Abstract
Engineered non-antibody scaffold proteins constitute a rapidly growing technology for diagnostics and modulation/perturbation of protein function. Here, we describe the rapid and systematic development of high-affinity 10FN3 domain inhibitors of the MDM2 and MDMX proteins. These are often overexpressed in cancer and represent attractive drug targets. Using facile in vitro expression and pull-down assay methodology, numerous design iterations addressing insertion site(s) and spacer length were screened for optimal presentation of an MDM2/X dual peptide inhibitor in the 10FN3 scaffold. Lead inhibitors demonstrated robust, on-target cellular inhibition of MDM2/X leading to activation of the p53 tumor suppressor. Significant improvement to target engagement was observed by increasing valency within a single 10FN3 domain, which has not been demonstrated previously. We further established stable reporter cell lines with tunable expression of EGFP-fused 10FN3 domain inhibitors, and showed their intracellular location to be contingent on target engagement. Importantly, competitive inhibition of MDM2/X by small molecules and cell-penetrating peptides led to a readily observable phenotype, indicating significant potential of the developed platform as a robust tool for cell-based drug screening.
Keywords: 10FN3 scaffold, bivalent binder, cancer therapy, MDM2, MDMX inhibitor, protein engineering
Introduction
Cell fate relies on a complex, yet well orchestrated web of protein–protein interactions. A change in any of these interactions governing normal function can lead to a diseased state, sometimes with devastating consequences as in the case of cancer. An important exemplar is the dysregulation of p53 by MDM2 and MDMX (murine double minute 2 or X—also known as HDM2 or HDMX in humans). p53 is a transcription factor crucial for modulation of target genes that regulate various cellular pathways including cell cycle arrest, DNA repair or apoptosis in response to stress signals (Lane, 1992; Vogelstein et al., 2000). Uncontrolled p53 activity is detrimental to growing stem cells and developing tissues (Jones et al., 1995; Montes de Oca Luna et al., 1995) while inactivation of p53 contributes to the development of malignancies (Hollstein et al., 1991). Consequently, the activity of p53 is tightly regulated in healthy cells by MDM2 and MDMX (Momand et al., 1992; Chen et al., 1995; Shvarts et al., 1996). Both negatively modulate p53 activity by interfering with its transcriptional activity. The E3 ubiquitin ligase activity of MDM2 can further polyubiquitinate and target p53 for proteosomal degradation (Haupt et al., 1997; Honda et al., 1997; Kubbutat et al., 1997). MDMX does not have E3 ligase function but stimulates MDM2-mediated ubiquitination and degradation of p53 via heterodimerization (Linares et al., 2003). MDM2 and MDMX are often overexpressed in cancer cells harboring wild-type p53 (Wade et al., 2013). Targeting the p53–MDM2/X interaction is therefore of great significance to cancer research.
The interaction between p53 and MDM2/X primarily involves the N-terminal domain of MDM2/X and an α-helix present in the N-terminal transactivation domain of p53 (amino acids 18–26) (Kussie et al., 1996). The N-terminal domain of MDM2/X comprises a pronounced hydrophobic cleft that engages three key hydrophobic residues (Phe19, Trp23 and Leu26) protruding from a single face of the p53 α-helix. In spite of the overall high structural similarities between the MDM2/X N-terminal domains most small molecule inhibitors show preferential inhibition of MDM2 (ElSawy et al., 2014; Joseph et al., 2014). In contrast, numerous equipotent dual peptide inhibitors of MDM2 and MDMX have been identified, predominantly from peptide phage libraries (Hu et al., 2007; Pazgier et al., 2009). Stabilization of the α-helical structure to minimize conformational entropy loss upon binding by incorporation of non-natural amino acids or peptide stapling has been shown to be important for efficient target binding and induction of p53 activity in vivo (Garcia-Echeverria et al., 2000; Banerjee et al., 2002; Bernal et al., 2007; Brown et al., 2013). An alternative stabilization strategy is to graft peptides onto a robust protein scaffold. These scaffolded peptides can readily facilitate detection and modulation of aberrant interactions, crucial for understanding underlying biological processes and also have great therapeutic potential (Binz and Plückthun, 2005; Weidle et al., 2013; Reverdatto et al., 2015; Tiede et al., 2017; Martin et al., 2018). Protein scaffolds under active development have expanded beyond conventional antibody fragments to include other non-antibody alternatives such as affimers, lipocalins, DARPins, kunitz domains, knottins and fibronectins. Ziconotide (irritable bowel syndrome) and Ecallantide (hereditary angioedema) are FDA approved drugs respectively based on the knottin and kunitz domains. Many more engineered protein scaffolds are in preclinical trials for treatment of cancers or inflammatory diseases (Vazquez-Lombardi et al., 2015; Simeon and Chen, 2018). The development of scaffold-based drugs stems from the need to overcome limitations of conventional antibodies. These include the ease and cost of production due to their smaller size, lack of cysteine residues and non-requirement of post-translational modification(s). A smaller size also promises improved tissue penetration and potential for modular architectures. Generation of libraries using protein scaffolds also permits shorter discovery times for hits to targets of interest. Libraries generated using the 10th fibronectin type III (10FN3) domain for example have allowed the selection of tight binders to various targets including the estrogen receptor (Koide et al., 2002), lysozyme (Lipovšek et al., 2007), SHP2 SH2 domain (Sha et al., 2013) and TNF-α (Xu et al., 2002).
Here, we describe systematic optimization of the 10FN3 domain as a protein scaffold for a highly potent dual peptide inhibitor of MDM2 and MDMX (Pazgier et al., 2009; Brown et al., 2013). Modulation of spacer lengths and insertion sites led to development of robust MDM2/X antagonists that showed excellent utility as fluorescent probes to monitor cellular distribution of MDM2/X and antagonism by drugs. We also successfully designed a bivalent binder in a single 10FN3 protein that displayed higher affinity to MDM2 and concomitant p53 activation in a cellular assay. Our results further highlight the versatility of the 10FN3 domain as a scaffold for the design of molecular probes for various applications.
Materials and Methods
Vector construction
Oligonucleotides used (Table I) in the construction of 10FN3 domain inhibitors were synthesized by Integrated DNA Technologies Pte Ltd (Singapore). Restriction enzymes and chemical reagents were purchased from NEB and Sigma respectively. The vector pET22b(+)-10FN3-HA was produced by amplifying vector PZV183 carrying the OPT10FN3 gene using primers INF-10FN3-ndeF and INT-10FN3-HA-bamR for infusion (Clontech) into pET22b(+) via NdeI and BamHI sites. This vector encodes OPT10FN3 with a C-terminal HA tag. pET22b(+)-M2-GGGS and pET22b(+)-M2C-GGGS were created by inverse PCR of pET22b(+)-10FN3-HA with primers FNR1 and FNF3, and FNC-R1 and FNC-F3, respectively, followed by intramolecular ligation of the PCR products. Inverse PCR was then performed on pET22b(+)-M2-GGGS and pET22b(+)-M2C-GGGS using the same reverse primer SGGG-Fstrand-10FN3-R and forward primers PM1-F and PM1ala-F, respectively, and the products ligated intramolecularly to construct pET22b(+)-MOP3 and pET22b(+)-MOP3C. To produce vector pET22b(+)-MOP3+, primers PM2-CD-for2 and PM2-CD-rev were used to perform an inverse PCR on pET22b(+)-MOP3 and the PCR product ligated intramolecularly. For immunoprecipitation work, relevant vectors were amplified with petF2 and petR to make amplicons with T7 promoter and ribosome binding sites required for in vitro transcription/translation (IVT). To generate pet22b(+)-HDM2-FLAG vector, pet22b(+)-HDM2-HA (Wei et al., 2013a) was amplified with infus-Mdm2-F and infus-Mdm2FLAG-R for infusion into pet22b(+) via NdeI and BamHI sites.
Table I.
Oligonucleotide sequences used for construction of vectors
| Oligos | Nucleotide sequeces in 5′–3′ direction |
|---|---|
| INF-10FN3-ndeF | AAGGAGATATACATATGGTTTCTGATGTTCCGCGTAAGC |
| INF-10FN3-HA-bamR | GCTCGAATTCGGATCCTCAAGCGTAATCTGGAACATCGTATGGGTACTCGAGCGCGGTACGGTAGTTAATCG |
| FNR1 | GTACTCGGCGAAGCTAGTAACAGCGTATACAGTGATGGT |
| FNF3 | TGGGCCCTGCTGAGCGGAGGAGGATCTCCAATCTCGATTAACTACCGTAC |
| FNC-R1 | GTACTCGGCAGCGCTAGTAACAGCGTATACAGTGATGGT |
| FNC-F3 | GCTGCCCTGGCTAGCGGAGGAGGATCTCCAATCTCGATTAACTACCGTAC |
| SGGG-Fstrand-10FN3-R | TCCTCCTCCAGAAGTAACAGCGTATACAGTGATGGT |
| PM1-F | AGCTTCGCCGAGTACTGGGCCCTGCTGAGC |
| PM1ala-F | AGCGCTGCCGAGTACGCTGCCCTGGCTAGC |
| PM2-CD-for2 | ATATTGGGCGCTGCTGAGCGGAGGAGGATCTGTTCAGGAGTTCACTGTACCTGGTTCCA |
| PM2-CD-rev | TCCGCAAAGCTTCCTCCTCCAGAGGTTTCACCGTACGTGATACGGTAATAAC |
| petF2 | CATCGGTGATGTCGGCGAT |
| petR | CGGATATAGTTCCTCCTTTCAGCA |
| INF-OPT10FN3-XhoI-pEGFP-C1-F | GGACTCAGATCTCGAGCTATGGTGAGTGACGTGCCTCGGAAAC |
| INF-OPT10FN3-BamH1-pEGFP-C1-R | TAGATCCGGTGGATCCCTACTCCAGGGCTGTGCGGTAGTTTATTG |
| OPT10FN3-FN3-F | GGAGGAGGATCTCCTATTTCAATAAACTACCGCACA |
| OPT10FN3-SGGG-PMI-Rcor | GCTCAGCAGGGCCCAGTACTCGGCGAAGCTTCCTCCTCCAGATGTCACGGCGTAGACAGTGATGGT |
| OPT10FN3-SGGG-PMIala-Rcor | GCTGCTAGCCAGGGCAGCGTACTCGGCAGCGCTTCCTCCTCCAGATGTCACGGCGTAGACAGTGATGGT |
| 10FN3-HIS-BamR | GCTCGAATTCGGATCCTCAGTGATGGTGATGGTGATGTCCTCCTCCAGACTCGAGCGCGGTACGGTAGT TAATCGAGATTGG |
| INF-FIBpTET-EcoR1F | CCCTCGTAAAGAATTCATGGTGAGCAAGGGCGAGGAGCTGTTC |
| INF-FIBpTET-BamH1R | GCAGAGATCTGGATCCCTACTCCAGGGCTGTGCGGTAGTTTATTG |
| INF-pTET-EcoR1F | GAATTCTTTACGAGGGTAGG |
| INF-pTET-BamH1R | GGATCCAGATCTCTGCAGC |
| infus-Mdm2-F | AAGGAGATATACATATGTGCAATACCAACATG |
| infus-Mdm2FLAG-R | GCTCGAATTCGGATCCTTATTTATCATCATCATCTTTATAATCGGGGAAATAAGTTAGCACAATCATT |
| INF-pTet-attL/R4X-F | GCAGGAAAGAACATGTGAAGCCTGCTTTATTTCATTAAGTTGGCATTATAAAAAAGCATTGCTTATCAAT TTGTTGCAACGAACAGGTCACTATCAGTCAAAATAAAATCATTATTTGATTTCAATTTTGTCCCACTCCCT CCCGGACATGTGAGCAAAAGG |
| INF-pTet-attL/R4X-R | CCTTTTGCTCACATGTCCGGGAGGGAGTGGGACAAAATTGAAATCAAATAATGATTTTATTTTGACTGAT AGTGACCTGTTCGTTGCAACAAATTGATAAGCAATGCTTTTTTATAATGCCAACTTAATGAAATAAAGCA GGCTTCACATGTTCTTTCCTGC |
For mammalian work, primers INF-OPT10FN3-XhoI-pEGFP-C1-F and INF-OPT10FN3-BamHI-pEGFP-C1-R were used to generate pEGFP-C1-OPT10FN3 by cloning mammalian optimized wild-type fibronectin via infusion into BamHI and XhoI sites of pEGFP-C1 vector. This vector also encodes an OPT10FN3 with an N-terminally fused EGFP tag. Inverse PCR was carried out on pEGFP-C1-OPT10FN3 using the same forward primer OPT10FN3-FN3-F and reverse primers OPT10FN3-SGGG-PMI-Rcor and OPT10FN3-SGGG-PMIala-Rcor, and the PCR products ligated intramolecularly to construct pEGFP-C1-MOP3 and pEGFP-C1-MOP3C, respectively. pEGFP-C1-MOP3+ was generated by cloning OPT10FN3 with M2 sequence in both CD and FG loop via infusion using primers INF-OPT10FN3-XhoI-pEGFP-C1-F and INF-OPT10FN3-BamHI-pEGFP-C1-R.
For stable cell line work, EGFP-MOPs expression plasmids were generated by insertion of EGFP-MOP gene between EcoRI and BamHI sites of pTetOne attL/R4X, a modified pTetOne vector (Clonetech) to include a C3 integrase recognition site (Vijaya Chandra et al., 2016). pTetOne attL/R4X was produced via infusion of annealed INF-pTet-attL/R4X-F and INF-pTet-attL/R4X-F oligos into PciI site of pTetOne vector. Amplified EGFP-MOP genes using primers INF-FIBpTET-EcoRIF and INF-FIBpTET-BamHIR were digested and ligated to PCR amplified pTetOne attL/RP4X vector using primers INF-pTET-EcoRIF and INF-pTET-BamHIR. C3 integrase expression plasmid pCMV-C3_CNLS_HIS was kindly provided by Peter Dröge (Nanyang Technological University).
For protein expression work, vectors pET22b(+)-10FN3-HA, pET22b(+)-MOP3, pET22b(+)-MOP3C and pET22b(+)-MOP3+ were amplified with INF-10FN3-ndeF and 10FN3-HIS-BamR, and the PCR products cloned via infusion into pET22b(+). All constructs carry a C-terminal HIS-tag.
Immunoprecipitation of in vitro-expressed proteins and western blot analysis
Magnetic protein G beads were incubated with anti-HA antibodies (0.5 μg antibody per 5 μL beads) for 1 h in PBST-3%BSA and subsequently washed thrice in PBST–0.1% BSA. To test for direct interaction between HA-tagged 10FN3-based constructs and FLAG-tagged MDM2, IVT-expressed 10FN3-HA constructs were first incubated with the beads on a rotator for 30 min to allow pull-down by virtue of the HA tag, followed by addition of IVT-expressed MDM2-FLAG and incubation for another 30 min. Where required, blank IVT (no template DNA added) was used as control.
For competitive interaction between 10FN3s and p53 to MDM2, IVT-expressed MDM2-FLAG was first incubated on anti-FLAG coated protein G beads (prepared as mentioned above), followed by the addition of respective 10FN3-HA constructs. The mixture was allowed to incubate for 30 min before IVT-expressed p53 was added. After all incubations, the beads were washed thrice with PBST–0.1% BSA and thrice with PBS, and the proteins eluted by resuspension with 20 μL 1× LDS buffer and incubation at 95°C for 5 min. The samples were subjected to SDS-PAGE and transferred to nitrocellulose membranes and probed for p53 with horseradish peroxidase (HRP)-conjugated DO1 antibody (Santa Cruz) or for MDM2 with HRP-conjugated anti-FLAG antibody (Roche). HA-tagged 10FN3s were probed for using anti-HA antibody (Sigma) followed by rabbit anti-mouse (Dakocytomation).
Cell culture
T22 reporter cells (stably transfected with a p53-responsive reporter construct, pRGCd-Fos-lacZ) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% (v/v) fetal calf serum (FCS) and 1% (v/v) penicillin/streptomycin. The cells were seeded at 8.0 × 104 cells/well in 6-well plates, 24 h prior to transfection.
HCT116 p53+/+ and p53−/− cells were maintained in McCoy’s 5A medium with 10% (v/v) fetal calf serum (FCS) and 1% (v/v) penicillin/streptomycin. The cells were seeded at 5.6 × 105 cells/well in 6-well plates, 24 h prior to transfection.
Transfection was carried out with respective 10FN3 mammalian expression constructs using Lipofectamine 2000 or Lipofectamine 3000 transfection reagent (ThermoFisher Scientific) according to manufacturer’s instructions. For β-galactosidase assay in T22 reporter cells, luciferase expression plasmid was co-transfected with respective expression constructs at 1:1 ratio.
β-Galactosidase assay and western blot analysis
T22 reporter cells were harvested 24 h after transfection and β-galactosidase activities were assessed using the Dual-light System (Applied Biosystems) according to the manufacturer’s protocol. The β-galactosidase activity was normalized with luciferase activity for each sample. Expression levels of EGFP-tagged 10FN3 proteins were checked via western blot by probing 5 μg of cell lysates with 3H9 antibody (Chromotek) followed by HRP-conjugated goat anti-rat IgG antibody (BioLegend). Endogenous actin levels were probed using HRP-conjugated AC15 (Sigma).
Immunoprecipitation of cell lysates and western blot analysis
HCT116 p53+/+ and p53−/− cells were harvested 24 h after transfection and lysed with RIPA buffer (50 mM Tris–HCl pH 7.4–8.0, 150 mM NaCl, 1% NP-40) supplemented with both protease and phosphatase inhibitors. 25 μL of GFP-Trap® agarose beads (Chromotek) was used per reaction and washed twice with PBST–0.1% BSA prior to incubation at 4°C overnight with 400–500 μg of cell lysates. The beads were then washed twice with PBST–0.1% BSA and once with PBS before elution with 20 μL 1X LDS buffer and incubation at 95°C for 5 min. The proteins were then separated by SDS-PAGE and transferred to nitrocellulose. A 10 μg of cell lysates was also loaded to check for endogenous protein levels. Immunoblotting was carried out with the relevant antibodies (2A9 antibody for endogenous MDM2, 3H9 antibody for EGFP-tagged 10FN3s, and HRP-conjugated DO1 and AC15 antibody for p53 and actin, respectively).
Protein expression and purification
BL21(DE3) cells (Invitrogen) transformed with HIS-tagged 10FN3-expressing constructs were grown in LB medium at 37°C and induced at OD600 nm of 0.5 with 1.0 mM IPTG at room temperature. The cells were harvested via centrifugation and resuspended in lysis buffer (50 mM Tris–HCl pH 8.5, 300 mM NaCl, 20 mM Imidazole, 10% glycerol, supplemented with protease inhibitor). The suspension was sonicated, centrifuged and filtered through 0.45 μm filter. 500 μL Ni-NTA slurry (Qiagen) was used per sample and equilibrated using wash buffer (50 mM Tris–HCl pH 8.5, 300 mM NaCl, 20 mM Imidazole, 10% glycerol). The filtered supernatant was then applied to the beads and incubated overnight at 4°C with rotation. After incubation, the beads were washed thrice with wash buffer and eluted in 5 fractions of 1 mL using elution buffer (50 mM Tris–HCl pH 8.5, 300 mM NaCl, 250 mM Imidazole, 10% glycerol). Protein purity as assessed by SDS-PAGE was ≈ 95% (Supplementary Fig. S5), and the proteins were concentrated using Amicon-Ultra (3 kDa MWCO) concentrator (Millipore).
Competitive fluorescence anisotropy assay
Fluorescence anisotropy assay was carried out as previously described (Brown et al., 2013). Briefly, constant concentration of FAM labeled 12-1 peptide (FAM-RFMDYWEGL-NH2; 5 nM) and N-terminal domain of MDM2 (M6-N125; 200 nM) was titrated with increasing concentrations of purified HIS-tagged MOP3C, MOP3 or MOP3+. Anisotropy measurements were carried out using the Envision Multilabel Reader (PerkinElmer). All experiments were carried out in PBST-3% DMSO. Titrations were carried out in duplicate and non-linear regression-fit was carried out using Prism 4.0 (GraphPad).
Generation of stable T22 clones
The EGFP-MOPs expression plasmids (pTetOne attL/R4X) were co-transfected into T22 with the C3 integrase expression plasmid (pCMV-C3_CNLS_HIS). Stable clones were selected via two rounds of cell sorting using either BDFACSAria IIu (BD Biosciences) or Moflo XDP (Beckman Coulter) for EGFP-MOP3C and EGFP-MOP3 expressing stable clones respectively. After overnight induction with 1μg/mL doxycycline Live cells were gated based on forward (FSC) and side (SSC) values in the BD FACSDiva or Summit software. First round of bulk sorting was done 1 week after transfection followed by single sort 2–3 weeks after the bulk sorting.
Live cell spinning disk confocal fluorescence microscopy
For live cell imaging, T22 stable clones were grown on 27 mm diameter Nunc glass bottom dish (Thermofisher) and induced with 0.1 or 1.0 μg/mL doxycycline for 5 h before image acquisition. Images were acquired on Nikon Eclipse Ti inverted microscope, using a 40× oil immersion objective (NA 1.3). Time-lapse images were acquired every 2.5 min for up to a duration of 1 h. Z-stacks were acquired for all images and sum slices projection was applied to all the stack images using ImageJ Software.
Sample preparation and mass spectrometric analysis
Following detergent free wash of the immunoprecipitated complexes, excess liquid was removed and beads were resuspended in 8 M Urea/ 50 mM Tris–HCl pH 8.5. Reduction was performed in 20 mM TCEP (20 min, 25°C) followed by alkylation with 55 mM CAA (20 min, 25°C, protected from light). Prior to digestion with LysC and Trypsin, 8 M Urea was diluted with100 mM TEAB (Urea < 1 M). Protein digestion was performed with LysC enzyme (Wako, Japan) and Trypsin (Promega, USA) for 4 and 18 h in 25°C (1:100, enzyme:protein ratio), respectively. Reaction was stopped with 1% TFA. Peptides were desalted using C-18 SepPak columns (Waters). Desalted peptides were vacuum dried and subsequently labeled with TMT10plex isobaric tag (Thermo). Labeling was performed following TMT10plex manufacturer protocol. Afterwards, samples were combined in 10 mM Ammonia Formate and fractionated using step gradient (12, 15, 25, 50% of ACN) on C-18 High pH reverse phase self-packed column (C-18 ReproSil-Pur Basic, Dr Maisch, 10μm). Each fraction was separately analyzed in 120 min gradient (0.1% formic acid in water and 99.9% ACN with 0.1% formic acid) using 50 cm 75 μm id Easy-Spray RP column (C-18, 2 μm particles, Thermo) connected to Easy nLC1000 (Thermo) chromatography system coupled online with Orbitrap Fusion mass spectrometer (Thermo). For acquisition, data dependent mode was used in a speed mode −3 s cycle. Orbitrap analyzer was utilized for both MS and MS/MS scans with ion targets and resolutions (OT-MS 4xE5 ions, resolution 60k, OT-MS/MS 5E4 ions, resolution 45k).
For data analysis, peak lists were generated using Proteome Discoverer 2.1 software (Thermo). Searches were done with Mascot 2.6 (Matrix Science) against forward/decoy Human Uniprot database and Sequest HT (Thermo) against contaminants database with customized sequence contents. Following parameters were used for search: precursor mass tolerance (MS) 20 ppm, OT-MS/MS 0.05 Da, three missed cleavages; Static modifications: Carboamidomethyl (C), TMT10plex. Variable modifications: oxidation (M), deamidated (NQ), acetyl N-terminal protein. Forward/decoy searches were used for false discovery rate (FDR) estimation on peptide/PSM level, high confidence (FDR 1%) /medium confidence (FDR 5%) were accepted.
Results
Design of 10FN3-based MDM2/X inhibitors
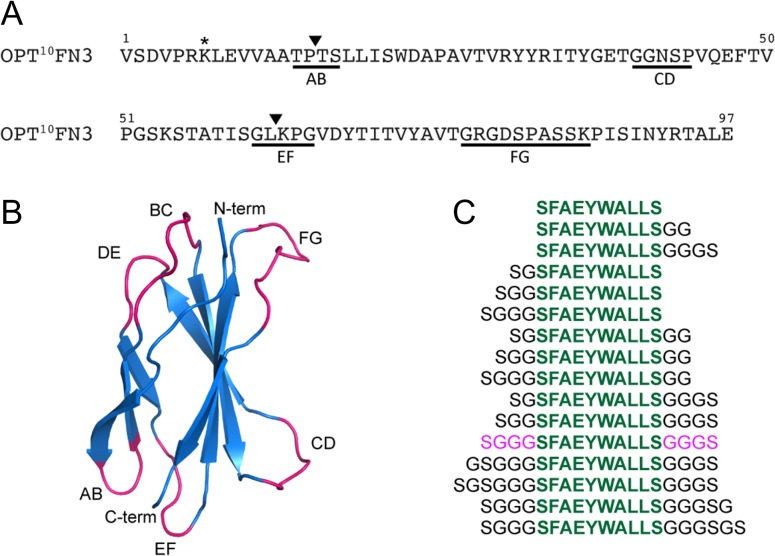
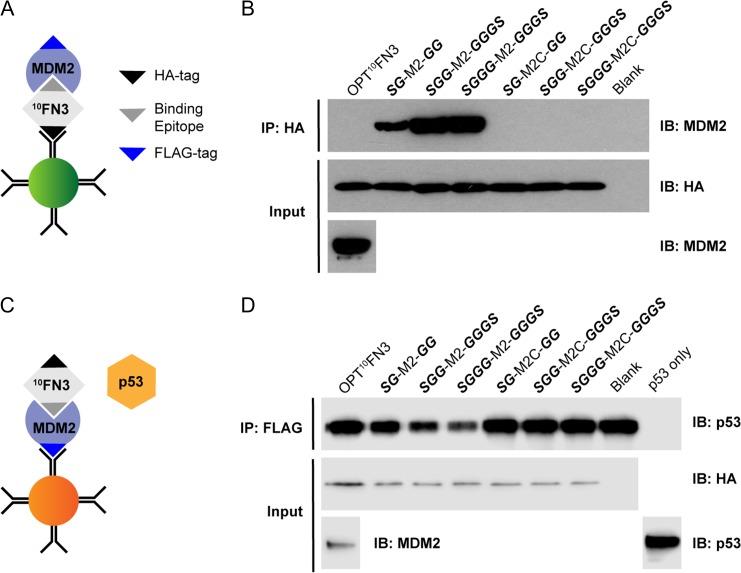
10FN3 is a well-validated protein scaffold for the design of specific and high-affinity binders (Koide et al., 1998; Xu et al., 2002; Hackel et al., 2008; Han et al., 2017). It is structurally similar to immunoglobulin domains, consisting of two β-sheets of seven anti-parallel β-strands connected by six exposed loops on two opposite poles (Fig. 1B). The BC, DE and FG loops correspond to CDR loops in immunoglobulin and have successfully been randomized to generate libraries from which tight binders to various target of interests have been identified (Bloom and Calabro, 2009; Lipovšek, 2010). In particular, the FG loop allows variable loop lengths (8–14 aa) and can tolerate amino acid insertions with very minimal loss of stability (Batori et al., 2003). Using an optimized 10FN3 (OPT10FN3) carrying the D7K mutation (Fig. 1A) for improved conformational stability (Koide et al., 2001), we replaced the FG loop region spanning residues G77 to K86 with the MDM2/X targeting peptide M2 (SFAEYWALLS), or a control peptide M2C where three key hydrophobic residues are mutated to alanine (SAAEYAALAS). M2 is a truncated version of MTide-02 (Brown et al., 2013), lacking an N-terminal threonine that is minimally involved in interaction with MDM2 (Kussie et al., 1996). The engineered 10FN3 domains were rapidly screened for expression and function using in-vitro transcription/translation, obviating the need for extensive protein purification. Intriguingly, no protein expression was detected for the pilot construct wherein the FG loop was replaced by M2 (Supplementary Fig. S1A, lane 2). We hypothesized that this resulted from steric clashes of the key F, W and L residues projecting from one face of M2 with the 10FN3 scaffold. A further panel of constructs comprising variant flanking spacer lengths and configurations (Fig. 1C) was therefore analyzed. The results indicated optimal expression when M2 was flanked on both sides by symmetrical spacers (SG-M2-GG, SGG-M2-GGGS and SGGG-M2-GGGS) (Supplementary Fig. S1B). These constructs were subsequently tested both for their ability to bind FLAG-tagged MDM2 and to inhibit binding of p53 to MDM2 using a pull-down assay with in vitro-expressed proteins (Fig. 2). OPT10FN3-M2 with flanking SGGG-GGGS spacer showed the most efficient binding to MDM2 and abrogation of p53 binding to MDM2 when compared to the other constructs (Fig. 2B and D, lane 4). This construct was termed MOP3 (MDM2/X-Obstructing Protein 3). All corresponding controls wherein the key F, W and L residues were mutated to alanine showed neither binding to MDM2 nor inhibition of p53 binding. Additional fine-tuning spacer iterations resulted in marginal improvements in expression but no major improved efficacy over MOP3 (Supplementary Fig. S2). These results highlight notable effects spacer lengths can have on engineered 10FN3 stability and performance, as seen in constructs with no spacer or short spacer sequences (Supplementary Fig. S1A and B), stressing the importance of diligent optimization.
Fig. 1.
Design of MDM2/X-Obstructing Protein (MOP) constructs. (A) Sequence of optimized 10FN3 (OPT10FN3). Position of the D7K mutation is indicated by *. Loop sequences that were replaced in our work are underlined. ▼ Represents the position within loop sequences where an insertion was made. (B) Ribbon representation of 10FN3. 10FN3 (PDB ID: 1FNF) shown in blue with exposed loops BC, DE, FG, AB, CD and EF colored in magenta and labeled accordingly. (C) Panel of tested peptide sequences comprised of M2 sequence in green with varying lengths of flanking GS-rich spacers. The optimal spacer combination is colored in pink.
Fig. 2.
Characterization of MDM2 binding to engineered FG loop 10FN3 domains. (A) Schematic diagram of a direct pull-down assay. (B) Western blot analysis of immunoprecipitated HA-tagged FG loop monobodies and FLAG-tagged MDM2 as shown in (A). Respective in-vitro transcription/translation (IVT) expressed HA-tagged FG loop 10FN3 domains were first immunoprecipitated with anti-HA antibody coated magnetic protein G beads followed by incubation with IVT expressed FLAG-tagged MDM2. (C) Schematic diagram of a competitive binding assay. (D) IVT expressed FLAG-tagged MDM2 was pulled down with anti-FLAG antibody coated magnetic protein G beads followed by co-incubation with respective IVT expressed HA-tagged FG loop 10FN3 domains and p53.
MOP3 activation of p53 in the T22 reporter cell line
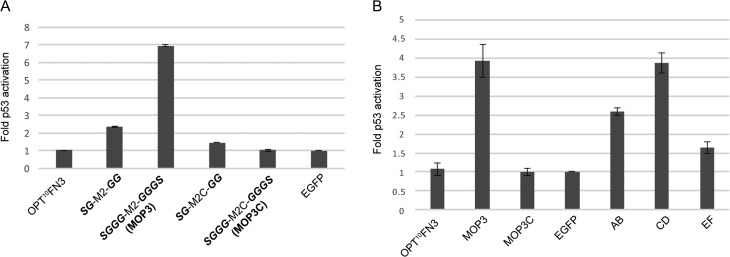
We next tested intracellular inhibition of MDM2/X by MOP3 using the well-established T22 reporter cell line. This comprises a stably integrated reporter gene (β-galactosidase) driven by a p53-dependent promoter (Lu et al., 1996; Berkson et al., 2005; Lain et al., 2008). Disruption of the MDM2/X-p53 interaction results in elevated p53 levels and β-galactosidase expression. Consistent with the pull-down assay, constitutively expressed EGFP-MOP3 resulted in notable upregulation of p53 activity when compared to corresponding EGFP-MOP3C control or empty vector (Fig. 3A). The SG-M2-GG construct that competed less efficiently with p53 for MDM2 binding (Fig. 2A) showed a correspondingly modest p53 activation in the T22 assay (Fig. 3A), further validating use of the in vitro expression and pull-down assay screening methodology.
Fig. 3.
Cellular inhibition of MDM2/X in T22 cell-based assay. (A) A comparison of the effect of lead engineered FG loop 10FN3 domain SGGG-M2-GGGS (MOP3) with SG-M2-GG on T22 reporter activity in transiently transfected T22 cells. SG-M2C-GG and SGGG-M2C-GGGS (MOP3C) are engineered negative control 10FN3 domains with mutation of key interacting residues. Reporter activity was determined 24 h after transfection. The results shown depict fold p53 reporter activation relative to EGFP alone. Data represent mean ± SD (n = 2). (B) Comparison of engineered AB/CE/EF loop 10FN3 domain to lead engineered FG loop 10FN3 domain (MOP3) in T22 reporter assay. Data represent mean ± SD (n = 2).
Single domain bivalent binder improves binding to MDM2/X and p53 activation
The 10FN3 scaffold presents numerous loops on opposite poles (Fig. 1B). This feature presents a unique opportunity for the design of bivalent or bispecific binders within a single domain while maintaining the small size of this scaffold. We therefore explored numerous bivalent iterations of MOP3, focusing attention on the AB (T14-S17), CD (G40-P44) and EF (G61-G65) loops displayed on the opposite face to the FG loop transplanted by M2. We first tested each loop in isolation for presentation of the optimized SGGG-M2-GGGS peptide. All three engineered 10FN3 domains were able to bind MDM2 and disrupt p53 binding to MDM2 in the pull-down assay (Supplementary Fig. S3). In the T22 cell reporter assay, the engineered CD loop 10FN3 domain induced a notable p53 response comparable to MOP3 followed by the engineered AB and EF loop 10FN3 domains (Fig. 3B). We also explored peptide insertion into existing loop sequences instead of direct replacement of the loop sequences (Table II) followed by double loop combinations. The best performing double loop constructs identified from the pull-down assays were AB2:FG, CD:FG and EF2:FG (Supplementary Fig. S3). In the T22 cell assay, only the CD:FG double loop construct (subsequently termed MOP3+) showed notable improvement over MOP3 (Supplementary Fig. S4). As with the in vitro assay (Supplementary Fig. S3), expression of AB2:FG and CD:FG was sub-optimal in T22 cells. Given the strong in vitro phenotypes associated with these variants despite poor expression, further engineering of the scaffold itself could be considered (Porebski et al., 2016).
Table II.
Loop sequences in engineered single or bivalent 10FN3 domain inhibitors
| Constructsa | AB loop | CD loop | EF loop | FG loop |
|---|---|---|---|---|
| WTb | TPTS | GGNSP | GLKPG | GRGDSPASSK |
| FG (MOP3) | SGGGSFAEYWALLSGGGSc | |||
| FGC (MOP3C) | SGGGSAAEYAALASGGGSd | |||
| AB | SGGGSFAEYWALLSGGGS | |||
| AB2 | TPSFAEYWALLSTS | |||
| AB:FG | SGGGSFAEYWALLSGGGS | SGGGSFAEYWALLSGGGS | ||
| AB2:FG | TPSFAEYWALLSTS | SGGGSFAEYWALLSGGGS | ||
| CD | SGGGSFAEYWALLSGGGS | |||
| CDC | SGGGSAAEYAALASGGGS | |||
| CD:FG (MOP3+) | SGGGSFAEYWALLSGGGS | SGGGSFAEYWALLSGGGS | ||
| CDC:FG | SGGGSAAEYAALASGGGS | SGGGSFAEYWALLSGGGS | ||
| EF | SGGGSFAEYWALLSGGGS | |||
| EF2 | GLSFAEYWALLSKPG | |||
| EF: FG | SGGGSFAEYWALLSGGGS | SGGGSFAEYWALLSGGGS | ||
| EF2:FG | GLSFAEYWALLSKPG | SGGGSFAEYWALLSGGGS |
aName of 10FN3 domain inhibitors based on loop where a replacement (AB, CD, EF or FG) or insertion was made (AB2, EF2).
bWild-type sequences of AB, CD, EF and FG loop region.
cSGGGSFAEYWALLSGGGS = SGGG-M2-GGGS.
dSGGGSAAEYAALASGGGS = SGGG-M2C-GGGS.
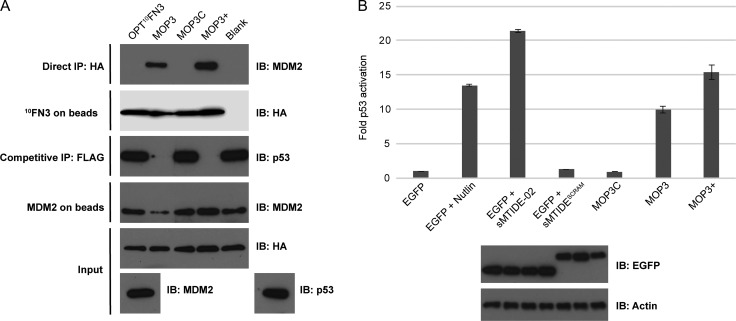
MOP3+ consistently displayed superior binding to and inhibition of MDM2, both in vitro and the cell-based assay (Fig. 4). The two binding interfaces on opposite poles of 10FN3 allowed the design of a single domain bivalent binder against MDM2 with improved avidity that led to a favorable increase in p53 activation. Our data shows that with proper optimization, loop elongation is tolerated in both our MOP3 and MOP3+ constructs. The 10-amino acid FG loop or the five-amino acid CD loop tolerated an 18-amino acid insert, illustrating the versatility of 10FN3 as a protein scaffold.
Fig. 4.
Characterization of engineered 10FN3-based MDM2/X inhibitors MOP3 and MOP3+. (A) Direct and competitive binding assay of MOP3C, MOP3 and MOP3+ to MDM2 as in Fig. 2. (B) Top: T22 reporter activity in transiently transfected cells expressing EGFP-fused MOP constructs compared to EGFP alone or EGFP expressing cells treated with Nutlin, sMTIDE-02 or sMTIDE-02SCRAM, a scrambled control stapled peptide. Bottom: Representative Western blot image of the expression levels of EGFP-fused MOP constructs (same order as in graph) in T22 and the loading control protein β-actin. Data represent mean ± SD (n = 3).
M2 peptide grafted onto 10FN3 binds MDM2 with similar affinity as free sMTIDE-02 stapled peptide
To determine the relative binding affinities of the MOP constructs to MDM2, we purified MOP proteins with C-terminal His-tags (Supplementary Fig. S5) and determined binding to MDM2 N-terminal domain (M6-N125) by competitive fluorescence polarization assay (Supplementary Fig. S6A). Purified MOP3 bound MDM2 N-terminal domain with an apparent Kd of 21.85 ± 0.56 nM, similar to the reported affinity of the sMTIDE-02 stapled peptide (Brown et al., 2013). Mutation of the three key hydrophobic residues in MOP3C significantly decreased affinity to MDM2 by >1000-fold (Supplementary Fig. S6B). In line with previous work detailing multivalent binders, MOP3+ showed 10-fold higher apparent affinity to MDM2 compared to MOP3 (apparent Kd = 2.43 ± 0.42 nM).
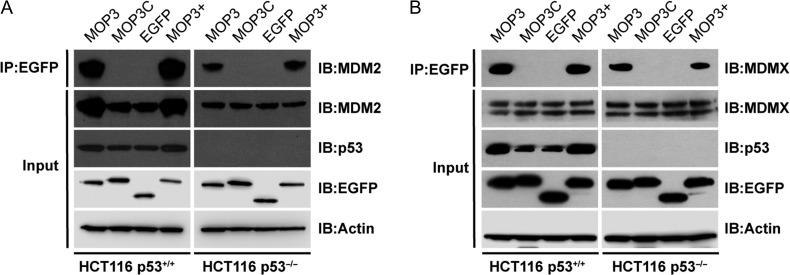
MOP3 binds endogenous MDM2 and MDMX and activates p53
We next tested engagement of MOP3 with endogenous MDM2/X in a human cell line using EGFP-fused MOP3 (EGFP-MOP3) or MOP3+ (EGFP-MOP3+) expressed in p53-positive (HCT116 p53+/+) and isogenic p53-null (HCT116 p53−/−) cells. EGFP-MOP3C and EGFP-alone were included as controls. Both MDM2 and MDMX were co-immunoprecipitated with EGFP-MOP3 and EGFP-MOP3+ but not control proteins in both cell lines, confirming cellular interaction of MOP3 and MOP3+ with MDM2 and MDMX (Fig. 5). Importantly, expression levels of EGFP-MOP3, EGFP-MOP3+ and EGFP-MOP3C were similar. Notably, increased MDM2 was pulled down by MOP3 and MOP3+ in p53-positive compared to p53-null cells (Fig. 5A, top row), due to the p53–MDM2 feedback loop, whereby MDM2 is a direct transcriptional target of p53 (Barak et al., 1993; Wu and Levine, 1997). Inhibition of MDM2 typically results in elevated p53 levels (due to reduced p53 ubiquitination) and subsequently increased MDM2 expression, the latter of which can be readily observed (Fig. 5A, row 2). MDMX, on the other hand, is not transcriptionally regulated by p53. Correspondingly, its levels remained unchanged in the two cell lines (Fig. 5B).
Fig. 5.
MOP3 and MOP3+ bind endogenous MDM2 and MDMX. p53-positive (HCT116 p53+/+) or p53-null (HCT116 p53−/−) cells were transfected with respective plasmids expressing EGFP-fusion and control constructs. Lysates were immunoprecipitated with anti-EGFP agarose beads and probed for (A) MDM2 or (B) MDMX.
We next determined the proteome-wide interactions of EGFP-MOP3 and EGFP-MOP3+ using pull-downs followed by mass spectrometry in HCT116 p53+/+ and p53−/− cell lines. As a stringent control, we used EGFP-MOP3C to filter out proteins interacting with the EGFP and fibronectin scaffold components. MDM2 ranked the most abundant interacting protein in HCT116 p53+/+ cells for both EGFP-MOP3 and EGFP-MOP3+ (Table SI). It also ranked highly in HCT116 p53−/− cells (Table SII) that are not subjected to the p53–MDM2 feedback loop and consequently express reduced amounts of MDM2. Intriguingly, whilst MDMX interaction was detected by immunoblotting (Fig. 5B), it was not identified by mass spectrometry. This could be due to comparatively low MDMX peptide abundance and/or significant post-translational modifications. Gene ontology analysis (Eden et al., 2009) of the top 100 proteins in the HCT116 p53+/+ MOP3/MOP3+ interactomes highlighted enrichment of genes involved in ubiquitin-dependent protein catabolism, consistent with MDM2 function. Importantly, known interactors of MDM2 were also identified including ribosomal proteins L5, L11, L23 and S27a (Marechal et al., 1994; Lohrum et al., 2003; Zhang et al., 2003; Dai et al., 2004; Sun et al., 2011; Czerwińska et al., 2017). These results show that MOP3/MOP3+ can efficiently bind their targets in complex cellular mixtures.
EGFP-MOP3 exhibits increased nuclear localization
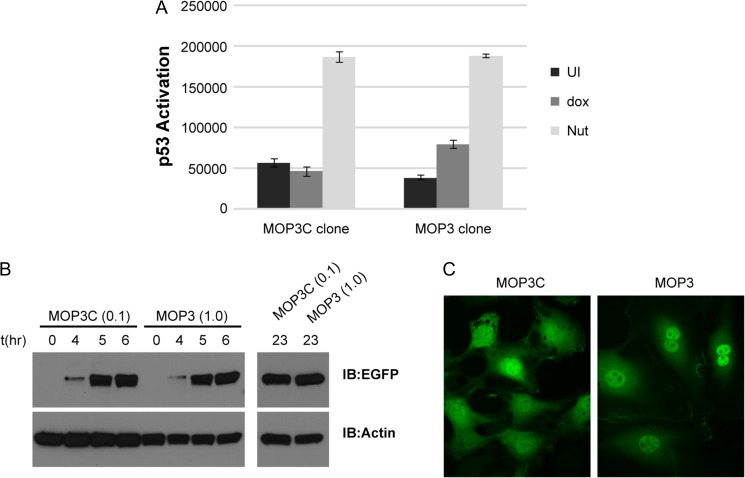
MDM2 and MDMX are predominantly located in the nucleus (Roth et al., 1998), mandating translocation of EGFP-MOP3/MOP3+ across the nuclear pore to effect target engagement. In order to reliably determine the cellular localization of EGFP-fused MOPs we generated stable T22 reporter cell lines expressing EGFP-MOPs under a doxycycline (dox) inducible promoter. Stable clones were selected via fluorescence assisted cell sorting (FACS) after dox induction. Whilst we succeeded in isolating stable cell lines capable of expressing low levels of EGFP-MOP3 (T22-MOP3), EGFP-MOP3+ cell lines could not be established. This phenotype is likely a result of negative selection pressure, commensurate with the relative potencies of MOP3/3+ in activating pro-apoptotic p53. In agreement, multiple cell lines were generated that readily expressed inactive EGFP-MOP3C (T22-MOP3C).
Consistent with transient transfection data (Fig. 4B), induction of EGFP-MOP3 expression but not EGFP-MOP3C resulted in p53 activation (Fig. 6A). For valid comparison, expression levels of EGFP-MOP3C and EGFP-MOP3 in the stable cell lines were equilibrated by adjusting dox concentrations (Fig. 6B). A 10-fold lower concentration of dox was required to induce the same level of EGFP-MOP3C as EGFP-MOP3. Live cell spinning disk confocal fluorescence microscopy was used to image stable T22 clones. As postulated, EGFP-MOP3 exhibited a predominantly nuclear localization, whereas EGFP-MOP3C that does not bind MDM2 or MDMX was evenly distributed throughout the cell (Fig. 6C). These results clearly showed that EGFP-MOP3 could engage its targets in the nucleus, leading to stabilization and activation of p53.
Fig. 6.
Characterization of T22 cell expressing EGFP-MOP3C or EGFP-MOP3. (A) T22 reporter activity of stable clones expressing either EGFP-MOP3C or EGFP-MOP3 after 48 h doxycycline (dox) induction. UI denotes uninduced samples. Nut denotes Nutlin induced samples. Data represents mean ± SD (n = 3). (B) Western blot analysis of protein levels after the indicated time of induction with dox. Concentrations of dox used for the respective EGFP-MOP3C or EGFP-MOP3 clones in μg/mL were indicated in parentheses ‘()’. (C) Live cell imaging of T22 clones after 5 h induction with dox using spinning disk confocal fluorescence microscope. Four z-stacks were taken. Each panel is a projection of sum slices of the z-stacks (ImageJ).
Enhancement of MOP3/MOP3+ activity by facilitated diffusion
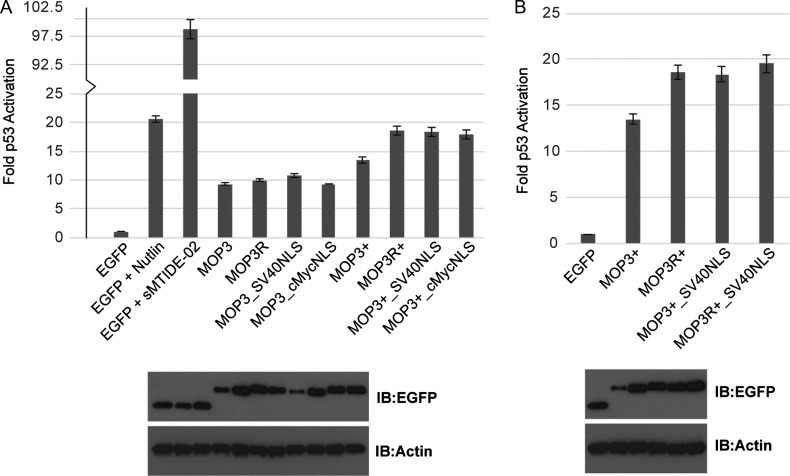
p53 activation by either stably expressed EGFP-MOP3 (Fig. 6A) or ectopically expressed protein (Fig. 4B) was notably reduced when compared to the small molecule MDM2 inhibitor Nutlin or the sMTIDE-02 stapled peptide. As the extent of p53 activation by Nutlins and stapled peptides is dose-dependent (Vassilev et al., 2004; Bernal et al., 2010; Brown et al., 2013), we postulated that increases in local concentration of MOP3 to MDM2/X would significantly boost p53 activation. We therefore generated EGFP-MOP3/MOP3+ constructs incorporating the nuclear localization signal (NLS) from either the SV40 large T antigen (SV40NLS; PKKKRKV) or c-Myc protein (cMycNLS; PAAKRVKLD). Inclusion of either NLS in EGFP-MOP3+ markedly improved p53 activation in T22 cells while only SV40NLS significantly increased in p53 activation in EGFP-MOP3 (Fig. 7A). Examination of expressed protein levels suggested that increased p53 activation corresponded with improved levels of expressed protein. The half-life of a protein is influenced by a combination of the localization of proteasomal degradation activity and its subcellular distribution (Lingbeck et al., 2003; Zheng et al., 2006). Enhanced shuttling of EGFP-MOPs into the nucleus may protect the protein from an active degradation mechanism in the cytoplasm, resulting in improved protein levels. Accordingly, protecting EGFP-MOPs from degradation could likewise increase the available pool of EGFP-MOPs that can gain access to the nucleus and improve p53 activation. We therefore mutated all surface lysines on the 10FN3 scaffold to arginines to reduce potential ubiquitination by bound MDM2 and proteosomal turn-over. This led to increased levels of MOP proteins in T22 cells (Fig. 7, compare levels of MOP3R with MOP3 and MOP3R+ with MOP3+). In the case of MOP3R+, the increased expression correlated with enhanced MDM2/X inhibition and p53 activation. For MOP3+, combination of lysine mutagenesis and addition of NLS signal (MOP3R+_SV40NLS) did not show any synergistic effect on p53 activation (Fig. 7B), most likely due to its expression level remaining the same as MOP3R+ and MOP3+_SV40NLS.
Fig. 7.
Characterization of MOP constructs. (A) Top: T22 reporter activity of transiently transfected cells expressing the indicated EGFP-fused MOP constructs. Bottom: Representative Western blot image of the expression levels of EGFP-fused MOP constructs (same order as in graph) in T22 and the loading control protein β-actin. (B) T22 reporter activity of EGFP-fused MOP3+ constructs as in (A).
MOP3 as a tool to measure pharmacological inhibition of MDM2
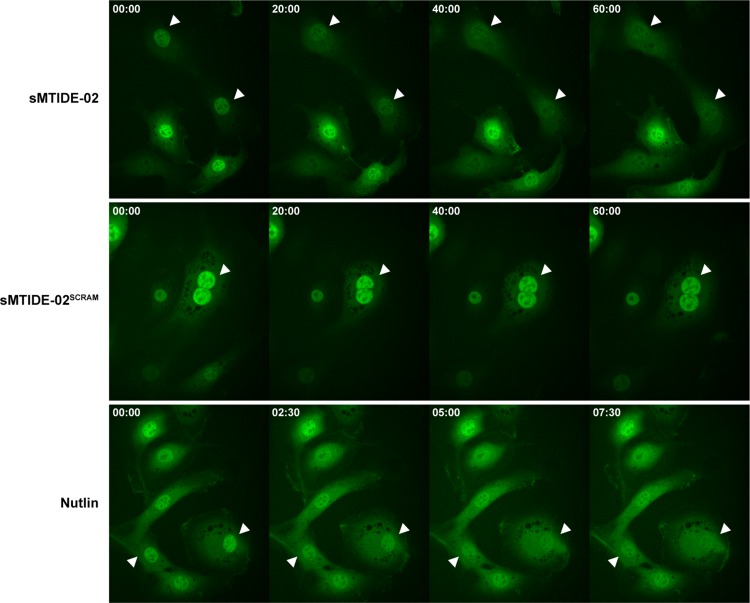
The enriched localization of EGFP-MOP3 in the nucleus via binding to its targets MDM2 and MDMX suggested that treatment with competitive inhibitors could free EGFP-MOP3 to re-equilibrate throughout the cells. We induced expression of EGFP-MOP3 in the stable cell lines and examined the localization of EGFP-MOP3 after treatment with sMTIDE-02, Nutlin or negative control stapled peptide sMTIDE-02SCRAM (Thean et al., 2017) by timelapse confocal microscopy (Fig. 8). We observed redistribution of EGFP-MOP3 within 10 min of Nutlin treatment, with sMTIDE-02 taking longer (~40 min) to achieve the same result (Fig. 8). Treatment with sMTIDE-02SCRAM did not result in equilibration of EGFP-MOP3 localization. Similar timeframes have been previously reported for the dissociation of nuclear RFP-HDM2 and GFP-p53 by Nutlin and sMTIDE-02 using the Fluorescent-2-Hybrid assay (Wei et al., 2013b).
Fig. 8.
Live cell spinning disk confocal fluorescence microscopy of stable T22 expressing EGFP-MOP3 in the presence of other MDM2 inhibitors. After 5 h of doxycycline induction, sMTIDE-02 (25 μM), sMTIDE-02SCRAM (25 μM) or Nutlin (10 μM) were added. Four z-stacks were immediately taken at time = 0 and every 2.5 min thereafter. Selected time points for the respective treatments were shown as indicated at the upper left corner of each panel in min:s. Each panel is a projection of sum slices of the z-stacks (ImageJ). White arrowheads were added to aid in visualization of selected cells through the time course.
Discussion
We have successfully generated highly specific 10FN3-based inhibitors against the p53–MDM2/X interaction. MOP3 contains a copy of the dual peptide inhibitor, M2, grafted onto the FG loop of 10FN3. The FG loop along with BC and DE loops of 10FN3 were widely explored for generating binders to new targets compared to AB, CD and EF loops on the opposite pole of 10FN3. Natural CD loops found in other FN3 domains, a common protein motif found in 2% of all human proteins (Bloom and Calabro, 2009), are involved in binding interactions (Somers et al., 1994; Wang et al., 2005), suggesting that the CD loop can be utilized for introducing additional binders. Bi- or multivalency, present in antibodies, nanobodies or protein binders can confer increases in avidity and potency (Zhang et al., 2004; Coppieters et al., 2006; Roovers et al., 2007; Jähnichen et al., 2010; Emanuel et al., 2011). This typically involves fusion of single domain binders to an oligomerization domain or linkage of separate single chain domains. The distinctive structural feature of 10FN3, presenting loops on opposite poles suggested that design of a bivalent molecule within a single domain format was possible. This was successfully demonstrated in the MOP3+ construct carrying two copies of M2 that showed significant improvements in binding affinity (Supplementary Fig. S6.) and p53 activation (Fig. 4B). To our knowledge, this is the first single domain bivalent 10FN3 binder reported. In a bivalent binder such as MOP3+, binding of a first scaffolded M2 peptide to MDM2 forces the second M2 to be in close proximity, effectively increasing its local concentration and target residence time (Vauquelin and Charlton, 2013). Multivalent ligands typically designed for targeting cell surface or multimeric antigens show improvements in avidity and slower dissociation rates (Holliger and Hudson, 2005). MDM2, which forms homo- and hetero-oligomers with MDMX (Sharp et al., 1999; Tanimura et al., 1999; Leslie et al., 2015), likely contributed to multimeric interactions with MOP3+.
Purified MOP3 bound MDM2 N-terminal domain with an apparent Kd similar to the affinity of the sMTIDE-02 stapled peptide (Brown et al., 2013). However, p53 activation by either stably expressed EGFP-MOP3 (Fig. 6A) or ectopically expressed protein (Fig. 4B) was notably reduced when compared to sMTIDE-02. One contributing factor to the observed differences between EGFP-MOP3 and sMTIDE-02 despite having similar binding affinities to MDM2 (Fig. 4B) may be presence of a nuclear barrier. Despite having access to the nucleus, EGFP-MOP3 is ~20-fold bigger in size compared to sMTIDE-02 (1.5 kDa). Molecules with larger molecular mass cross the NPC at a slower diffusion rate and have to overcome a larger free-energy barrier (ΔGpassive) (Timney et al., 2016), likely resulting in lower effective concentration of expressed EGFP-MOP3 compared to sMTIDE-02. As sMTIDE-02 was added exogenously, its actual concentration in cells is difficult to determine. Even though we cannot rule out the possibility that steric hindrances from EGFP can affect MOP3 binding in our mammalian cell assays, we postulated that increases in local concentration of MOP3 to MDM2/X would significantly boost p53 activation. Modifications aimed at improving access to the nuclear compartment or stability of the MOP proteins enhanced potency, suggesting that p53 activation may be delimited by their effective nuclear concentrations. Size may be an important factor to consider when designing a ligand that has to compete for limited space in the nucleus. In this regard, the compact structure of 10FN3 with two large interfaces on opposite poles allows the design of bivalent or bispecific binders in a single domain without large increments in size.
MDM2 and MDMX are predominantly located in the nucleus (Roth et al., 1998). The EGFP-MOPs (~40 kDa) are well within the diffusion limit of the nuclear pore complex (NPC) and thus should freely equilibrate across the nucleus and cytoplasm via passive diffusion (Wang and Brattain, 2007; Wühr et al., 2015). Retention in a single compartment would indicate formation of larger molecular complexes (>100 kDa) (Wühr et al., 2015). Binding of EGFP-MOP3/MOP3+ to MDM2 (55.2 kDa) or MDMX (54.9 kDa) (monomer or dimer/heterodimer) would result in larger complexes (~100–150 kDa) that would most likely be retained in the nucleus. Consistent with this, EGFP-MOP3 exhibited predominantly nuclear localization in confocal microscopy studies (Fig. 6C). Disruption of EGFP-MOP3 binding to MDM2/X should permit re-equilibration across the nuclear membrane. In agreement, target engagement was reversible upon treatment with competitive MDM2/X inhibitors, as evidenced by increased cytoplasmic localization of EGFP-MOP3. The real-time observation of small molecule and stapled peptide engagement with MDM2/X afforded by the EGFP-MOP3 stable cell line indicates significant potential in drug screening utilizing plate/microscopy/FACS-based high-content imaging platforms (Starkuviene and Pepperkok, 2007; Dudgeon et al., 2010; Black et al., 2011). We also foresee use in the study of p53 pathways, with tractable 10FN3-based MDM2/X inhibition providing an alternative to pharmaceutical abrogation or siRNA-based knockdown.
The MOP proteins represent well-characterized alternatives to conventional antibodies and are in keeping with recent calls for standardized recombinant affinity reagents (Bradbury and Plückthun, 2015). On-target engagement was demonstrated by in vitro/ex vivo immunoprecipitation, cell-based reporter assays and live-cell imaging studies.
In summary, we have developed and stringently validated a robust panel of MDM2/X interacting 10FN3-based inhibitors, along with appropriate controls and companion inducible cell lines. We foresee multiple applications of this toolbox in areas of basic research, diagnostics and drug-development.
Supplementary Material
Acknowledgements
The authors would like to thank the Singapore Immunology Network (SigN) FACS facility for assistance with FACS sorting and IMB Microscopy Unit for training and technical support with data acquisition and analysis.
Funding
This work was supported by the National Research Foundation [NRF2013-THE001-070]; Institute of Molecular and Cell Biology at the Agency for Science, Technology and Research [core fund]; Biomedical Research Council at the Agency for Science, Technology and Research [Young Investigator Grant YIG 2015]; and the National Medical Research Council MS-CETSA platform grant [MOHIAFCAT2/004/2015 to R.M.S.].
References
- Banerjee R., Basu G., Chène P. and Roy S. (2002) J. Pept. Res., 60, 88–94. [DOI] [PubMed] [Google Scholar]
- Barak Y., Juven T., Haffner R. and Oren M. (1993) EMBO J., 12, 461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batori V., Koide A. and Koide S. (2003) Protein Eng., 15, 1015–1020. [DOI] [PubMed] [Google Scholar]
- Berkson R.G., Hollick J.J., Westwood N.J., Woods J.A., Lane D.P. and Lain S. (2005) Int. J. Cancer, 115, 701–710. [DOI] [PubMed] [Google Scholar]
- Bernal F., Tyler A.F., Korsmeyer S.J., Walensky L.D. and Verdine G.L. (2007) J. Am. Chem. Soc., 129, 2456–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal F., Wade M., Godes M., Davis T.N., Whitehead D.G., Kung A.L., Wahl G.M. and Walensky L.D. (2010) Cancer Cell, 18, 411–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binz H.K. and Plückthun A. (2005) Curr. Opin. Biotechnol., 16, 459–469. [DOI] [PubMed] [Google Scholar]
- Black C.B., Duensing T.D., Trinkle L.S. and Dunlay R.T. (2011) Assay Drug Dev. Technol., 9, 13–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom L. and Calabro V. (2009) Drug Discov. Today, 14, 949–955. [DOI] [PubMed] [Google Scholar]
- Bradbury A. and Plückthun A. (2015) Nature, 518, 27–29. [DOI] [PubMed] [Google Scholar]
- Brown C.J., Quah S.T., Jong J. et al. (2013) ACS Chem. Biol., 8, 506–512. [DOI] [PubMed] [Google Scholar]
- Chen J., Lin J. and Levine A.J. (1995) Mol. Med., 1, 142–152. [PMC free article] [PubMed] [Google Scholar]
- Coppieters K., Dreier T., Silence K. et al. (2006) Arthritis Rheum., 54, 1856–1866. [DOI] [PubMed] [Google Scholar]
- Czerwińska P., Mazurek S. and Wiznerowicz M. (2017) J. Biomed. Sci., 24, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M.S., Zeng S.X., Jin Y., Sun X.X., David L. and Lu H. (2004) Mol. Cell. Biol., 24, 7654–7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudgeon D.D., Shinde S., Hua Y. et al. (2010) J. Biomol. Screen., 15, 766–782. [DOI] [PubMed] [Google Scholar]
- Eden E., Navon R., Steinfeld I., Lipson D. and Yakhini Z. (2009) BMC Bioinformatics, 10, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElSawy K., Verma C.S., Joseph T.L., Lane D.P., Twarock R. and Caves L. (2014) Cell Cycle, 12, 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuel S.L., Engle L.J., Chao G. et al. (2011) MAbs., 3, 38–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Echeverria C., Chène P., Blommers M.J. and Furet P. (2000) J. Med. Chem., 43, 3205–3208. [DOI] [PubMed] [Google Scholar]
- Hackel B.J., Kapila A. and Wittrup K.D. (2008) J. Mol. Biol., 381, 1238–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X., Cinay G.E., Zhao Y., Guo Y., Zhang X. and Wang P. (2017) Mol. Ther., 25, 2466–2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haupt Y., Maya R., Kazaz A. and Oren M. (1997) Nature, 387, 296–299. [DOI] [PubMed] [Google Scholar]
- Holliger P. and Hudson P.J. (2005) Nat. Biotechnol., 23, 1126–1136. [DOI] [PubMed] [Google Scholar]
- Hollstein M., Sidransky D., Vogelstein B. and Harris C.C. (1991) Science, 253, 49–53. [DOI] [PubMed] [Google Scholar]
- Honda R., Tanaka H. and Yasuda H. (1997) FEBS Lett., 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Hu B., Gilkes D.M. and Chen J. (2007) Cancer Res., 67, 8810–8817. [DOI] [PubMed] [Google Scholar]
- Jähnichen S., Wacker D., Kapoor M. et al. (2010) Proc. Natl. Acad. Sci. USA, 107, 20565–20570.21059953 [Google Scholar]
- Jones S.N., Roe A.E., Donehower L.A. and Bradley A. (1995) Nature, 378, 206–208. [DOI] [PubMed] [Google Scholar]
- Joseph T.L., Madhumalar A., Brown C.J., Lane D.P. and Verma C.S. (2014) Cell Cycle, 9, 1167–1181. [DOI] [PubMed] [Google Scholar]
- Koide A., Abbatiello S., Rothgery L. and Koide S. (2002) Proc. Natl. Acad. Sci. USA, 99, 1253–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide A., Bailey C.W., Huang X. and Koide S. (1998) J. Mol. Biol., 284, 1141–1151. [DOI] [PubMed] [Google Scholar]
- Koide A., Jordan M.R., Horner S.R., Batori V. and Koide S. (2001) Biochemistry, 40, 10326–10333. [DOI] [PubMed] [Google Scholar]
- Kubbutat M.H., Jones S.N. and Vousden K.H. (1997) Nature, 387, 299–303. [DOI] [PubMed] [Google Scholar]
- Kussie P.H., Gorina S., Marechal V., Elenbaas B., Moreau J., Levine A.J. and Pavletich N.P. (1996) Science, 274, 948–953. [DOI] [PubMed] [Google Scholar]
- Lain S., Hollick J.J., Campbell J. et al. (2008) Cancer Cell, 13, 454–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.P. (1992) Nature, 358, 15–16. [DOI] [PubMed] [Google Scholar]
- Leslie P.L., Ke H. and Zhang Y. (2015) J. Biol. Chem., 290, 12941–12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares L.K., Hengstermann A., Ciechanover A., Müller S. and Scheffner M. (2003) Proc. Natl. Acad. Sci. USA, 100, 12009–12014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingbeck J.M., Trausch-Azar J.S., Ciechanover A. and Schwartz A.L. (2003) J. Biol. Chem., 278, 1817–1823. [DOI] [PubMed] [Google Scholar]
- Lipovšek D. (2010) Protein Eng. Des. Sel., 24, 3–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipovšek D., Lippow S.M., Hackel B.J., Gregson M.W., Cheng P., Kapila A. and Wittrup K.D. (2007) J. Mol. Biol., 368, 1024–1041. [DOI] [PubMed] [Google Scholar]
- Lohrum M.A.E., Ludwig R.L., Kubbutat M.H.G., Hanlon M. and Vousden K.H. (2003) Cancer Cell, 3, 577–587. [DOI] [PubMed] [Google Scholar]
- Lu X., Burbidge S.A., Griffin S. and Smith H.M. (1996) Oncogene, 13, 413–418. [PubMed] [Google Scholar]
- Marechal V., Elenbaas B., Piette J., Nicolas J.C. and Levine A.J. (1994) Mol. Cell. Biol., 14, 7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin H.L., Bedford R., Heseltine S.J., Tang A.A., Haza K.Z., Rao A., McPherson M.J. and Tomlinson D.C. (2018) N. Biotechnol. pii: S1871-6784(17)30541-1. doi: 10.1016/j.nbt.2018.02.008. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Momand J., Zambetti G.P., Olson D.C., George D. and Levine A.J. (1992) Cell, 69, 1237–1245. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R., Wagner D.S. and Lozano G. (1995) Nature, 378, 203–206. [DOI] [PubMed] [Google Scholar]
- Pazgier M., Liu M., Zou G. et al. (2009) Proc. Natl. Acad. Sci. USA, 106, 4665–4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porebski B.T., Conroy P.J., Drinkwater N. et al. (2016) Protein Eng. Des. Sel., 29, 541–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverdatto S., Burz D.S. and Shekhtman A. (2015) Curr. Top. Med. Chem., 15, 1082–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roovers R.C., Laeremans T., Huang L., De Taeye S., Verkleij A.J., Revets H., De Haard H.J. and van Bergen en Henegouwen P.M.P. (2007) Cancer Immunol. Immunother., 56, 303–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J., Dobbelstein M., Freedman D.A., Shenk T. and Levine A.J. (1998) EMBO J., 17, 554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha F., Gencer E.B., Georgeon S., Koide A., Yasui N., Koide S. and Hantschel O. (2013) Proc. Natl. Acad. Sci. USA, 110, 14924–14929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.A., Kratowicz S.A., Sank M.J. and George D.L. (1999) J. Biol. Chem., 274, 38189–38196. [DOI] [PubMed] [Google Scholar]
- Shvarts A., Steegenga W.T., Riteco N. et al. (1996) EMBO J., 15, 5349–5357. [PMC free article] [PubMed] [Google Scholar]
- Simeon R. and Chen Z. (2018) Protein Cell, 9, 3–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers W., Ultsch M., De Vos A.M. and Kossiakoff A.A. (1994) Nature, 372, 478–481. [DOI] [PubMed] [Google Scholar]
- Starkuviene V. and Pepperkok R. (2007) Br. J. Pharmacol., 152, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X.X., DeVine T., Challagundla K.B. and Dai M.S. (2011) J. Biol. Chem., 286, 22730–22741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimura S., Ohtsuka S., Mitsui K., Shirouzu K., Yoshimura A. and Ohtsubo M. (1999) FEBS Lett., 447, 5–9. [DOI] [PubMed] [Google Scholar]
- Thean D., Ebo J.S., Luxton T., Lee X.C., Yuen T.Y., Ferrer F.J., Johannes C.W., Lane D.P. and Brown C.J. (2017) Sci Rep., 7, 1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiede C., Bedford R., Heseltine S.J. et al. (2017) Elife, 6, 515. [Google Scholar]
- Timney B.L., Raveh B., Mironska R., Trivedi J.M., Kim S.J., Russel D., Wente S.R., Sali A. and Rout M.P. (2016) J. Cell Biol., 215, 57–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B. et al. (2004) Science, 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Vauquelin G. and Charlton S.J. (2013) Br. J. Pharmacol., 168, 1771–1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Lombardi R., Phan T.G., Zimmermann C., Lowe D., Jermutus L. and Christ D. (2015) Drug Discov. Today, 20, 1271–1283. [DOI] [PubMed] [Google Scholar]
- Vijaya Chandra S.H., Makhija H., Peter S. et al. (2016) Nucleic Acids Res., 44, e55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Lane D. and Levine A.J. (2000) Nature, 408, 307–310. [DOI] [PubMed] [Google Scholar]
- Wade M., Li Y.C. and Wahl G.M. (2013) Nat. Rev. Cancer, 13, 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R. and Brattain M.G. (2007) FEBS Lett., 581, 3164–3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Rickert M. and Garcia K.C. (2005) Science, 310, 1159–1163. [DOI] [PubMed] [Google Scholar]
- Wei S.J., Joseph T., Chee S. et al. (2013. a) PLoS One, 8, e81068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei S.J., Joseph T., Sim A.Y.L., Yurlova L., Zolghadr K., Lane D., Verma C. and Ghadessy F. (2013. b) PLoS One, 8, e62564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidle U.H., Auer J., Brinkmann U., Georges G. and Tiefenthaler G. (2013) Cancer Genomics Proteomics, 10, 155–168. [PubMed] [Google Scholar]
- Wu L. and Levine A.J. (1997) Mol. Med., 3, 441–451. [PMC free article] [PubMed] [Google Scholar]
- Wühr M., Güttler T., Peshkin L. et al. (2015) Curr. Biol., 25, 2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Aha P., Gu K. et al. (2002) Chem. Biol., 9, 933–942. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tanha J., Hirama T., Khieu N.H., To R., Tong-Sevinc H., Stone E., Brisson J.R. and MacKenzie C.R. (2004) J. Mol. Biol., 335, 49–56. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Wolf G.W., Bhat K., Jin A., Allio T., Burkhart W.A. and Xiong Y. (2003) Mol. Cell. Biol., 23, 8902–8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Ruas J.L., Cao R., Salomons F.A., Cao Y., Poellinger L. and Pereira T. (2006) Mol. Cell. Biol., 26, 4628–4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.