Abstract
Technologies for the production of bispecific antibodies need to overcome two major challenges. The first one is correct heavy chain assembly, which was solved by knobs-into-holes technology or charge interactions in the CH3 domains. The second challenge is correct light chain assembly. This can be solved by engineering the Fab-arm interfaces or applying the immunoglobulin domain crossover approach. There are three different crossovers possible, namely Fab-arm, constant domain and variable domain crossovers. The CrossMabCH1–CL exchange does not lead to the formation of unexpected side products, whereas the CrossMabFab and the CrossMabVH–VL formats result in the formation of typical side products. Thus, CrossMabCH1–CL was initially favored for therapeutic antibody development. Here, we report a novel improved CrossMab design principle making use of site-specific positional exchanges of charged amino acid pairs in the constant domain of these CrossMabs to enable the correct light chain assembly in the CrossMabVH–VL and improvements for the CrossMabFab design.
Keywords: Angiopoietin-2, bispecific antibody, CrossMab, VEGF
Introduction
The idea of bispecific antibodies and their potential to efficiently treat diseases attracted researchers almost since the invention of monoclonal antibodies. The first approvals of the bispecific antibodies like catumaxomab (Chelius et al., 2010), blinatumomab (Nagorsen et al., 2012), emicizumab (Kitazawa et al., 2017; Oldenburg et al., 2017) and various bispecific molecules in advanced clinical development (Brinkmann and Kontermann, 2017; Verdino et al., 2018), underline the maturity of this field of research. The underlying technologies to design and manufacture bispecific antibodies are manifold and have enabled industrial scale production (Spiess et al., 2015; Brinkmann and Kontermann, 2017; Krah et al., 2017; Verdino et al., 2018). This is reflected in the increasing number of first in man trials with this class of molecules (Brinkmann and Kontermann, 2017; Mullard, 2017).
In 2011, we reported the principle of domain crossover IgG-format antibodies as a generic principle to produce bispecific antibodies (Schaefer et al., 2011; Klein et al., 2012). The principle is based on the domain exchange/positional exchange of one complete Fab (Fragment antigen binding) arm, or individual domains within one of the Fab binding arms. These simple positional exchanges aimed to exclude the possibility of light chain mispairing by an alteration of the symmetry of the bispecific antibody. There are three general possibilities for the domain crossover: The most obvious exchange is the translocation of the light chain in the former position of the heavy chain Fab on one side of the antibody, termed CrossMabFab. Alternatively, the variable domains (variable heavy: VH, variable light: VL) or the constant domains (constant heavy 1: CH1, constant light: CL) can be exchanged resulting in the CrossMabVH–VL or CrossMabCH1–CL, respectively. For these two CrossMab versions the elbow region differs from the natural amino acid sequence (Schaefer et al., 2011; Klein et al., 2012). Consequently, it is required to design the new elbow region as its length differs on the light and heavy side of the originating Fab. There are many elbow sequences possible and optimized elbow sequences were recently published (see also Table I) (Klein et al., 2016). Typically, the crossover principle is independent of the elbow sequences, while alterations in these sequences may sometimes impact stability and the light chain pairing selectivity. Our experimental data from all three CrossMab designs indicate that the recommended sequences show a beneficial side product profile and less non-cognate light chain pairings than a bispecific antibody simply using knobs-into-holes and two unmodified light chains (Schaefer et al., 2011, 2016). Nevertheless, based on the theoretical domain interactions only for the CrossMabCH1–CL, no relevant amount of non-cognate light chain pairings can be expected. Consequently, this CrossMab design has already been used for currently four clinical stage bispecific CrossMabs of 1 + 1, 2 + 1 and 2 + 2 valencies (Kienast et al., 2013; Brunker et al., 2016; Lehmann et al., 2016; Regula et al., 2016, 2017; Bacac et al., 2016a, 2016b) and also been applied by academic groups for the development of bispecific molecules (Klein et al., 2016). Since the first conceptual CrossMab design experiments we tried to understand the driving forces for correct light chain assembly and the root cause of side product formation to enable efficient use of the Fab-arm exchange and the variable domain exchange as an alternative to the CH1–CL crossover. The main side-product of the CrossMabFab is a non-functional one-armed monovalent antibody (MoAb), which forms by heterodimerization of the two different heavy chains. For the CrossMabVH–VL, a Bence-Jones like product can be assembled by adding the same non-crossed light chain twice to the formed heavy chain heterodimer (Klein et al., 2016), resulting in both cases above in a non-cognate light chain assembly. The understanding of the root-cause of their assembly enables the improvements based on rational design.
Table I.
Typical and recommended elbow sequences for kappa and lambda domain cross-overs
| Wildtype elbow sequences | |
| VH-CH1 | LVTVSSASTKGPSV |
| Vκ-Cκ | KVEIK-RTVAAPSV |
| Vλ-Cλ | KVTVLGQPKAAPSV |
| Kappa crossover | |
| Vκ-CH1 | KVEIK-SSAS-TKGPSV |
| VH-Ck | LVVTV-SSAS-VAAPSV |
| Vκ-CH1 | KVESK-SSAS-TKGPSV |
| VH-Ck | LVVTV-SSRT-VAAPSV |
| Lambda crossover | |
| Vλ-CH1 | KVTVL-SSAS-TKGPSV |
| VH-Cλ | LVVTV-SGQP-KAAPSV |
The introduction or the modification of electrostatic or hydrophobic interactions, as well as steric hindrance is key elements in the modern protein engineering toolbox. These modifications can be applied to the variable and constant domains. The most well-known approach is the knobs-into-holes modification which ensures correct heavy chain heterodimers in bispecific antibodies (Merchant et al., 1998). Igawa et al. reported the improved expression of a single chain diabody directed against the thrombopoietin receptor by the mutation of glutamine at VH(39) and VL(38) into VH(39E) and VL(38 K) in one scFv and vice versa in the second one to ensure the diabody assembly and not the tandem scFv conformation (Igawa et al., 2010). Liu et al. reported the modification of the VH-CH1 and VL-CK interface with opposite charges in each Fab arm (Liu et al., 2015). The combination of computational and rational design helped Lewis et al. to develop orthogonal Fab interfaces to express correctly assembling bispecific antibodies with four individual chains (Lewis et al., 2014). As a matter of fact, today, there are many different solutions to generate heterodimeric bispecific antibodies utilizing the IgG scaffold. Here, we report improvements on the CrossMab technology for the CrossMabFab and VH–VL to prevent non-cognate light chain pairings via inversion of existing charged residue pairs in the constant Fab domains.
Material and Methods
Ang-2-VEGF CrossMabs
The Ang-2-VEGF CrossMabs for this publication were previously described (Schaefer et al., 2011; Fenn et al., 2013). Briefly, CrossMabs were generated by the appropriate domain cross-over in the VEGF binding Fab arm combined with knob-into-holes modification with hole on the VEGF binding heavy chain, whereas the Ang-2 side remained unmodified in the Fab region and contained the knob modification (Kuglstatter et al., 2017). The sequences including highlighted sites of modification were attached in the supplement.
Recombinant expression of CrossMab antibodies
All antibody HC and LC genes were ordered as synthetic gene fragments and cloned via unique restriction sites using standard cloning procedures into separate expression vectors for each chain enabling secretory expression in HEK cells growing in suspension. The KiH mutations described by Carter and colleagues were used (Knob: T366W; Hole: T366S, L368A, and Y407V) (Merchant et al., 1998). In addition, two Cys residues were introduced in the CH3 domains (S354C in the knob chain and Y349C in the hole chain) that form a stabilizing disulfide bridge (Carter, 2001; Kuglstatter et al., 2017). Transfection (1:1:1:1 plasmid ratios) into HEK293-F cells (Invitrogen, 510029) was performed according to the cell supplier’s instructions using Maxiprep (Qiagen, 12163) preparations of the antibody vectors, Opti-MEM I medium (Invitrogen, 31985) 293fectin (Invitrogen, 31985070), and an initial cell density of 1–2 × 106 viable cells/ml in serum free FreeStyle 293 expression medium (Invitrogen, 12338018). Antibody containing cell culture supernatants were harvested after 7 days of cultivation in shake flasks by centrifugation at 14 000 × g for 30 min and filtered through a 0.22 μm sterile filter (Thermo Scientific, 566–0020). The antibodies were purified directly from the supernatant, or the supernatant was stored at −80°C until purification.
Protein purification
Proteins were purified from filtered cell culture supernatants referring to standard protein A protocols. The antibodies were captured by affinity chromatography using HiTrap MabSelect SuRe (GE Healthcare, 11–0034–93) equilibrated with PBS. Elution of antibodies was achieved at pH 3.0 followed by immediate neutralization of the sample. Aggregated protein, or in the case of the CrossMabFab a light chain heterodimer, was separated from monomeric antibodies by size exclusion chromatography (Superdex 200; GE Healthcare, 17–5175–01) in 20 mM histidine, 140 mM NaCl, pH 6.0. Monomeric antibody fractions were pooled, concentrated if required using a 30 kDa molecular weight cut-off Millipore Amicon Ultra (Millipore, UFC803096) centrifugal concentrator, and stored at −80°C.
Protein concentration determination
The protein concentration was determined by the optical density (OD) at 280 nm, using the molar extinction coefficient calculated on the basis of the amino acid sequence.
Capillary electrophoresis
Purity and antibody integrity were analyzed by CE-SDS using microfluidic Labchip technology (PerkinElmer, USA). Five microliter of protein solution was prepared for CE-SDS analysis using the HT Protein Express Reagent Kit according to the manufacturer’s instructions and analyzed on a LabChip GXII system using a HT Protein Express Chip. Data were analyzed using the LabChip GX Software.
Aggregation onset temperature/dynamic light scattering
Samples were prepared at 1 mg/ml in 20 mM Histidine chloride, 140 mM NaCl, pH 6.0. In a 384-well plate, 40 μl sample was filtered through a 0.4 μm filter, overlaid with 20 μl of paraffin oil and heated in a DynaPro plate reader (Wyatt Inc., Santa Barbara, USA) from 25°C to 80°C at a rate of 0.02°C/min. DLS data were recorded continuously and plotted against the temperature. The aggregation onset temperature (Tagg) is defined as the temperature at which the average hydrodynamic radius begins to increase.
Analytical size exclusion chromatography
Size exclusion chromatography (SEC) for the determination of the aggregation and oligomeric state of antibodies was performed by HPLC chromatography. Briefly, Protein A purified antibodies were applied to a Tosoh TSKgel G3000SW column in 300 mM NaCl, 50 mM KH2PO4/K2HPO4, pH 7.5 on an Dionex Ultimate® system (Thermo Fischer Scientific) or to a Superdex 200 column (GE Healthcare) in 2 × PBS on a Dionex HPLC-System. The eluted protein was quantified by UV absorbance and integration of peak areas. BioRad Gel Filtration Standard 151–1901 served as a standard.
Thermal stability (aggregation onset temperature)
Samples were prepared at 1 mg/ml in 20 mM histidine chloride, 140 mM NaCl, pH 6.0 and transferred in a 9 μl multi-cuvette array. The multi-cuvette array was heated from 35°C to 90°C at a constant rate of 0.1°C/min in an Optim1000 instrument (Avacta Analytical Inc.). The instrument continuously records the intensity of scattered light of a 266 nm laser with a data point approximately every 0.5°C. Light scattering intensities were plotted against the temperature. The aggregation onset temperature (Tagg) is defined as the temperature at which the scattered light intensity begins to increase.
The melting temperature Tm, was assessed by recording the intrinsic Tryptophan fluorescence with an Optim1000 instrument (Avacta Analytical Inc.). Samples were prepared at ~1 mg/ml in 20 mM His, 140 mM NaCl pH 6.0 and transferred to a 9 μl multi-cuvette array. The multi-cuvette array was heated from 30°C to 90°C at a constant rate of 0.1°C/min. The instrument continuously records fluorescence emission spectra after excitation with a 266 nm laser, providing a data point approximately every 0.6°C. The melting temperature, Tm, is determined by plotting the fluorescence intensity against the temperature and Tm is defined as the inflection point in these curves.
UHR-ESI-QTOF mass spectrometry
The correct assembly of the CrossMab antibodies was analyzed by electrospray ionization mass spectrometry of deglycosylated and deglycosylated/limited LysC digested molecules. 100 μg antibody was deglycosylated with N-Glycosidase F (Roche, 11 836 552 001) at 37°C for 16 h in a 100 mM phosphate buffer and subsequently denatured with 2 M Guanidinium-HCl at 37°C for 30 min. The limited LysC (Roche) digestions were performed with 100 μg deglycosylated CrossMabs in a Tris buffer pH 8 at 37°C for 40 min. Subsequently, the samples were desalted by HPLC on a Sephadex G25 column (GE Healthcare, 17–0032–02) using 40% acetonitrile with 2% formic acid (v/v). The total mass was determined by UHR-ESI-QTOF mass spectrometry on a maXis 4 G UHR-QTOF MS system (Bruker Daltonik) equipped with a TriVersa NanoMate source (Advion). Calibration was performed with sodium iodide (Tof G2-Sample Kit 2; Waters). Data acquisition was done at 1000–4000m/z (ISCID: 130.0 eV) and 600–2000m/z (ISCID: 0.0 eV) for the deglycosylated and deglycosylated/limited LysC digested CrossMabs, respectively. The raw mass spectra were evaluated and transformed into individual relative molar masses using an in-house developed software tool. The quantitative evaluation of the mass spectra was performed by summing up contributions of m/z ion intensities of all charge states forming the dominant part (larger than 20%) of the charge state envelope as observed for the most abundant individual product mass. Then all peak contributions (fitted as Gaussians) of all signals in these charge states were used to calculate the relative contents of the individual species.
Functional characterization
Determination of binding and binding affinity of multispecific antibodies to the respective antigens using surface plasmon resonance (SPR) (BIACORE®, GE Healthcare).
Assessment of VEGF binding
Binding of indicated antibodies to human VEGFA-121 was investigated by surface plasmon resonance using a BIACORE® T200 instrument (GE Healthcare). Around 10 000 (RU) of anti His antibody (1 μg/ml anti His antibody; Order Code: 28995056; GE Healthcare Bio-Sciences AB, Sweden) were coupled on a Series S CM5 chip (GE Healthcare BR-1005-30) at pH 5.0 by using an amine coupling kit supplied by the GE Healthcare. HBS-N (10 mM HEPES, 150 mM NaCl pH 7.4, GE Healthcare) was used as running buffer during the immobilization procedure. For the following kinetic characterization, sample and running buffer was PBS-T (10 mM phosphate buffered saline including 0.05% Tween20) at pH 7.4. The flow cell was set to 25°C – and the sample block set to 12°C – and primed with running buffer twice prior to kinetic characterization.
VEGF-A-121-His was captured by injecting a 0.5 μg/ml solution for 30 s at a flow of 5 μl/min. The association was measured by injection of the indicated antibodies in various concentrations in solution for 180 s at a flow of 30 μl/min starting with 1000 nM in 1:3 serial dilutions. The dissociation phase was monitored for up to 600 s and triggered by switching from the sample solution to running buffer. The surface was regenerated by 60 s washing with a Glycine pH 1.5 solution at a flow rate of 30 μl/min. Sensorgrams were double referenced by subtracting both anti His antibody and buffer only responses. For calculation of KD and other kinetic parameters the Langmuir 1:1 model was used.
Assessment of Ang-2 binding
Binding of indicated antibodies to human Ang-2-RBD-Fc was investigated by surface plasmon resonance using a BIACORE® T200 instrument (GE Healthcare). Around 8000 (RU) of goat anti human F(ab’)2 (10 μg/ml anti human F(ab’)2; Order Code: 28958325; GE Healthcare Bio-Sciences AB, Sweden) were coupled on a Series S CM5 chip (GE Healthcare BR-1005-30) at pH 5.0 by using an amine coupling kit supplied by the GE Healthcare. HBS-N (10 mM HEPES, 150 mM NaCl pH 7.4, GE Healthcare) was used as running buffer during the immobilization procedure. For the following kinetic characterization, sample and running buffer was PBS-T (10 mM phosphate buffered saline including 0.05% Tween20) at pH 7.4. The flow cell was set to 25°C – and the sample block set to 12°C – and primed with running buffer twice prior to kinetic characterization.
The bispecific antibody was captured by injecting a 5 nM solution for 25 s at a flow of 5 μl/min. The association was measured by injection of human Ang-2-RBD-Fc at various concentrations in solution for 120 s at a flow of 30 μl/min starting with 100 nM in 1:3 serial dilutions. The dissociation phase was monitored for up to 180 s and triggered by switching from the sample solution to running buffer. The surface was regenerated by 60 s washing with a Glycine pH 2.1 solution at a flow rate of 30 μl/min. Bulk refractive index differences were corrected by subtracting the response obtained from a goat anti human F(ab’)2 surface. Blank injections are also subtracted (= double referencing). For calculation of apparent KD the Langmuir 1:1 model was used.
Crystallization
Prior to crystallization the Fab protein was concentrated to 20–26 mg/ml in a buffer containing 200 mM CHES pH 9.0, 150 mM NaCl. Initial crystallization trials were performed in sitting drop vapor diffusion setups at 21°C. Crystals appeared within 2 days out of 0.1 M MES, pH 6.5, 20% PEG2000 MME after cross-seeding of droplets. Plate shaped crystals grew within 1 week to a final size of 10 × 200 × 300 μm.
Structure determination and refinement
For data collection crystals were flash cooled at 100 K in precipitant solution containing 10% ethylene glycol. Diffraction data were collected at a wavelength of 1.0000 Å using a PILATUS 6 M detector at the beamline X10SA of the Swiss Light Source (Villigen, Switzerland). Data were processed and scaled with XDS (Kabsch, 2010) and SADABS (Bruker AXS Inc, Madison, 2009). The data+ belong to the space group P1 with cell axes of a = 59.56 Å, b = 59.80 Å, c = 79.09 Å. The resolution limit as obtained was 1.88 Å. The structure was determined by molecular replacement with PHASER (McCoy et al., 2007) using coordinates of an in-house Fab fragment structure as search model. The asymmetric unit contains two Fab molecules. Programs from the CCP4 suite (1994) and Buster (Bricogne et al., 2011) were used to subsequently refine the model. Manual rebuilding of the structure using difference electron density was done with COOT (Emsley et al., 2010). Data collection and refinement statistics for both structures are summarized in Supplemental Table II. Coordinates are deposited in RCSB under the PDB ID 6GHG.
Molecular modeling
Based on the crystal structure of the KK–EE (light chain K123, K124, heavy chain E147, K213; positions given in EU numbering) charge variant non-crossed Fab, the following intermediate charge variants were modeled: KQ–EK, KQ–KE, EK–EK and EK–EK. First, the sidechain types were exchanged where necessary. Afterwards, the sidechain conformers of residues at positions 123, 124 (light chain) and 147, 213 (heavy chain) were optimized simultaneously to attain a maximum number of favorable and a minimal number of unfavorable interactions, the latter including steric clashes. Rotamer optimization was performed using the Dunbrack rotamer library (Shapovalov and Dunbrack, 2011), plus the original rotamers as found in the crystal structure. The rotamer-optimized models were used as starting coordinates for unrestrained energy minimization using the CHARMM36 (Huang and MacKerell, 2013) force field in combination with the GBSW implicit solvent model (Im et al., 2003) and the ‘Smart Minimizer’ protocol (steepest descent followed by conjugate gradient (Luenberger, 1973) for 500 integration steps. All molecular modeling and simulation was done with BIOVIA Discovery Studio 4.5 (Dassault Systèmes BIOVIA Discovery Studio 4.5, San Diego).
Results
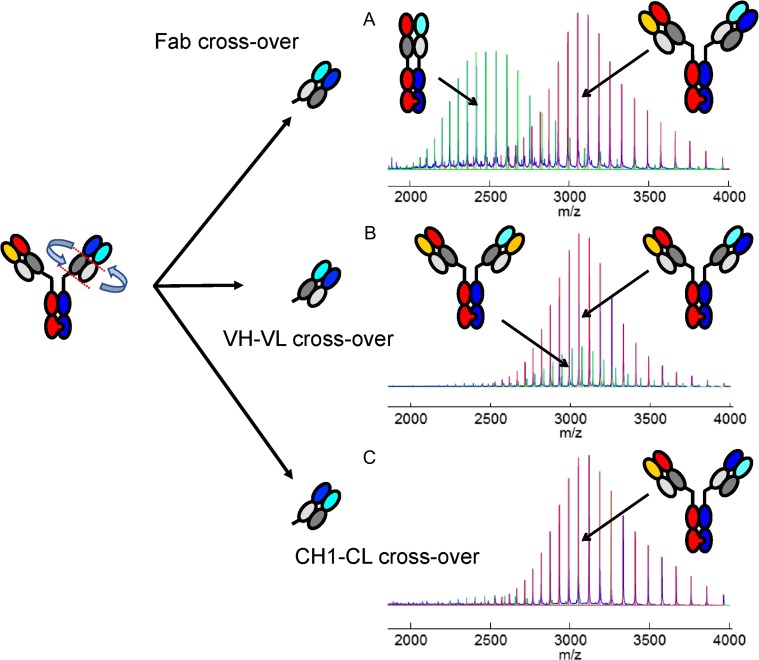
For proof-of-concept, we generated the three possible domain crossover antibodies (as described (Schaefer et al., 2011; Klein et al., 2016) with target specificities directed against Angiopoietin 2 (Ang-2) and Vascular Endothelial Growth Factor (VEGF-A) based on the parental antibodies Ang-2i-LC06 (Thomas et al., 2013; Scheuer et al., 2016) and bevacizumab (Presta et al., 1997; Ferrara et al., 2004) on a human IgG1 framework with knobs-into-holes including an additional stabilizing disulfide bond between the CH3 (constant heavy 3) domains (Merchant et al., 1998; Carter, 2001; Kuglstatter et al., 2017). The domain crossover was introduced into the VEGF binding arm using the described elbow sequence (Klein et al., 2016) together with the knob (T366W) and an additional cysteine (S345C), whereas the hole (T366S, L368A, Y407V) was introduced into the CH3 domain of the Ang-2 binder together with an additional cysteine (Y349C). The antibodies containing the Fab, VH–VL and CH1–CL domain exchange were transiently expressed using four plasmids encoding the respective light chain, the ‘crossed light chain’, the ‘crossed’ knob heavy chain and the hole heavy chain as previously described (Schaefer et al., 2011). The different CrossMabs were purified using Protein A chromatography and preparative SEC and analyzed by CE-SDS, SEC and mass spectrometry after each purification step. All three CrossMabs expressed well and represent the main product/peak as demonstrated in this experiment. The analysis of the side products for the Ang-2-VEGF Fab domain crossover antibody revealed a one-armed monovalent non-functional antibody (knob-hole antibody without light chains; Fig. 1A) and a non-functional Fab (VEGF HC-Fab and Ang-2 LC) which together amounted to 49% (sum). In addition to the correct bispecific antibody, the VH–VL domain crossover antibody expression yielded a side product with twice the non-crossed Ang-2 light chain on both arms of the antibody based on a ‘Bence-Jones like’ non-cognate pairing of the uncrossed light chain with the crossed VEGF heavy chain (VL–VL interaction). This side product occurred in amounts of up to 20% (Fig. 1B). For the CrossMabCH1–CL no non-cognate light chain pairing was observed (Fig. 1C). An observed side product was an antibody lacking the crossed light chain despite the used equimolar ratios of the plasmids encoding the four different chains.
Fig. 1.
Possible domain crossovers to generate bispecific antibodies and the observed side products. A bispecific antibody consists of two light and two heavy chains. The knobs-into-holes technology ensures correct heavy chain assembly. The domain crossover ensures correct light chain assembly. The gray colors indicate the similarity of constant domains in both Fab arms. There are three possibilities for this cross-over, as indicated by the red dotted cutting lines. The CrossMabFab cuts N-terminally of the hinge region and translocates the light blue light chain into the former position of the heavy chain. The CrossMabVH–VL cuts between the constant and variable domains (elbow) and translocates the VH domain into the former position of the VL domain and vice versa. Total mass determination by UHR-ESI-QTOF mass spectrometry of the cross-over bispecific antibody formats (A) CrossMabFab, (B) CrossMabVH–VL, (C) CrossMabCH1–CL with observed major side products. The CrossMabFab and CrossMabVH–VL formats result in a non-functional monovalent antibody, and an antibody with two identical non-crossed light chains (orange/light gray) formed by a Bence-Jones like interaction, respectively. In many cases no side products are observed using the CrossMabCH1–CL format.
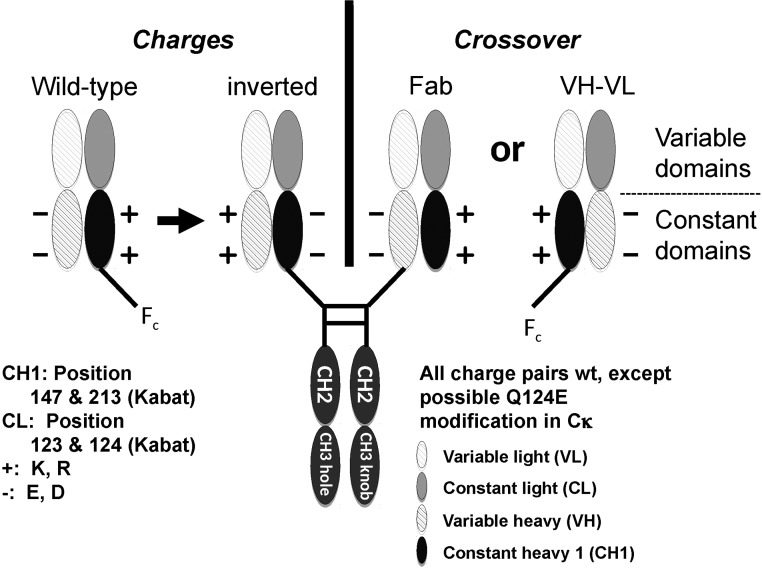
The existence of side products of the CrossMabFab and CrossMabVH–VL suggested a need for further improvements to specificity by making the CH1–CL interactions selective for the knob and hole sides of the antibody. One way to achieve correct chain pairing is by the introduction of charged amino acids which make an interaction attractive in case of opposite charges and repulsive in case of identical charges. Previously, this was approached by introducing novel charged residue pairs (Lewis et al., 2014; Liu et al., 2015) in the CH1–CL interface in suitable positions. The approach described herein uses existing charge pairs and selectively is achieved by inverting the existing charge pairs. For this purpose, we searched for existing pairwise electrostatic interactions between residues in the CH1–CL interface and identified the positions E123 and Q124 for Cκ and E123 and E124 for Cλ in the light chain and K147 and K213 in the CH1 (Fig. 2 and Supplemental Fig. 1). An inversion of these charge pairs on the hole side of the antibody by introducing K or R as positively charged amino acids and E or D as negatively charged amino acid or the introduction of a new charge pair in the case of Q124, respectively (on the knob side) should lead to attractive interactions in case of cognate light chain assembly and to repulsive interactions in case of incorrect assembly. Thus, these modifications are intended to enforce correct light chain assembly.
Fig. 2.
The approach used herein to make the strong CH1–CL interactions selective for each heavy chain or side of the antibody is the inversion of existing salt bridges or opposite charge pairs (inversion of +(K) to –(E)). The selected charge pairs are depicted on the left. The numbering is according to Kabat. The domain cross-over changes the symmetry of one Fab arm and enables a selective fit (right drawing). Inverted charge-pairs in CH1–CL induce attractive and repulsive forces. Both modifications together enforce correct light chain assembly on each side of the bispecific antibody. Additional exchanges (e.g. D instead of E, R instead of K) are listed in Tables II–IV and VI.
Charge exchanged CrossMabVH–VL
To test this hypothesis, the respective Ang-2-VEGF CrossMabVH–VL was generated with the inverted charged pairs on the Ang-2 side (knob): E123→K, Q124→K in the CL domain and K147→E and K213→E in the CH1 domain, whereas the crossed VEGF Fab arm (hole) remained unchanged. Expression yields and the side products were compared with a parallel expression of the Ang-2-VEGF CrossMabCH1–CL without charge exchanges. The yield increased twofold and no side products or missing light chains could be detected (data not shown).
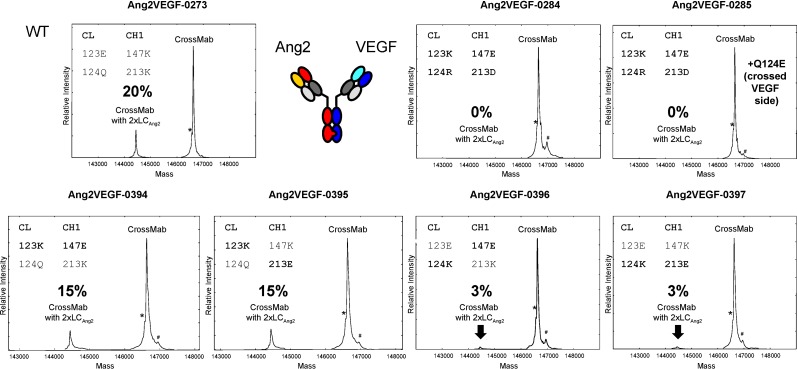
Based on this promising initial result, the next logical step was to assess the influence of each individual charge (Table II). The basis for the evaluation was the CrossMabVH–VL without any charged residue exchanges which shows a ‘Bence-Jones-like’ non-cognate light chain pairing with about 20% (wt Ang-2 light chain in two copies, described above) (Fig. 3, Table II).
Table II.
Overview of Ang-2-VEGF CrossMabVH–VL with 0, 1 and 2 charge pair inversions and their influence on correct light chain assembly
| Molecule # (Ang2-VEGF-) | Ang-2-Fab | Crossed VEGF Fab | ‘Bence Jones like’ non-cognate LC pairings by MS [%] | ||
|---|---|---|---|---|---|
| LC (123/124) | HC (147/213) | LC (123/124) | HC (147/213) | ||
| 0273 (wt) | EQ | KK | EQ | KK | 20 |
| 0394 | KQ | EK | EQ | KK | 15 |
| 0395 | KQ | KE | EQ | KK | 15 |
| 0396 | EK | EK | EQ | KK | 3 |
| 0397 | EK | KE | EQ | KK | 3 |
| 0282 | KR | EE | EE | KK | 0 |
| 0283 (lambda) | KK | EE | EQ | KK | 0 |
| 0284 | KR | ED | EQ | KK | 0 |
| 0285 (Q124E in crossed VEGF Fab) | KR | ED | EE | KK | 0 |
| 0286 | KK | EE | EE | KK | 0 |
Fig. 3.
Side product quantification for the CrossMabVH–VL (Bence-Jones like side product) in dependency of individual charge pair exchanges. The influence of each individual charge exchange, its position and pairing preference can be visualized by comparing and quantifying the MS signals for the cognate light chain assembly and the Bence-Jones like assembly, i.e. two Ang2 light chains (% in bold). The charge pair inversion is always on the wt Fab arm. For Ang2VEGF-0285 two charge pairs were exchanged on the Ang2 arm and one additional charge (Q124E) is added on the crossed VEGF arm. *HC wo C-term. glycine; #phosphate adduct.
Data from constructed homology models or available X-ray structures at that time did not provide clear hints to which of the selected positions in the light chain interacts with the corresponding charged residue in the heavy chain (Supplemental Fig. 1). Probing each combination individually and assessing its influence on the side product profile aimed to identify the key interactions and their contributions. The first charge pair to be inverted was the position E123→K in the CL which could potentially form a salt bridge with K147→E or K213→E in the CH1. These modifications reduced the amount of the CrossMab with two Ang-2 light chains from 20% to about 15% as quantified by mass spectrometry (0394 & 0395 in Fig. 3). Modifying position Q124→K in the light chain and probing it with the K147→E and or K213→E mutations reduced the side product amount down to 3% (0396 & 0397 in Fig. 3). The combination of E123→K and Q124→R in the light chain and K147→E and K213→D in the heavy chain reduced the amount of side products below the detection limit by mass spectrometry (0284 & 0285 in Fig. 3). Also, combinations of E123→K and Q/E124→K or R (including lambda), and K147→E and K213→E did not show any side products (Table II). The beneficial impact of the position Q124 in kappa light chains and E124 in lambda light chains suggests modifying position 124 in the crossed VEGF CL domain for kappa light chains with a Q→E exchange. An additional benefit for the Ang-2-VEGF CrossMabVH–VL could not be shown as already no side products occurred in the case of two inverted charge pairs in the wt Ang-2 chain (0285 in Fig. 3; Table II). Nevertheless, there was no loss of selectivity or stability observed with it. Finally, no impact on target binding was observed in the correctly assembled CrossMab as determined by SPR-based binding assays. The affinity towards both targets was not altered within the precision of these measurements. Likewise, there was no significant impact on the thermal stability as measured as Tagg (°C) or Tm (°C) for all generated variants. All CrossMabVH–VL with one or two charge pair inversions and the optional Q124E modification in CL of the crossed Fab arm showed Tagg in a range between 56°C and 57°C and Tm in a range between 61°C and 62°C. The data of the charge pair exchanges and their impact on non-cognate light chain pairing are summarized in Table II and Supplemental Table I.
To demonstrate the general suitability of the newly identified charged residue bispecific CrossMabVH–VL antibodies with different target specificities were evaluated. As a first example, a bispecific antibody targeting the two soluble ligands Tweak (Lammens et al., 2013) and IL-17 (Fischer et al., 2015) for use in inflammatory indications was generated. The initial CrossMabVH–VL showed ~20% ‘Bence Jones like’ non-cognate light chain as quantified by mass spectrometry (Table III). The introduction of two inverted charged residue pairs in position CL E123→R or K and D124→ K and CH1 K147→E and K213→E or D resulted in a correctly assembled bispecific antibody without formation of detectable side products. An additional charge correction in the crossed light chain (Q124E) did not show additional benefits (Table III).
Table III.
Overview on Tweak-IL17 CrossMabVH–VL with 0 and 2 charge pair inversions and its influence on correct light chain assembly
| Molecule # (TweakIL17-) | IL17 Fab | Crossed Tweak Fab | ‘Bence Jones like’ non-cognate LC pairings by MS [%] | ||
|---|---|---|---|---|---|
| LC (123/124) | HC (147/213) | LC (123/124) | HC (147/213) | ||
| 0096 (wt) | ED | KK | EQ | KK | ~20 |
| 0097 (Q124E in crossed Tweak Fab) | RK | EE | EE | KK | 0 |
| 0098 | RK | ED | EQ | KK | 0 |
| 0099 (Q124E in crossed Tweak Fab) | RK | ED | EE | KK | 0 |
| 0100 (Q124E in crossed Tweak Fab) | KK | EE | EE | KK | 0 |
As a second example, a trispecific c-Met/HER1/3 Cross-DAF (DAF = dual action Fab) for potential use in oncology was tested. In this trispecific antibody (Klein et al., 2016) one target binding Fab is directed against c-Met (Merchant et al., 2013) whereas the other is a dual acting Fab capable of binding either HER1 or HER3 (Eigenbrot and Fuh, 2013). The corresponding CrossMabVH–VL showed only trace impurities of a non-cognate light chain, nevertheless, the introduction of the charged residues in one or both Fab arms removed any incorrect light chain assembly (Table IV).
Table IV.
Overview on c-Met-Her1/3 CrossDAFVH–VL with 0, and 2 charge pair inversions and their influence on correct light chain assembly
| Molecule # (MetHER1(3)DAF-) | Her DAF | Crossed cMet Fab | ‘Bence Jones like’ non-cognate LC pairings by MS [%] | ||
|---|---|---|---|---|---|
| LC (123/124) | HC (147/213) | LC (123/124) | HC (147/213) | ||
| 0004 (wt) | ED | KK | ED | KK | Trace amounts |
| 0005 | RK | ED | ED | KK | 0 |
| 0006 (Q124E in crossed cMet Fab) | RK | ED | EE | KK | 0 |
All deglycosylated CrossMabs were analyzed in their intact state by mass spectrometry and were also evaluated following an additional limited digestion with endoprotease LysC to generate Fab fragments. The purpose of this additional evaluation was to exclude the presence of any non-cognate side products with both the crossed and uncrossed LCs misassembled with the HCs within one CrossMab molecule. In that case, the correctly and incorrectly assembled intact CrossMabs would be isobaric. The analysis of all LysC generated Fabs excluded the presence of this kind of side product (data not shown).
Charge exchanged CrossMabFab
The use of the CrossMabFab is impeded by the formation of a heavy chain dimer containing a non-functional binding domain comprised of two variable domains with different target specificity. Reversing the charge interaction of existing charge pairs in the CH1–CL domains as described for the CrossMabVH–VL could improve this design by reducing the amount of HC-dimer. Therefore, we evaluated the influence of two charge-pairs in a kappa–kappa and a lambda–kappa CrossMabFab. While the results of this approach evidently showed a beneficial influence, the overall effect was not big enough to completely eliminate the HC-dimer formation (Table V). The HC-dimer occurred in amounts of 40% in the wt kappa–kappa CrossMabFab and could be reduced to about 20% as measured by CE-SDS under non-reducing conditions. By introduction of two inverted charge pairs in the non-crossed CH1–CL interface, the amount of this side product could be reduced to 14% (Table V). Taken together, the overall effect on the formation of the correct CrossMabFab products was not big enough to eliminate the undesired heavy chain dimer side product.
Table V.
Overview of Ang-2-VEGF CrossMabFab with 0 and 2 charge pair inversions and their influence on correct light chain assembly
| Molecule # (Ang2VEGF-) | Ang-2-Fab | Crossed VEGF Fab | ‘HC-dimer’ formation by CE-SDS [%] | ||
|---|---|---|---|---|---|
| LC (123/124) | HC (147/213) | LC (123/124) | HC (147/213) | ||
| 289 (wt) | EQ | KK | EQ | KK | 40 |
| 280 | KK | EE | EQ | KK | 27 |
| 111 (Q124E in crossed VEGF Fab) | KK | EE | EE | KK | 19 |
| 290 (lambda, wt) | EE | KK | EE | KK | 30 |
| 279 (lambda) | KK | EE | EE | KK | 14 |
X-ray structure of the KK–EE charge variant non-crossed Fab and modeling of intermediate charge variants
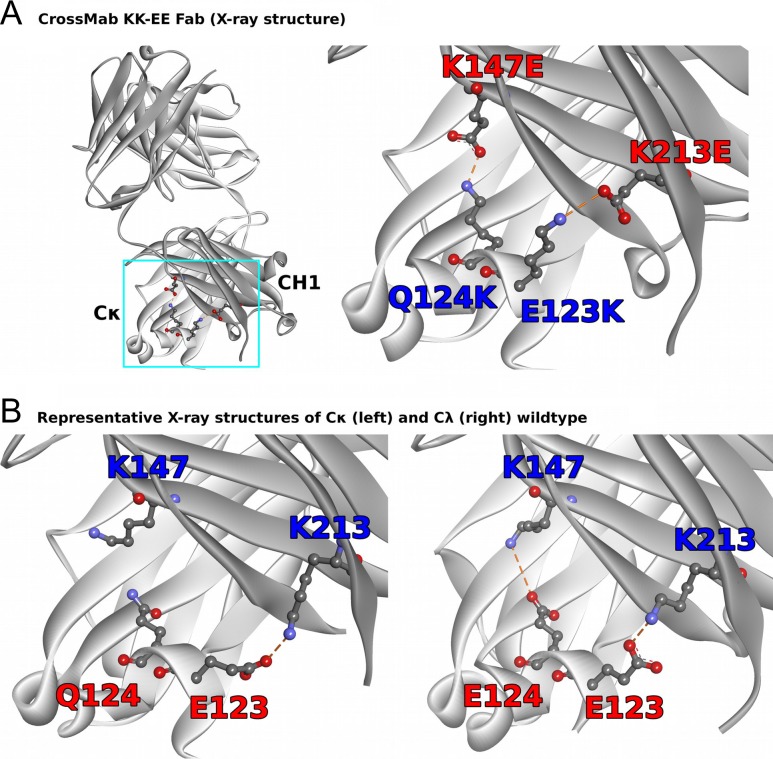
To establish a structure function relationship further and to correlate the experimental results to structural information, the Fab fragment of a non-binding germline antibody with GS motif containing CDR residues in the heavy chain CDR3 termed DP47 with the charged residue inversion KK–EE described above was crystallized and the X-ray structure solved. Fig. 4 shows the respective crystal structure with the mutated charge interactions (Fig. 4A, PDB code 6GHG) in context with two high-resolution CH1–Cκ and CH1–Cλ X-ray structures representing the respective wildtypes (Fig. 4B, left: PDB code 4XAW at 1.47 Å resolution (Irimia et al., 2016); Fig. 4B right: PDB code 4LLD at 1.19 Å resolution (Lewis et al., 2014)).
Fig. 4.
(A) X-ray structure of the DP47 Fab with the inverted charge pairs E123K, Q124K, K147E and K213E (left) and close-up on the mutated residues in the CH1 and Cκ domains (right). (B) Cross-chain interactions involving residues at the same positions in two representative high-resolution X-ray structures of CH1–Cκ wildtype (PDB code 4XAW, left) (Irimia et al., 2016) and CH1–Cλ wildtype (PDB code 4LLD) (Lewis et al., 2014).
To investigate the canonical state of the pairwise interactions between E123 of the light chain and K213 heavy chain and E/Q214 of the light chain and K147 of the heavy chain, we collected a number of publicly available high-resolution X-ray structures of CH1–Cκ (n = 50, resolution ≤1.8 Å) and CH1–Cλ (n = 26, resolution ≤2.0 Å) from the Protein Data Bank and monitored the non-covalent interactions using BIOVIA Discovery Studio. The results are shown in Table VI.
Table VI.
Non-covalent canonical state of the pairwise interactions between E123 of the light chain and K213 heavy chain and E/Q214 of the light chain and K147 of the heavy chain based on publicly available high-resolution X-ray structures of CH1–Cκ (n = 50, resolution ≤1.8 Å) and CH1–Cλ (n = 26, resolution ≤2.0 Å) from the Protein Data Bank
| Residue pair (light chain-heavy chain) | Interaction found (Discovery Studio) | Salt bridge | Attractive charge | |
|---|---|---|---|---|
| CH1–Cκn = 50 | E123–K213 | 46/50 | 29/46 | 17/46 |
| Q124–K147 | 0/50 | – | – | |
| CH1–Cλn = 26 | E123–K213 | 19/26 | 11/26 | 8/26 |
| E124–K147 | 24/26 | 7/24 | 17/24 |
In the case of CH1–Cκ, the interaction E123–K213 is a highly conserved salt bridge or (in case of longer distances) attractive charge pair interaction. By contrast, there seems to be no conserved interaction between residues Q124 and K147. In the case of CH1–Cλ, the number of high-resolution X-ray structures is limited, but the available structural data suggests that there are conserved interactions between the pairs E123–K213 and E124–K147, either in the form of salt bridges or longer distance attractive charge pairs.
In the X-ray structure of the CrossMab KK–EE charge variant of the DP47 Fab (PDB ID 6GHG, Fig. 5B) the two pairs 123–213 and 124–147 form salt bridges. This holds for both copies of the Fab that can be found in the asymmetric unit of the crystal structure. The configuration is thus more similar to the two interacting glutamic acid-lysine pairs in the CH1–Cλ wildtype Fab (Fig. 4B right) than to the CH1–Cκ wildtype Fab, where only the E123–K213 salt bridge seems to be conserved (Fig. 5A).
Fig. 5.
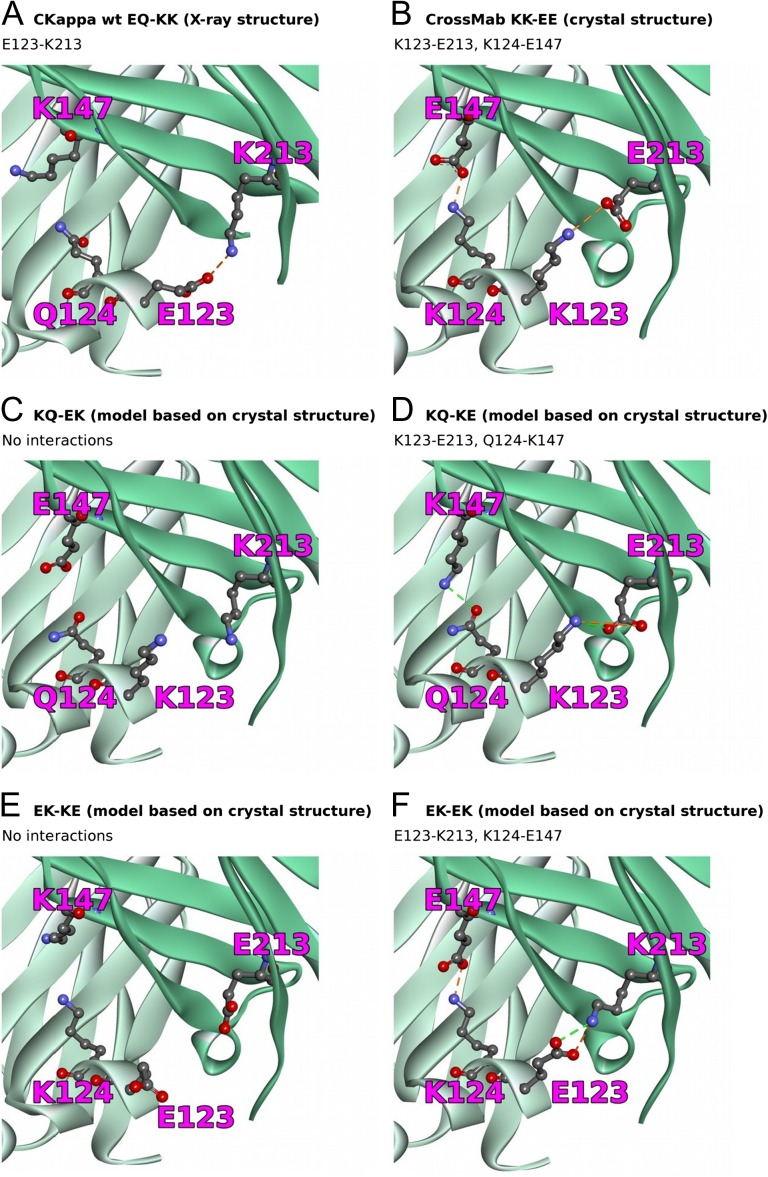
Electrostatic interactions between the residues at light chain positions 123 and 124 and heavy chain positions 147 and 213 in the X-ray structure of the canonically paired CH1–Cκ wildtype Fab (A, PDB code 4XAW) (Irimia et al., 2016) and in the crystal structure of the KK–EE charge variant Fab (B, PDB code 6GHG). The results of the modeling of individual charge-pair interactions (using structure 6GHG as starting configuration) are depicted in panels C–F. The light chain is shown in light green, the heavy chain in dark green. Residue numbers are given in EU numbering.
In comparison to the wildtype, the polarity is reversed and accentuated: The positively charged residues are moved from the heavy chain to the light chain side, while the negatively charged residues are moved from the light chain to the heavy chain side, and in this process are made more negative by replacing the glutamine with glutamic acid (in Cκ). Cκ–CH1 pairing with chains that carry the canonical charge configuration is thus rendered energetically unfavorable. This could apply not only with regard to putative direct interactions between the two residue pairs, but also affect general electrostatic steering during the assembly of the encounter complex.
To evaluate the structural implications of partial charge exchange (only one residue per chain), we modeled the intermediate variants KQ–EK, KQ–KE, EK–KE, and EK–EK (always residue 123, 124–147, 213) as assessed experimentally and studied the potential pairing interactions. These variants are shown in Fig. 5C–F). For the variants KQ–KE and EK–EK, we found favorable pairwise interactions between both 123–213 and 124–147. By contrast, for the variants KQ–EK and EK–KE, our modeling predicts no inter-chain interactions involving the four residues. In the case of EK–KE (Fig. 5E), the obvious reason is that the pairs 123–213 and 124–147 possesses the same charge and hence lead to mutual repulsion. In the case of KQ–EK (Fig. 5C), hydrogen bonding between Q124 and E147 is conceivable; however, the model suggests that the opposing glutamine and glutamic acid side chains are not long enough to form the inter-chain hydrogen bond. Instead, the side chain amine of Q124 prefers to interact with its own backbone carbonyl group. The remaining K123 and K213 pair is repulsive. In all of the cases, we only observed interactions between the residues at positions 123 and 213 and between the residues at positions 124 and 147, and not the hypothetical crosswise interactions 123–147 or 124–213.
Discussion
Human bispecific heterodimeric IgG-format antibodies have paved their way from early research to clinical development and recently with the approval of emicizumab (Kitazawa et al., 2017; Oldenburg et al., 2017). One of the key challenges for large scale pharmaceutical production of similar bispecific antibodies is the correct assembly of all chains. Currently, there are very few technologies available which ensure this prerequisite. The common light chain approach as applied for emicizumab utilizes two heavy and one common light chain to generate the bispecific molecule. This approach is straightforward and works well. Nevertheless, it does not allow the use of existing antibodies pairs, and the identification of a suitable common light chain can be challenging (Sampei et al., 2013). For antibodies consisting of four individual chains the challenge remains to enable correct light chain assembly. Recently, novel orthogonal Fab interfaces were generated by a combined in silico and screening approach (Lewis et al., 2014; Liu et al., 2015; Bonisch et al., 2017). Challenges in case of non-cognate light chain pairings during cell-line and process development may occur as the similarity with regards to the biophysical properties of the two different Fab arms can be very high. Nevertheless, this approach is predicted to work well and the potential challenges maybe circumvented by novel approaches in cell-line development like targeted integration. A common and well-established work around is a sophisticated and tailored purification strategy to enable pharmaceutical grade quality even for non-completely self-assembling bispecific antibodies (Smith et al., 2015).
We chose a novel strategy combining the CrossMab technology with selected charge pairs in order to disable the formation of undesired side products for the VH–VL and the Fab arm crossover (Schaefer et al., 2015; Imhof-Jung et al., 2016). For the CrossMabCH1–CL non-cognate light chain pairing does not occur. The challenge can be the weaker expression of the crossed light chain, leading to an antibody lacking the crossed light chain. This might be explained by the missing BiP chaperone interaction during the secretion of the antibody out of the cell (Feige et al., 2009). The correct domain order for interaction with BiP is given for the remaining two CrossMab designs as the CH1 domain is part of the heavy chain. For the CrossMabVH–VL, the main side product is a ‘Bence-Jones like’ VL–VL interaction, which is hard to separate from the bispecific antibody with chromatographic methods. In theory, the CrossMabFab is the most preferred CrossMab design, as it utilizes only one non-natural domain exchange/transition, which does not impact stability and assembly and no modified elbow linker regions. Even though we did not manage to find a way to completely prevent the non-cognate VH–VL interaction in the CrossMabFab, this side product of a monovalent non-functional antibody can be easily removed by chromatographic methods due to the difference in molecular weight.
Notably, all side products possess the wildtype CH1–CL interaction, whereas a Bence-Jones like VL–VL interaction is observed for the VH–VL crossover and a non-cognate VH–VL is observed for the CrossMabFab. Based on this experimental result, it can be concluded that the driving force for correct light chain localization is the strong CH1–CL interaction (at least for antibodies without stability engineered VH–VL domains).
Some additional conclusions can be drawn from the results of the described experiment. First, the interaction between VL–VL is not repulsive enough to enforce correct light chain assembly in the CrossMabVH–VL. Second, the interaction between VH–VL of non-cognate domains originating from different target binders/germlines is not repulsive enough to enforce cognate light chain assembly or to overcome the CH1–CL pairing preference in the CrossMabFab. Third, the driving force for correct light chain assembly is in all three CrossMabs the strong CH1–CL interaction.
The focus of the work described here was the optimization and development of a completely self-assembling bivalent bispecific antibody expressed by four individual chains, two light and two heavy chains. The CrossMabVH–VL design including inverted charge pairs in CH1–CL of the non-crossed chain represents such a system ensuring error-free self-assembly in a modular system based on available antibody pairs, allowing the use of notably, any preexisting antibody. The solved X-ray structure illustrates the effect of the two charge pair exchanges/inversions and shows the corresponding electrostatic interactions, namely 123–213 and 124–147 independent of the applied charges. The effect of each individual charge pair exchange is difficult to interpret, as the effects on the complete self-assembly are manifold. In our experiments we observed a higher expression level of the non-crossed light chain, resulting only in non-cognate light chain assembly on the crossed Fab arm. The amount and impact of these kinetic effects is not understood. The influence of attractive interactions like salt bridges and hydrogen-bond interactions on the stabilization of a cognate Fab arm as well as repulsive effects originating from similar charges need to be considered on both Fab arms including cognate and non-cognate pairing of each possible Fab arm combination. Importantly, the optimal effect can be achieved by introduction of only two charge pair inversions. This may be caused by a beneficial impact due to similar charges on neighboring residues, resulting in a larger charged surface area, strengthening attractive and repulsive forces. In addition, the two charge pair exchanges result in only attractive interactions for cognate pairing and always repulsive interactions for non-cognate pairing in the CrossMabVH–VL. This observation is the main difference to the CrossMabVH–VL without charge pair exchanges, where all wild-type charge pair interactions (123–213, 124–147, cognate and non-cognate) are attractive.
According to the structural model and the experimental results we can provide the following recommendations for preferred CrossMabVH–VL design: First, include the domain crossover on the side of the more stable Fab arm. Second, the inverted charge pairs are preferred to be put on the non-crossed Fab arm. Third, it is favored to apply the knobs-into-holes approach in combination with an additional disulfide bridge, while the knob-side is located on the crossed half of the antibody. It is recommended to correct the position Q124 in Cκ to E in addition.
In summary, the CrossMabVH–VL with inverted charge pairs is a mature bispecific design, allowing the combination of individual target binding domains in an IgG-like molecule utilizing 1 + 1, 1 + 2 or 2 + 2 target binding sites as required from the biological point of view. The potential of the CrossMab technology is reflected by the number of bispecific molecules in preclinical and clinical development (Klein et al., 2016; Brinkmann and Kontermann, 2017; Verdino et al., 2018). Currently there are already two trivalent heterodimeric 2 + 1 T-cell bispecific antibodies utilizing the CrossMabVH–VL design with charge pair inversions targeting CD20 (Bacac et al., 2018) and BCMA (Seckinger et al., 2017) in Phase 1 early clinical development. This demonstrates the maturity of the CrossMabVH–VL design with inverted charge pairs and CrossMab technology. The ability to combine 1 + 1, 1 + 2 and 2 + 2 target binding sites in one molecule enables to make use of affinity or avidity driven target binding to enable control over (cell-) selectivity and biodistribution.
Supplementary Material
Acknowledgements
The authors thank their management for continuous support and sponsorship. We thank Anton Jochner, Xaver Reiser, Manuel Endesfelder, Achim Gärtner and Alexandra Kaczmarek for material supply and experimental support.
References
- Bacac M., Colombetti S., Herter S. et al. (2018) Clin. Cancer Res. doi: 10.1158/1078-0432.CCR-18-0455 [DOI] [PubMed] [Google Scholar]
- Bacac M., Fauti T., Sam J. et al. (2016. a) Clin. Cancer Res., 22, 3286–3297. [DOI] [PubMed] [Google Scholar]
- Bacac M., Klein C. and Umana P. (2016. b) Oncoimmunology, 5, e1203498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonisch M., Sellmann C., Maresch D., Halbig C., Becker S., Toleikis L., Hock B. and Ruker F. (2017) Protein Eng. Des. Sel., 30, 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bricogne G., Blanc E., Brandl M. et al. (2011) Buster version 295. Global Phasing Ltd, Cambridge, United Kingdom. [Google Scholar]
- Brinkmann U. and Kontermann R.E. (2017) MAbs, 9, 182–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunker P., Wartha K., Friess T. et al. (2016) Mol. Cancer Ther., 15, 946–957. [DOI] [PubMed] [Google Scholar]
- Carter P. (2001) J. Immunol. Methods, 248, 7–15. [DOI] [PubMed] [Google Scholar]
- Chelius D., Ruf P., Gruber P., Ploscher M., Liedtke R., Gansberger E., Hess J., Wasiliu M. and Lindhofer H. (2010) MAbs, 2, 309–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigenbrot C. and Fuh G. (2013) Curr. Opin. Chem. Biol., 17, 400–405. [DOI] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W.G. and Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr., 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige M.J., Groscurth S., Marcinowski M., Shimizu Y., Kessler H., Hendershot L.M. and Buchner J. (2009) Mol. Cell, 34, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenn S., Schiller C.B., Griese J.J. et al. (2013) PLoS One, 8, e61953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrara N., Hillan K.J., Gerber H.P. and Novotny W. (2004) Nat. Rev. Drug Discov., 3, 391–400. [DOI] [PubMed] [Google Scholar]
- Fischer J.A., Hueber A.J., Wilson S. et al. (2015) Arthritis Rheumatol., 67, 51–62. [DOI] [PubMed] [Google Scholar]
- Huang J. and MacKerell A.D. Jr. (2013) J. Comput. Chem., 34, 2135–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T., Tsunoda H., Kikuchi Y. et al. (2010) Protein Eng. Des. Sel., 23, 667–677. [DOI] [PubMed] [Google Scholar]
- Im W., Lee M.S. and Brooks C.L. 3rd (2003) J. Comput. Chem., 24, 1691–1702. [DOI] [PubMed] [Google Scholar]
- Imhof-Jung S., Klein C., Klostermann S., Regula J.T. and Schaefer W. (2016) WO2016016299A1: Multispecific antibodies. Patent application.
- Irimia A., Sarkar A., Stanfield R.L. and Wilson I.A. (2016) Immunity, 44, 21–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. (2010) Acta Crystallogr. D Biol. Crystallogr., 66, 133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kienast Y., Klein C., Scheuer W. et al. (2013) Clin. Cancer Res., 19, 6730–6740. [DOI] [PubMed] [Google Scholar]
- Kitazawa T., Esaki K., Tachibana T. et al. (2017) Thromb. Haemost., 117, 1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Schaefer W. and Regula J.T. (2016) MAbs, 8, 1010–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Sustmann C., Thomas M. et al. (2012) MAbs, 4, 653–663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krah S., Sellmann C., Rhiel L. et al. (2017) N. Biotechnol., 39, 167–173. [DOI] [PubMed] [Google Scholar]
- Kuglstatter A., Stihle M., Neumann C., Muller C., Schaefer W., Klein C. and Benz J. (2017) Protein Eng. Des. Sel., 30, 649–656. [DOI] [PubMed] [Google Scholar]
- Lammens A., Baehner M., Kohnert U., Niewoehner J., von Proff L., Schraeml M., Lammens K. and Hopfner K.P. (2013) PLoS One, 8, e62697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S., Perera R., Grimm H.P. et al. (2016) Clin. Cancer Res., 22, 4417–4427. [DOI] [PubMed] [Google Scholar]
- Lewis S.M., Wu X., Pustilnik A. et al. (2014) Nat. Biotechnol., 32, 191–198. [DOI] [PubMed] [Google Scholar]
- Liu Z., Leng E.C., Gunasekaran K. et al. (2015) J. Biol. Chem., 290, 7535–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luenberger D.G. (1973) Introduction to Linear and Nonlinear Programming. Addison-Wesley Reading, Reading, MA. [Google Scholar]
- McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C. and Read R.J. (2007) J. Appl. Crystallogr., 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant M., Ma X., Maun H.R. et al. (2013) Proc. Natl Acad. Sci. U S A, 110, E2987–E2996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merchant A.M., Zhu Z., Yuan J.Q., Goddard A., Adams C.W., Presta L.G. and Carter P. (1998) Nat. Biotechnol., 16, 677–681. [DOI] [PubMed] [Google Scholar]
- Mullard A. (2017) Nat. Rev. Drug Discov., 16, 810. [DOI] [PubMed] [Google Scholar]
- Nagorsen D., Kufer P., Baeuerle P.A. and Bargou R. (2012) Pharmacol. Ther., 136, 334–342. [DOI] [PubMed] [Google Scholar]
- Oldenburg J., Mahlangu J.N., Kim B. et al. (2017) N. Engl. J. Med., 377, 809–818. [DOI] [PubMed] [Google Scholar]
- Presta L.G., Chen H., O’Connor S.J., Chisholm V., Meng Y.G., Krummen L., Winkler M. and Ferrara N. (1997) Cancer Res., 57, 4593–4599. [PubMed] [Google Scholar]
- Regula J.T., Lundh von Leithner P., Foxton R. et al. (2016) EMBO Mol. Med., 8, 1265–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regula J.T., Lundh von Leithner P., Foxton R. et al. (2017) EMBO Mol. Med., 9, 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampei Z., Igawa T., Soeda T. et al. (2013) PLoS One, 8, e57479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer W., Klein C., Imhof-Jung S., Klostermann S., Molhoj M. and Regula J.T. (2015) WO2015150447A1: Multispecific antibodies. Patent application
- Schaefer W., Regula J.T., Bahner M. et al. (2011) Proc. Natl Acad. Sci. U S A, 108, 11187–11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer W., Volger H.R., Lorenz S., Imhof-Jung S., Regula J.T., Klein C. and Molhoj M. (2016) MAbs, 8, 49–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuer W., Thomas M., Hanke P. et al. (2016) MAbs, 8, 562–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seckinger A., Delgado J.A., Moser S. et al. (2017) Cancer Cell, 31, 396–410. [DOI] [PubMed] [Google Scholar]
- Shapovalov M.V. and Dunbrack R.L. Jr. (2011) Structure, 19, 844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.J., Olson K., Haber L.J. et al. (2015) Sci. Rep., 5, 17943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiess C., Zhai Q. and Carter P.J. (2015) Mol. Immunol., 67, 95–106. [DOI] [PubMed] [Google Scholar]
- Thomas M., Kienast Y., Scheuer W. et al. (2013) PLoS One, 8, e54923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdino P., Atwell S. and Demarest S.J. (2018) Curr. Opin. Chem. Eng., 19, 107–123. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.