then 2.5 min in xylene. The stained tissue was viewed using an Evos XL Core (Life Technologies) and to obtain the final image.
Sample preparation
Frozen, intact brain was cryosectioned at −20°C using a Microm HM 525 (Thermo Fisher Scientific). Serial tissue sections of 16 μm thickness were thaw-mounted directly on MALDI stainless steel target plates and optical microscope slides for H&E staining and then were placed under vacuum for 10 min, then stored at −80°C until further analysis. The MALDI stainless-steel plate was dried under vacuum for 10 min to remove residual moisture. The plate containing the tissue section was then submerged in 50 mM ammonium formate solution (Alfa Aesar) for 20 s (47). Matrix application was conducted using an artist-model airbrush to produce an even coating. The matrix 9-aminoacridine was prepared at 10 mg/ml concentration in acetone (Thermo Fisher Scientific). In an effort to have reproducible matrix application, the density of matrix applied was calculated by weighing the target plates before and after the application with the desired amount of matrix being 0.15–0.25 mg/cm2. The analysis of matrix deposition is provided in supplemental Fig. S1. The mass spectrum for the on-tissue analysis (supplemental Fig. S1A–D) is provided for each replicate to evaluate m/z 612.3, a matrix ion of 1,5-diaminonapthalene (48). An example image of a matrix ion is provided to demonstrate reproducible matrix application (supplemental Fig. S1E). Slight differences are observed from the tissue compared with the stainless-steel plate, which is confirmed by recent reports of ion suppression in MALDI-MSI experiments (49). Biological replicate analyses (n = 5 Npc1+/+, n = 6 Npc1−/−) were conducted with n = 3–4 technical replicates (serial sections) collected for each animal. Matrix sublimation of dihydroxybenzoic acid (DHB; Sigma-Aldrich) was conducted for cholesterol imaging using a homemade sublimation apparatus. DHB matrix was dissolved in acetone (30 mg/ml) and transferred into the bottom of the sublimation apparatus, then evaporated using a stream of nitrogen gas. The preweighed target plate was then attached to the flat bottom of the apparatus using copper tape. The sand bath temperature was set to 120°C, and the trap was filled with ice slush. The internal vacuum was kept at 80 mTorr before starting the sublimation which lasts for 2 min. The target plate was weighed again and measured to obtain the density. Initial lipid assignments were made by comparing accurate mass measurements with the LIPID MAPS database (http://www.lipidmaps.org). Second, tandem MS data was acquired to yield signature fragment ions of each lipid species.
MALDI-MSI
MSI was performed using a model 4800 Plus MALDI TOF/TOF Analyzer (Sciex) equipped with a 200 Hz Nd-YAG pulsed laser (355 nm). The approximate laser spot size was 90 µm. Data were acquired in both the positive and negative ion reflectron modes as separate experiments. Red phosphorus was used for external mass calibration as previously described (50). The number of laser shots per pixel was set at 255 and the raster distance between each pixel was set to 50 μm using the 4800 Imaging Tool (version 3.2; www.maldi-msi.org, M. Stoeckli). Data processing for the MS ion images was conducted using TissueView software (Version 1.1; Sciex) or MSiReader (51). Regions-of-interest were determined based on ion intensities compared with the MALDI plate which was then compared with a representative H&E-stained section. Data analysis was carried out using the R-Programming Language (https://www.r-project.org/about.html).
LC/MS/MS
Cerebellar tissue from three Npc1+/+ and Npc1−/− animals at 7 weeks of age were homogenized in 1X PBS. Protein concentration was measured using the Pierce BCA Protein Assay (Thermo Scientific). Extraction of cerebellar lipids from aliquots of each homogenized tissue was carried out using a modified chloroform/methanol method to extract gangliosides originally developed by Folch et. al (52). The upper aqueous layer from the chloroform/methanol protocol was collected, and the lipid extracts were resuspended in 9:1 (v/v) methanol:toluene with volumes normalized according to the protein concentration. Lipids were separated using an Agilent EclipsePlus C18 RRHD C18 column (100 × 2.1 mm, 1.8 μm) coupled to an Agilent 1290 Infinity II system. The column was maintained at 50°C at a flow rate of 250 μl/min. The mobile phases consisted of solvent A, 60:40 (v/v) acetonitrile:water with 10 mM ammonium acetate, and solvent B, 90:10 (v/v) isopropanol:acetonitrile with 10 mM ammonium acetate. MS analysis was carried out in “auto MS/MS mode” (8MS/s and 6 MS/MS/s) in triplicate analysis with an Agilent 6545 quadrupole TOF mass spectrometer in negative ion mode.
RESULTS AND DISCUSSION
The goal of this study was to address variability in MALDI-MSI experiments while incorporating technical and biological measurements to obtain strong biomarker candidates in the neurodegenerative disease NPC1. To carry this out, we used the Npc1 null mouse model, which is characterized by accumulation of unesterified cholesterol and glycosphingolipids in the late endosomal/lysosomal systems as well as displaying progressive neuron loss prominent in cerebellar Purkinje neurons. The mechanism detailing the interplay of cholesterol and sphingolipid accumulation and neuronal death is still under investigation. Therefore, this study demonstrates a useful tool for all MSI studies, while providing additional information about NPC1 disease specifically.
The general experimental workflow is similar to a traditional MSI, except that this study included technical and biological replicates with these replicates being used to develop the data analysis pipeline. The strategy here addresses the challenges in obtaining spatially mapped analytes in multiple datasets while employing statistically robust methods to evaluate such data. To accomplish this, we performed MSI of lipid species in cerebellar tissue from the Npc1-null mouse model. This model is ideal for such a development because known accumulating lipid species, such as gangliosides, may serve as internal controls while simultaneously allowing the discovery of new potential lipid biomarkers.
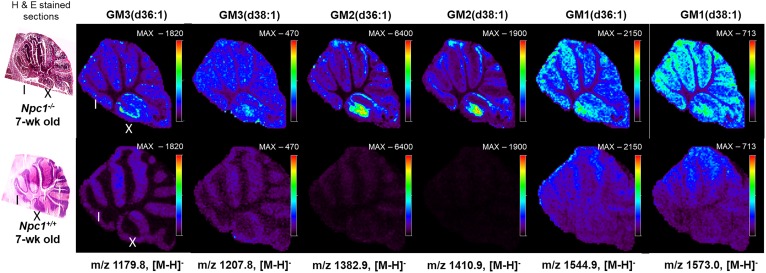
Four experiments were carried out to generate molecular maps of lipids in NPC1. First, positive-ion mode was used to detect basic lipids, such as phosphatidylcholines. Second, negative-ion mode was used to obtain data for acidic lipids such as sulfatides, phosphatidylinositols, phosphatidylglycerols, and phosphatidylethanolamines. Third, negative-mode data was collected at a higher mass-to-charge ratio range to obtain data for gangliosides, and, finally, we evaluated cholesterol. As observed in Fig. 1 (Top), gangliosides accumulate in the mutant animal tissues analyzed compared with controls and regional specificity was observed. Interestingly, we discern primary accumulation of several gangliosides in lobule X of the cerebellum for the Npc1 mutant animal tissue. It is important to note that images are shown as a total count scale for the specific ion of interest.
Fig. 1.
Representative MSI images of gangliosides in the cerebellum acquired from Npc1−/− (upper) and Npc1+/+ (lower) mice at 7 weeks of age with corresponding H&E-stained sections (left). Cerebellar lobules noted I and X. Raster distance between pixels is set at 50 μm. Scale bar indicates maximum intensity for each ion observed in both the control and mutant images. In all cases, the intensity of the ganglioside was higher in the mutant. The negative ion mass to charge for each ganglioside species is included.
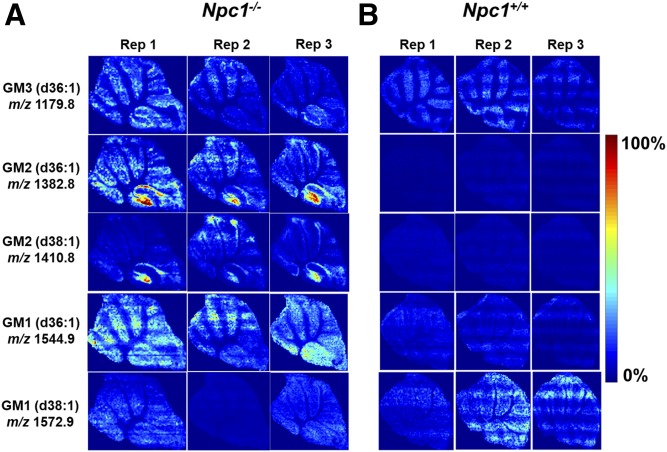
An added advantage of using the MSI technique over traditional staining methodologies, such as antibodies, is the specificity achieved for detection of different fatty acid compositions of the same molecular family. Here, we are able to differentiate and simultaneously detect multiple chain length forms of GM1, GM2, and GM3 (Fig. 1). Based on accurate mass of each molecule, they most likely contain stearic or arachidic acids as their fatty acyls. The spatial distributions of the possible GM2 and GM3 stearic forms are different. GM2 (stearic and arachidic forms) reside mainly in regions that appear to be the granular layer of the lobule, whereas GM3 (stearic acid) is found in what we propose is the Purkinje layer. This supports previous findings that GM2 and GM3 are not sequestered together and may have differing roles involving different biochemical mechanisms (28, 53, 54). Interestingly, the two GM3 species (steric and arachidonic acid) appear to localize differently within lobule X. Although gangliosides have been shown to be elevated in NPC1 disease, these new data indicate the specific distribution at a symptomatic time point in the neurodegeneration progression. Interestingly, lobule X typically does not show loss of Purkinje neurons at this time point in the disease progression in this mouse model; therefore, to observe this strong accumulation is of significant interest. As opposed to high spatial resolution microscopy studies that provide single cell or subcellular imaging, this approach addresses a means to look across the entire tissue for trends. Furthermore, the molecular specificity obtained regarding fatty acid composition is new given that antibody-based staining has only reported total GM1, GM2, or GM3 for NPC1 (28). To provide insight into biological and technical variability, we evaluated images from three serial sections obtained from the same animal. Figure 2 displays several ganglioside replicate images in both mutant and control animals. It should be noted that these images were obtained by manually investigating the lipids of interest, using MS spectra and then generating maps for each genotype and replicate. Therefore, initial assignments are made by accurate mass measurements, followed by on-tissue tandem MS experiments. The exact location of double bonds and length of the chains is inferred from database notation and cannot be precisely determined via this method. As a secondary verification, we then evaluated relative differences in ganglioside abundance between control and mutant animals using LC/MS/MS in which elevated amounts were confirmed in cerebellar tissue (supplemental Fig. S2). That is to say that increased levels of GM1, GM2, and GM3 were observed in NPC1 mutant cerebellar via both methods.
Fig. 2.
MALDI-MS images of major gangliosides found to be increased in 7-week-old Npc1−/− (A) and Npc1+/+ (B) cerebellum with serial sections for each mouse at a pitch of 50 μm. Substantial accumulation is detected for GM3 (d36:1) (m/z 1,179.8) in the Purkinje layer, specifically in lobule X, whereas the accumulation of GM2 (d36:1) (m/z 1,382.8) and GM2 (d38:1) (m/z 1,410.8) is determined to be localized in lobule X. GM1 (m/z 1,544.9) does not reveal localization. On the other hand, GM1 (m/z 1,572.9) does not appear to be accumulating or localizing in the KO mouse compared with the WT. Triplicate analysis shows reproducible ion intensities and lipid distributions, and images were normalized by the TIC of each section using MSiReader.
Unbiased detection of cerebellar lipid changes
A challenge with analyzing MSI datasets is having the ability to analyze multiple replicates and identifying analytes with differing abundance or spatial distribution in an unbiased and timely manner. To address this challenge, we developed an algorithmic-based method for clustering replicates, peak selection, and normalization. The following scheme depicts the algorithm executed: Centroided spectra → Peak selection → Normalize → Assess Similarity → Generate Consensus → Peak match to image. The three steps “Normalize,” “Assess Similarity,” and “Generate Consensus” are embedded in the program used for quality threshold (QT) clustering described below and in previous work evaluating MALDI linear and tandem MS spectra (55, 56). For the purposes of this work, however, we have developed the peak selection step and modified the basis on which consensus spectra are compared.
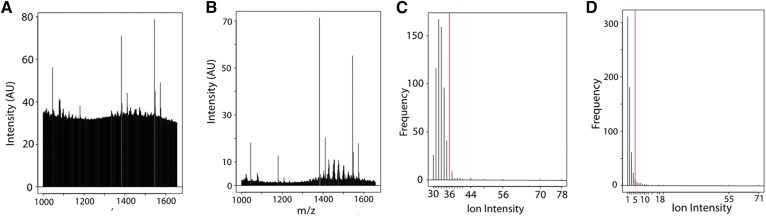
MALDI-MSI spectra tend to have great variation in both absolute intensity and in the baseline noise level due to variations in matrix application and amounts of material present in any given pixel (57). Although every attempt is in place to minimize these effects, recognizing the possibility for heterogeneous sample preparation is crucial. In order to analyze such spectra, we devised an approach based on an analysis of the frequency distribution of peak intensities in a spectrum (Fig. 3). That is, plotting peak intensities independent of the mass-to-charge-ratio at which they occur yields an intensity frequency distribution that can be processed subsequently to yield the ions of most significance in a spectrum. Manual inspection of a number of spectra suggested that incorporating the 40 most intense peaks into a “filtered spectrum” yielded the ions of most interest in that spectrum. Fig. 3A, B are examples of high and low noise baseline in a dataset, respectively. The intensity frequency distributions of these two spectra are shown below each of them (Fig. 3C, D), where the vertical red lines indicate the cut-off for the 40 most intense peaks, i.e., those peaks appearing to the right of the vertical red line. Note that for the spectrum in Fig. 3A, the origin of the intensity frequency distribution is ∼30 (shown in Fig. 3C), whereas the intensity frequency distribution for the spectrum in Fig. 3B has an origin of one. The number of peaks to be accepted, i.e., the position of the vertical red line, could be modified as required for particular analyses.
Fig. 3.
Example of selecting ions from spectra for each sample for the newly developed algorithm. Centroid mass spectrum of a typical noisy spectrum (A) and clean spectrum (B). Frequency distribution analysis of intensities for the corresponding spectra collected (C, D) to illustrate the approach to remove noise. The red lines show implementation of an algorithm to select the 40 most intense peaks from each spectrum. Note that the use of 40 peaks was determined for these dataset and can be changed for any given analysis. AU, arbitrary units.
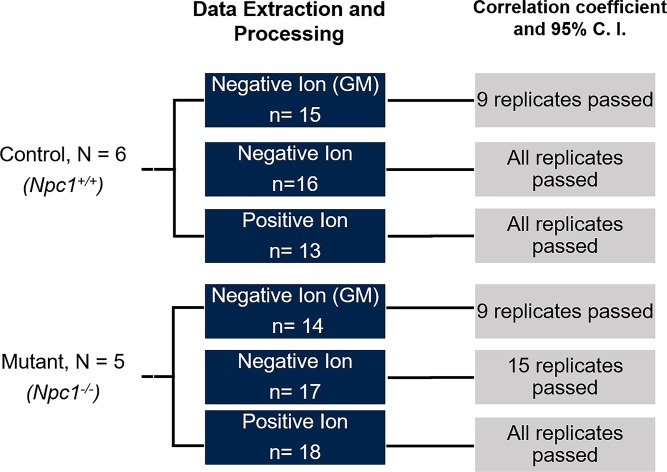
The QT clustering algorithm normalizes spectra to unit vectors and then forms clusters of peaks within the multiple spectra that depend on a sensitivity threshold related to the mass measurement precision. Following the clustering, a consensus spectrum for the dataset is formed, and each replicate is compared with the consensus using a dot product analysis. The dot products of each replicate to the consensus are used as a similarity metric; statistical confidence intervals can be calculated and used to evaluate the quality of each replicate spectrum with the aim of including only the most robust replicates into the consensus (55). Outlier spectra can be removed, and the algorithm can be rerun until only the most robust replicates are used to form the consensus spectrum. The overall algorithm workflow in the context of the present study is represented in Fig. 4. Multiple animals were studied for both genotypes (n = 5 and 6 for control and mutant, respectively) and technical replicates (n = 3 or 4 serial sections) for each animal were collected. Each box represents the number of total spectra collected for the MALDI-MSI ion mode of interest. Upon correlation analysis by dot product, and considering the 95% confidence interval, the final column of boxes depicts the number of replicates that passed and were used to generate the final consensus spectrum for the dataset. The resulting consensus spectra for each dataset are a normalized spectrum that represents all replicates for a given condition.
Fig. 4.
General workflow for collection and processing of multiple biological and technical replicates in MALDI-MSI datasets. Biological replicates were processed by investigating n = 6 (control) and n = 5 (mutant) animals. Cerebellar tissue was imaged using MALDI-MSI, and spectra were collected in three experiments to detect GM, negative, and positive lipid ions. Using technical replicates from each animal, as indicated by “n,” the newly developed alignment, normalization, and consensus spectra algorithm was utilized. To evaluate spectral reproducibility, a dot product analysis was carried out and evaluated based on correlation. Outlier spectra were removed, and those that “passed” are indicated by having a correlation above the 95% CI.
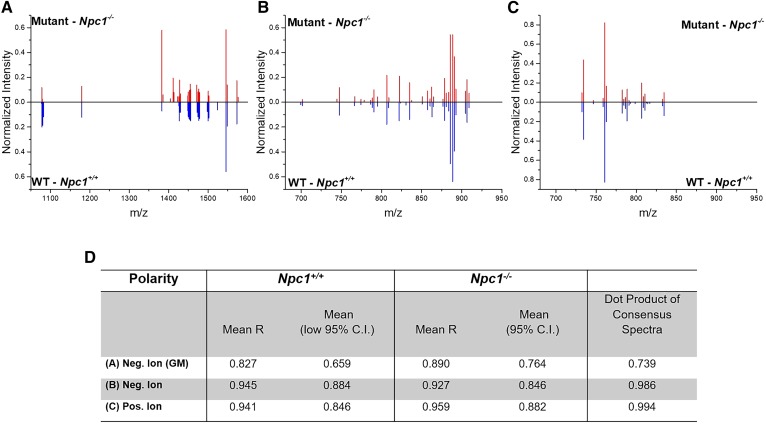
Figure 5A-C shows the consensus spectra for each of the three MS datasets collected with a focus on lipids in control and Npc1 mutant animals. Comparison of the spectra in Fig. 5A, showing the ganglioside mass region, we observed differences in normalized intensity for m/z 1,383 and 1,411 corresponding to the two GM2 species previously discussed (see Fig. 1). Tandem MS spectra for each are provided in the supplemental data (supplemental Figs. S3 and S4), where the presence of a sialic acid moiety (m/z 290) confirms the identity as ganglioside. Unfortunately, the exact lipid chain length and degree of saturation cannot be determined by the on-tissue tandem MS spectra. Additionally, isobaric interference in GM2(d38:1) (supplemental Fig. S4) shows a fragment ion at m/z 673.5. A common limitation in most MALDI-MSI experiments is the lack of separation of isobaric or near-isobaric species prior to detection as well as issues owing to instrumental limitations mainly in the timed ion selector. Interestingly, the GM1 species are not drastically elevated by this metric, thus demonstrating the power of the normalization strategy and unbiased detection tool, although manual inspection suggests a slight increase in the NPC1 mutant. In fact, the ratios of normalized peak intensity for GM1(d36:1) and GM(38:1) are 1.05 and 0.98, respectively, for the mutant over the control. Compared with LC/MS/MS data, these ratios are slightly decreased, thereby suggesting a conservative approach to identifying differences. To supplement the lipid assignments initially based upon accurate mass measurement, on tissue tandem MS measurements were collected for GM1(d36:1) at m/z 1,544.8 and GM1(d38:1) at m/z 1,572.8 (supplemental Figs. S5 and S6, respectively). The GM3 lipids assigned also included accurate mass measurement and tandem MS spectra (supplemental Figs. S7 and S8).
Fig. 5.
Consensus spectra and summary of MALDI-MSI replicate analysis in NPC1. Consensus mass spectra are provided for the GM mass region (A), negative ion (B), and positive ion (C), respectively. Data for WT is indicated in blue (below) and for the mutant in red (above). D: Data table shows average correlation coefficient between replicates for each dataset and the mean value of the 95% CI. The dot product analysis of the consensus spectra is also provided between the control and disease datasets (far right column). Dot products >0.9 are indicative of high similarity and, therefore, do not suggest spectral differences between control and mutant datasets.
Fig. 5B, C display the consensus spectra for the low mass negative ion mode and the positive ion mode in which significant differences were not observed. It is important to note at this point that the positive ion mode spectra were subjected to an additional step of data processing to remove all ions less than m/z 705, as these mass-to-charge-ratio values represent “chemical noise” that we associated with the matrix background. A summary of the consensus results is provided in Fig. 5D for all MS modes collected. Mean values for each of the consensus spectra and the dot product analysis between the two genotypes is included. Notably, the dot product analysis of control versus Npc1 mutant cerebellar imaging datasets indicated differences with a value of 0.739 for the negative-ion-mode ganglioside region; however, notable spectral differences were not observed in the negative ion mode (0.986) and positive ion mode (0.994) by the dot product analysis. Although this tool drives the further investigation of differences, manual interrogation is still needed to map and evaluate each image.
Total unesterified cholesterol mapping in the cerebellum
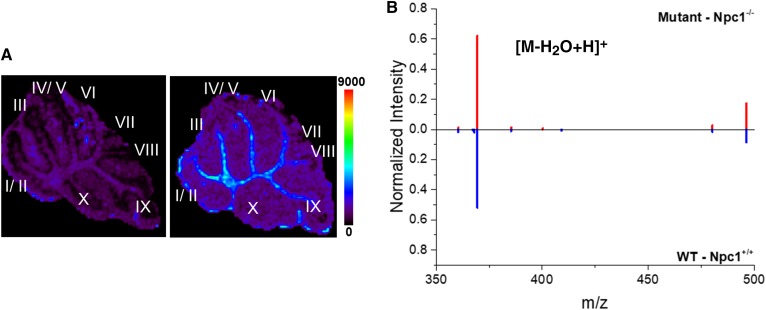
Given that NPC is a cholesterol storage disease, we utilized MALDI-MSI to map unesterified cholesterol in the cerebellum of control and Npc1 mutant mice (Fig. 6). In control animals, a slightly more abundant localization of cholesterol is observed in the white matter region, whereas this is less pronounced in the Npc1 mutants (Fig. 6A). This observation is consistent with reduced myelin in NPC1 (58, 59). Owing to the spatial resolution limits of our instrument, we are unable to localize intra-lysosomal storage in the Npc1 mutant animals. Using our consensus algorithm (Fig. 6B), we evaluated the normalized levels of cholesterol across the cerebellum. The consensus spectra reveal similar levels of cholesterol from biological replicates (n = 3 each), which included technical analyses (n = 7 for Npc1+/+ and n = 8 Npc1−/−). The indistinguishable difference in cholesterol levels agrees with previous work in which liver, kidney, spleen, and brain tissue from the Npc1 null mouse model, which were analyzed by GC/MS for cholesterol and oxysterol levels. Compared with controls, the brain did not have elevated levels of cholesterol despite altered levels in other tissues (60).
Fig. 6.
MALDI-MSI analysis of cholesterol. A: Representative MSI images of cholesterol m/z 369.3 (dehydrated ion, [M-H2O+H]+) in the cerebellum collected from Npc1−/− (left) and Npc1+/+ (right) mice at 7 weeks of age. Spatially, MSI images reveal an overall similar distribution between the genotypes however cholesterol may be slightly higher in the white matter of control animals. Scale bar represents 0–9,000 counts. B: Consensus spectra alignment following normalization of the top most intense 20 ions is provided for the cholesterol mass range for the whole cerebellum. The consensus spectra reveals similar normalized abundance of cholesterol for mutant (top, red) and WT (bottom, blue) tissue. Three animals of each genotype were analyzed, which included n = 8 and 7 technical replicates for mutant and WT, respectively.
Spatially resolved lipids in lobules
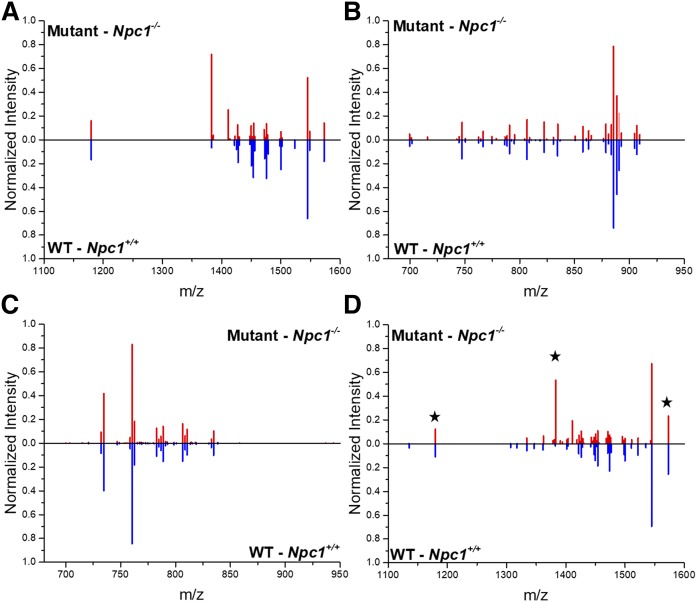
The age of the animals in the present study represents a symptomatic point in the NPC1 disease progression. With that knowledge, and based upon our preliminary data, we further analyzed the dataset to investigate changes we observed in only lobule X of the cerebellum. The original analysis considered data across the entire cerebellum, where at the 7-week time point, significant cerebellar degeneration has occurred (19, 61). Therefore, when observing differences by manual mapping, spatial, and quantitative differences for the GM3 species, as an example, were not found in the consensus spectra comparison. We attribute this to the possibility that in the earlier lobules, Purkinje neurons are known to degenerate, and, therefore, the accumulation in the later lobules is less pronounced by this method. To address this observation, further refinement of the approach was carried out. First, rather than analyzing the entire cerebellar tissue, only lobule X was analyzed (Fig. 7A–C). This is appropriate, given that this region of the cerebellum is preserved in the late stages of NPC1 disease and may provide insight into the neurodegeneration mechanism of NPC1 (19). Second, the analysis included the 60 most intense ions, which was determined after manual evaluation of the spectra. Similar differences were observed in the consensus spectra for lobule X, as was observed in the whole cerebellar tissue analysis.
Fig. 7.
Lobule analysis in NPC1. Consensus spectra alignment following normalization of the top most intense 60 ions is provided for the GM mass range (A), negative ion mode (B), and positive ion mode (C) with a focus on lobule X, which does not show Purkinje cell loss in NPC1 disease. Significant spectral intensity differences are observed in the ganglioside region. D: Consensus spectrum obtained from evaluating the ganglioside mass range for lobules IV/V in control and Npc1 mutant cerebella. Example gangliosides presented in Fig. 1 are denoted with stars including m/z 1,179.8; 1,382.9; and 1,573.0 representing variants of GM3, GM2, and GM1, respectively. Similar to whole cerebellar analysis, these sphingolipids are altered in Npc1 mutant lobule IV/V compared with control.
Based upon our imaging and lobule X analysis, we sought to determine whether the differences in gangliosides were only present in lobule X. We then evaluated the ganglioside mass range in cerebellar lobule IV/V to evaluate differences compared with lobule X in control and NPC1 mutant animals. This region of the cerebellum was affected earlier in the disease in terms of Purkinje neuron loss. Because the abundance differences in lobule X were so pronounced, we sought to evaluate a region of the cerebellum that may have differences, yet those differences may not be quite as obvious. Fig. 7D displays the consensus spectrum for the ganglioside mass range of lobule IV/V in control and Npc1 mutant cerebella. Ions corresponding to examples of ganglioside molecules GM1, GM2, and GM3 are denoted with stars. Consistent with our analysis in both the complete cerebellum and lobule X alone, elevation of gangliosides was observed in the mutant animal compared with control. From these data, we can conclude that the spectrum-based QT clustering normalization tool provides unbiased evaluating of biological and technical replicate imaging experiments to reveal alterations in disease models.
CONCLUSIONS
Using MALDI-MSI, lipid mapping, and visualization of different gangliosides species including fatty acidspecific composition of GM3, GM2, and GM1 in the cerebellum of a 7-week-old Npc1−/− mouse model was performed. Using this label-free technique, we were able to detect variants of different fatty acid chain length and determine the localizations of gangliosides. The use of unbiased peak selection and QT clustering for calculation of consensus spectra was able to validate spectral-expected features expressed differentially between the mutant and control datasets. Proof-of-concept experiments included MALDI-MSI analysis of cholesterol in the cerebellum. Validation by MS/MS and LC/MS analysis confirmed alterations spatially across the entire cerebellum and within specific lobule regions. With these tools set in place, continued studies on can be performed to evaluate the lipid changes during the entire disease progression of NPC1.
Supplementary Material
Acknowledgments
The authors thank Dr. Alfred L. Yergey for technical expertise and guidance with R-programming.
Footnotes
This work was supported by the University of Illinois at Chicago and the Ara Parseghian Medical Research Foundation.
The online version of this article (available at http://www.jlr.org) contains a supplement.
REFERENCES
- 1.Colsch B., Jackson S. N., Dutta S., and Woods A. S.. 2011. Molecular microscopy of brain gangliosides: illustrating their distribution in hippocampal cell layers. ACS Chem. Neurosci. 2: 213–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jackson S. N., Wang H. Y., and Woods A. S.. 2005. In situ structural characterization of phosphatidylcholines in brain tissue using MALDI-MS/MS. J. Am. Soc. Mass Spectrom. 16: 2052–2056. [DOI] [PubMed] [Google Scholar]
- 3.Jackson S. N., and Woods A. S.. 2009. Direct profiling of tissue lipids by MALDI-TOFMS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 877: 2822–2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stoeckli M., Chaurand P., Hallahan D. E., and Caprioli R. M.. 2001. Imaging mass spectrometry: a new technology for the analysis of protein expression in mammalian tissues. Nat. Med. 7: 493–496. [DOI] [PubMed] [Google Scholar]
- 5.Chaurand P., Schwartz S. A., Billheimer D., Xu B. J., Crecelius A., and Caprioli R. M.. 2004. Integrating histology and imaging mass spectrometry. Anal. Chem. 76: 1145–1155. [DOI] [PubMed] [Google Scholar]
- 6.Ly A., Buck A., Balluff B., Sun N., Gorzolka K., Feuchtinger A., Janssen K. P., Kuppen P. J., van de Velde C. J., Weirich G., et al. . 2016. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat. Protoc. 11: 1428–1443. [DOI] [PubMed] [Google Scholar]
- 7.Benabdellah F., Touboul D., Brunelle A., and Laprevote O.. 2009. In situ primary metabolites localization on a rat brain section by chemical mass spectrometry imaging. Anal. Chem. 81: 5557–5560. [DOI] [PubMed] [Google Scholar]
- 8.Caprioli R. M., Farmer T. B., and Gile J.. 1997. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69: 4751–4760. [DOI] [PubMed] [Google Scholar]
- 9.Berry K. A., Hankin J. A., Barkley R. M., Spraggins J. M., Caprioli R. M., and Murphy R. C.. 2011. MALDI imaging of lipid biochemistry in tissues by mass spectrometry. Chem. Rev. 111: 6491–6512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patterson M. C., Vanier M. T., Suzuki K., Morris J. A., Carstea E., Neufeld E. B., Blanchette-Mackie J. E., and Pentchev P.. 2001. Niemann-Pick disease type C: a lipid trafficking disorder. In The Metabolic and Molecular Bases of Inherited Disease. C. R. Scriver, A. L. Beaudet, W. S. Sly, D. Valle, B. Childs, K. W. Kinzler, and B. Vogelstein, editors. Mc Graw Hill, New York. [Google Scholar]
- 11.Naureckiene S., Sleat D. E., Lackland H., Fensom A., Vanier M. T., Wattiaux R., Jadot M., and Lobel P.. 2000. Identification of HE1 as the second gene of Niemann-Pick C disease. Science. 290: 2298–2301. [DOI] [PubMed] [Google Scholar]
- 12.Ko D. C., Binkley J., Sidow A., and Scott M. P.. 2003. The integrity of a cholesterol-binding pocket in Niemann–Pick C2 protein is necessary to control lysosome cholesterol levels. Proc. Natl. Acad. Sci. USA. 100: 2518–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohgami N., Ko D. C., Thomas M., Scott M. P., Chang C. C. Y., and Chang T-Y.. 2004. Binding between the Niemann–Pick C1 protein and a photoactivatable cholesterol analog requires a functional sterol-sensing domain. Proc. Natl. Acad. Sci. USA. 101: 12473–12478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Infante R. E., Abi-Mosleh L., Radhakrishnan A., Dale J. D., Brown M. S., and Goldstein J. L.. 2008. Purified NPC1 protein. I. Binding of cholesterol and oxysterols to a 1278-amino acid membrane protein. J. Biol. Chem. 283: 1052–1063. [DOI] [PubMed] [Google Scholar]
- 15.Higashi Y., Murayama S., Pentchev P. G., and Suzuki K.. 1993. Cerebellar degeneration in the Niemann-Pick type C mouse. Acta Neuropathol. 85: 175–184. [DOI] [PubMed] [Google Scholar]
- 16.Pentchev P. G., Brady R. O., Blanchette-Mackie E. J., Vanier M. T., Carstea E. D., Parker C. C., Goldin E., and Roff C. F.. 1994. The Niemann-Pick C lesion and its relationship to the intracellular distribution and utilization of LDL cholesterol. Biochim. Biophys. Acta. 1225: 235–243. [DOI] [PubMed] [Google Scholar]
- 17.Zervas M., Dobrenis K., and Walkley S. U.. 2001. Neurons in Niemann-Pick disease type C accumulate gangliosides as well as unesterified cholesterol and undergo dendritic and axonal alterations. J. Neuropathol. Exp. Neurol. 60: 49–64. [DOI] [PubMed] [Google Scholar]
- 18.Sun X., Marks D. L., Park W. D., Wheatley C. L., Puri V., O’Brien J. F., Kraft D. L., Lundquist P. A., Patterson M. C., Pagano R. E., et al. . 2001. Niemann-Pick C variant detection by altered sphingolipid trafficking and correlation with mutations within a specific domain of NPC1. Am. J. Hum. Genet. 68: 1361–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sarna J. R., Larouche M., Marzban H., Sillitoe R. V., Rancourt D. E., and Hawkes R.. 2003. Patterned Purkinje cell degeneration in mouse models of Niemann-Pick type C disease. J. Comp. Neurol. 456: 279–291. [DOI] [PubMed] [Google Scholar]
- 20.Fu R., Yanjanin N. M., Bianconi S., Pavan W. J., and Porter F. D.. 2010. Oxidative stress in Niemann–Pick disease, type C. Mol. Genet. Metab. 101: 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lloyd-Evans E., Morgan A. J., He X., Smith D. A., Elliot-Smith E., Sillence D. J., Churchill G. C., Schuchman E. H., Galione A., and Platt F. M.. 2008. Niemann-Pick disease type C1 is a sphingosine storage disease that causes deregulation of lysosomal calcium. Nat. Med. 14: 1247–1255. [DOI] [PubMed] [Google Scholar]
- 22.Cologna S. M., Cluzeau C. V. M., Yanjanin N. M., Blank P. S., Dail M. K., Siebel S., Toth C. L., Wassif C. A., Lieberman A. P., and Porter F. D.. 2014. Human and mouse neuroinflammation markers in Niemann-Pick disease, type C1. J. Inherit. Metab. Dis. 37: 83–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanier M. T., Wenger D. A., Comly M. E., Rousson R., Brady R. O., and Pentchev P. G.. 1988. Niemann-Pick disease group C: clinical variability and diagnosis based on defective cholesterol esterification. A collaborative study on 70 patients. Clin. Genet. 33: 331–348. [DOI] [PubMed] [Google Scholar]
- 24.Nicoli E-R., Al Eisa N., Cluzeau C. V. M., Wassif C. A., Gray J., Burkert K. R., Smith D. A., Morris L., Cologna S. M., Peer C. J., et al. . 2016. Defective cytochrome P450-catalysed drug metabolism in Niemann-Pick type C disease. PLoS One. 11: e0152007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cluzeau C. V. M., Watkins-Chow D. E., Fu R., Borate B., Yanjanin N., Dail M. K., Davidson C. D., Walkley S. U., Ory D. S., Wassif C. A., et al. . 2012. Microarray expression analysis and identification of serum biomarkers for Niemann–Pick disease, type C1. Hum. Mol. Genet. 21: 3632–3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tharkeshwar A. K., Trekker J., Vermeire W., Pauwels J., Sannerud R., Priestman D. A., te Vruchte D., Vints K., Baatsen P., Decuypere J-P., et al. . 2017. A novel approach to analyze lysosomal dysfunctions through subcellular proteomics and lipidomics: the case of NPC1 deficiency. Sci. Rep. 7: 41408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy J. V., Ganley I. G., and Pfeffer S. R.. 2006. Clues to neuro-degeneration in Niemann-Pick type C disease from global gene expression profiling. PLoS One. 1: e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou S., Davidson C., McGlynn R., Stephney G., Dobrenis K., Vanier M. T., and Walkley S. U.. 2011. Endosomal/lysosomal processing of gangliosides affects neuronal cholesterol sequestration in Niemann-Pick disease type C. Am. J. Pathol. 179: 890–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulinski A., and Vance J. E.. 2007. Lipid homeostasis and lipoprotein secretion in Niemann-Pick C1-deficient hepatocytes. J. Biol. Chem. 282: 1627–1637. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y., Wu Y-P., Wada R., Neufeld E. B., Mullin K. A., Howard A. C., Pentchev P. G., Vanier M. T., Suzuki K., and Proia R. L.. 2000. Alleviation of neuronal ganglioside storage does not improve the clinical course of the Niemann–Pick C disease mouse. Hum. Mol. Genet. 9: 1087–1092. [DOI] [PubMed] [Google Scholar]
- 31.Fan M., Sidhu R., Fujiwara H., Tortelli B., Zhang J., Davidson C., Walkley S. U., Bagel J. H., Vite C., Yanjanin N. M., et al. . 2013. Identification of Niemann-Pick C1 disease biomarkers through sphingolipid profiling. J. Lipid Res. 54: 2800–2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Praggastis M., Tortelli B., Zhang J., Fujiwara H., Sidhu R., Chacko A., Chen Z., Chung C., Lieberman A. P., Sikora J., et al. . 2015. A murine Niemann-Pick C1 I1061T knock-in model recapitulates the pathological features of the most prevalent human disease allele. J. Neurosci. 35: 8091–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pettazzoni M., Froissart R., Pagan C., Vanier M. T., Ruet S., Latour P., Guffon N., Fouilhoux A., Germain D. P., Levade T., et al. . 2017. LC-MS/MS multiplex analysis of lysosphingolipids in plasma and amniotic fluid: a novel tool for the screening of sphingolipidoses and Niemann-Pick type C disease. PLoS One. 12: e0181700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dufresne M., Guneysu D., Patterson N. H., Marcinkiewicz M. M., Regina A., Demeule M., and Chaurand P.. 2017. Multimodal detection of GM2 and GM3 lipid species in the brain of mucopolysaccharidosis type II mouse by serial imaging mass spectrometry and immunohistochemistry. Anal. Bioanal. Chem. 409: 1425–1433. [DOI] [PubMed] [Google Scholar]
- 35.Vens-Cappell S., Kouzel I. U., Kettling H., Soltwisch J., Bauwens A., Porubsky S., Muthing J., and Dreisewerd K.. 2016. On-tissue phospholipase C digestion for enhanced MALDI-MS imaging of neutral glycosphingolipids. Anal. Chem. 88: 5595–5599. [DOI] [PubMed] [Google Scholar]
- 36.Sikora J., Dworski S., Jones E. E., Kamani M. A., Micsenyi M. C., Sawada T., Le Faouder P., Bertrand-Michel J., Dupuy A., Dunn C. K., et al. . 2017. Acid ceramidase deficiency in mice results in a broad range of central nervous system abnormalities. Am. J. Pathol. 187: 864–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones E. E., Zhang W., Zhao X., Quiason C., Dale S., Shahidi-Latham S., Grabowski G. A., Setchell K. D. R., Drake R. R., and Sun Y.. 2017. Tissue localization of glycosphingolipid accumulation in a Gaucher disease mouse brain by LC-ESI-MS/MS and high-resolution MALDI imaging mass spectrometry. SLAS Discov. 22: 1218–1228. [DOI] [PubMed] [Google Scholar]
- 38.Snel M. F., and Fuller M.. 2010. High-spatial resolution matrix-assisted laser desorption ionization imaging analysis of glucosylceramide in spleen sections from a mouse model of Gaucher disease. Anal. Chem. 82: 3664–3670. [DOI] [PubMed] [Google Scholar]
- 39.Marsching C., Jennemann R., Heilig R., Grone H. J., Hopf C., and Sandhoff R.. 2014. Quantitative imaging mass spectrometry of renal sulfatides: validation by classical mass spectrometric methods. J. Lipid Res. 55: 2343–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marsching C., Eckhardt M., Grone H. J., Sandhoff R., and Hopf C.. 2011. Imaging of complex sulfatides SM3 and SB1a in mouse kidney using MALDI-TOF/TOF mass spectrometry. Anal. Bioanal. Chem. 401: 53–64. [DOI] [PubMed] [Google Scholar]
- 41.Rabe J-H., Sammour D. A., Schulz S., Munteanu B., Ott M., Ochs K., Hohenberger P., Marx A., Platten M., Opitz C. A., et al. . 2018. Fourier transform infrared microscopy enables guidance of automated mass spectrometry imaging to predefined tissue morphologies. Sci. Rep. 8: 313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Norris J. L., Cornett D. S., Mobley J. A., Andersson M., Seeley E. H., Chaurand P., and Caprioli R. M.. 2007. Processing MALDI mass spectra to improve mass spectral direct tissue analysis. Int. J. Mass Spectrom. 260: 212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tracy M. B., Chen H., Weaver D. M., Malyarenko D. I., Sasinowski M., Cazares L. H., Drake R. R., Semmes O. J., Tracy E. R., and Cooke W. E.. 2008. Precision enhancement of MALDI-TOF-MS using high resolution peak detection and label-free alignment. Proteomics. 8: 1530–1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ràfols P., Vilalta D., Brezmes J., Cañellas N., del Castillo E., Yanes O., Ramírez N., and Correig X.. 2016. Signal preprocessing, multivariate analysis and software tools for MA(LDI)-TOF mass spectrometry imaging for biological applications. Mass Spectrom. Rev 37: 281–306. [DOI] [PubMed] [Google Scholar]
- 45.Eijkel G. B., Kükrer Kaletaş B., van der Wiel I. M., Kros J. M., Luider T. M., and Heeren R. M. A.. 2009. Correlating MALDI and SIMS imaging mass spectrometric datasets of biological tissue surfaces. Surf. Interface Anal. 41: 675–685. [Google Scholar]
- 46.Pentchev P. G., Comly M. E., Kruth H. S., Patel S., Proestel M., and Weintroub H.. 1986. The cholesterol storage disorder of the mutant BALB/c mouse. A primary genetic lesion closely linked to defective esterification of exogenously derived cholesterol and its relationship to human type C Niemann-Pick disease. J. Biol. Chem. 261: 2772–2777. [PubMed] [Google Scholar]
- 47.Angel P. M., Spraggins J. M., Baldwin H. S., and Caprioli R.. 2012. Enhanced sensitivity for high spatial resolution lipid analysis by negative ion mode matrix assisted laser desorption ionization imaging mass spectrometry. Anal. Chem. 84: 1557–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaya I., Michno W., Brinet D., Iacone Y., Zanni G., Blennow K., Zetterberg H., and Hanrieder J.. 2017. Histology-compatible MALDI mass spectrometry based imaging of neuronal lipids for subsequent immunofluorescent staining. Anal. Chem. 89: 4685–4694. [DOI] [PubMed] [Google Scholar]
- 49.Taylor A. J., Dexter A., and Bunch J.. 2018. Exploring ion suppression in mass spectrometry imaging of a heterogeneous tissue. Anal. Chem. 90: 5637–5645. [DOI] [PubMed] [Google Scholar]
- 50.Sládková K., Houska J., and Havel J.. 2009. Laser desorption ionization of red phosphorus clusters and their use for mass calibration in time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 23: 3114–3118. [DOI] [PubMed] [Google Scholar]
- 51.Robichaud G., Garrard K. P., Barry J. A., and Muddiman D. C.. 2013. MSiReader: an open-source interface to view and analyze high resolving power MS imaging files on Matlab platform. J. Am. Soc. Mass Spectrom. 24: 718–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Folch J., Lees M., and Sloane Stanley G.. 1957. A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226: 497–509. [PubMed] [Google Scholar]
- 53.Walkley S. U. 2004. Secondary accumulation of gangliosides in lysosomal storage disorders. Semin. Cell Dev. Biol. 15: 433–444. [DOI] [PubMed] [Google Scholar]
- 54.Walkley S. U., and Vanier M. T.. 2009. Secondary lipid accumulation in lysosomal disease. Biochim. Biophys. Acta. 1793: 726–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olson M. T., Epstein J. A., Sackett D. L., and Yergey A. L.. 2011. Production of reliable MALDI spectra with quality threshold clustering of replicates. J. Am. Soc. Mass Spectrom. 22: 969–975. [DOI] [PubMed] [Google Scholar]
- 56.Heyer L. J., Kruglyak S., and Yooseph S.. 1999. Exploring expression data: identification and analysis of coexpressed genes. Genome Res. 9: 1106–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stoyanovsky D. A., Sparvero L. J., Amoscato A. A., He R. R., Watkins S., Pitt B. R., Bayir H., and Kagan V. E.. 2014. Improved spatial resolution of matrix-assisted laser desorption/ionization imaging of lipids in the brain by alkylated derivatives of 2,5-dihydroxybenzoic acid. Rapid Commun. Mass Spectrom. 28: 403–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie C., Burns D. K., Turley S. D., and Dietschy J. M.. 2000. Cholesterol is sequestered in the brains of mice with Niemann-Pick type C disease but turnover is increased. J. Neuropathol. Exp. Neurol. 59: 1106–1117. [DOI] [PubMed] [Google Scholar]
- 59.Kodachi T., Matsumoto S., Mizuguchi M., Osaka H., Kanai N., Nanba E., Ohno K., and Yamagata T.. 2017. Severe demyelination in a patient with a late infantile form of Niemann-Pick disease type C. Neuropathology. 37: 426–430. [DOI] [PubMed] [Google Scholar]
- 60.Tint G. S., Pentchev P., Xu G., Batta A. K., Shefer S., Salen G., and Honda A.. 1998. Cholesterol and oxygenated cholesterol concentrations are markedly elevated in peripheral tissue but not in brain from mice with the Niemann–Pick type C phenotype. J. Inherit. Metab. Dis. 21: 853–863. [DOI] [PubMed] [Google Scholar]
- 61.Walkley S. U., and Suzuki K.. 2004. Consequences of NPC1 and NPC2 loss of function in mammalian neurons. Biochim. Biophys. Acta. 1685: 48–62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.