ABSTRACT
Studies of the ovine prion-related protein (testis-specific) gene (PRNT), including studies of genetic diversity, have highlighted its potential relationship to scrapie infection and economically important ovine traits. PRNT was previously reported to be highly polymorphic in Portuguese sheep. To characterize genetic polymorphisms in this gene in Asian sheep, a direct sequencing method was used to detect polymorphic loci in PRNT in 285 individual sheep from four Chinese and one Mongolian breeds. Seven SNP variants in PRNT were identified, including three novel variants (g.93G>A, g.162G>T, and g.190A>G) and four previously reported variants (g.17 C>T, g.112G>C, g.129C>T, and g.144A>G). In the five breeds that we analyzed, the mutation frequencies of g.190A>G in Lanzhou Fat-tail sheep (LFTS) and g.129C>T in the other four varieties were high (F>0.5). Moreover, thirteen different haplotypes that had a comparable distribution in the tested breeds were also identified; ‘C-G-G-C-A-G-A’ occurred at the highest frequency in the five sheep breeds. Additionally, we previously explored the significance of relationships between polymorphisms in PRNP or PRND and ovine growth performance. Here, we also performed correlation analysis in all tested loci. These loci polymorphisms were significantly associated with ten different growth traits (P<0.05), except for g.93G>A. Meanwhile, in contrast to a previous study, there was no significant association between the seven SNP loci analyzed and our previously reported sheep PRND or PRNP insertion/deletion mutations. Our findings may provide new insights into polymorphic variation in ovine PRNT, which may contribute to genetic improvements in economic traits that are important for sheep breeding.
KEYWORDS: Sheep, PRNT gene, SNP, growth traits
1. Introduction
As a novel member of the prion gene family, the prion-related protein (testis-specific) gene (PRNT) was first detected in adult testis [1]. It was once thought to be the duplication product of the prion protein (PRNP) and prion-related protein doppel (PRND) genes [2]. Recent studies have confirmed the translation of PRNT gene in ovine [3] and goat [4], and corresponding gene structures have also been reported. Based on previous reports, the goat [4] and sheep [5] PRNT are both single-exon genes. Although the gene structure of ovine PRNT is simple, its coding region sequence (CDS) is highly polymorphic. In Portuguese sheep, four SNP loci [g.17C>T (p.Ser6Phe), g.112G>C (p.Gly38Arg), g.129T>C (synonymous), and g.144A>G (synonymous)] have been found and significant correlations between PRND heterozygote (c.78G>A) and three PRNT haplotypes have been identified [5].
Moreover, previous work developed by our group [6] identified significant associations between novel 20 bp insertion/deletion (indel) polymorphisms in PRND with the cannon circumference index and trunk index in Chinese Hu sheep. Moreover, considering the linkage of PRND and its homologous gene, PRNP [7–8], our previous work also explored high rate of polymorphic mutations in PRNP and whether these were correlated with growth traits in Chinese cattle [9–10] or native sheep [11]. For insertion/deletion mutations of ovine PRNP, polymorphisms of intron-2-insertion-15 bp were significantly associated with chest width in Small Tail Han sheep (ewe). Additionally, correlations of 3′-UTR-insertion-7 bp on chest circumference in Hu sheep and intron-2-insertion-19 bp on tail length in Tong sheep were highly significant (P ≤ 0.001) [11]. However, to date, available data for East Asian sheep breeds and PRNT polymorphisms have been limited and no study has examined the effect of these polymorphisms on ovine phenotypic traits. Furthermore, considering the linkage of PRND and PRNT [5], the potential for an association between PRNT polymorphisms and economically important sheep traits still needs to be investigated.
Considering the geographic characteristics and excellent economic traits of sheep, Lanzhou fat-tail sheep (LFTS), Tong sheep (TS), Small Tail Han sheep (STHS), Hu sheep (HS), and Sartuul sheep (SS), which are representative indigenous breeds in China and Mongolia, were selected for study. Using a Sanger direct sequencing approach, we examined sequence variants in PRNT in Asian sheep breeds and explored the potential impact of these polymorphisms on ovine growth performance.
2. Materials and methods
2.1. Ethics statement
All experiments implemented in this study were approved by the International Animal Care and Use Committee of the Northwest A&F University (IACUC-NWAFU) fully according to the local animal welfare guidelines, laws and policies.
2.2. Genomic DNA collection
A total of 285 healthy sheep samples from five diverse and proligerous breeds were used. Samples of Sartuul sheep (n = 61) were collected from Zhawkhan Province (Mongolia), while Hu sheep (n = 60) were collected from Henan Province (China). Small Tail Han sheep (n = 65) and Lanzhou Fat-Tail sheep (n = 38) were from Gansu Province (China), while the Tong sheep (n = 61) were from Shanxi Province (China). All of the tested sheep were 2 to 6 years old. According to these same standards [12], the body traits for all selected individuals were measured by one person; measurements included body height (BH), body length (BL), hip height (HH), chest width (ChW), chest circumference (ChC), head length (HL), head depth (HD), and teat number (TN). Subsequently, according to previous reports [13–14], the body length index (BLI), chest circumference index (ChCI), chest width index (ChWI), and cannon circumference index (CaCI) were also calculated.
Genomic DNA used in this study were extracted from sheep ear tissues (stored in 70% alcohol at −80°C) using the phenol-chloroform method, according to our previously reported protocol [15–17]. DNA samples were quantified using a Nanodrop 1000 (Thermo Scientific, Waltham, MA, USA) and all DNA samples were diluted to 50 ng/μL and temporarily stored at 4°C.
2.3. Novel SNP discovery and genotyping by direct sequencing
Referencing the whole sequence of ovine chromosome 13 and published sheep PRNT gene sequence (Gene ID: 100049065), specific primers for the complete PRNT sequence (forward: 5′-ACCTTGGTGGTTTTGGTTTTG-3′; reverse: 5′-CAGTGGTACATGGGATTGGGA-3′) were designed with Primer Premier Software 5.0 (Premier Biosoft International USA) and verified using the Basic Local Alignment Search Tool (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The PCR reaction volume and amplification procedure were in accordance with our previous study [6,18]: initial denaturation for 4 min at 95°C, followed by 18 cycles of denaturation for 30 s at 94°C, annealing for 30 s at 68°C (with a reduction of 1°C per cycle), and extension for 1000 bp/min at 72°C, followed by another 23 cycles of 30 s at 94°C, 30 s at 50°C, and 45 s at 72°C, and a final extension of 10 min at 72°C. After cooling to 4°C, all PCR products were detected by 1.5% agarose gel electrophoresis and directly sequenced by Sangon Company (Shanghai, China). Finally all individuals were genotyped [19]. Considering the limitations of Sanger sequencing, apart from 20 bases at the start of the sequence or at the termination of the PCR amplification, areas that both bind to primers, the sequencing results of the sequence in the middle were plausible.
2.4. Sequence alignment and statistical analyses
Sequence alignment and Neighbor-Joining tree analyses were performed using MEGA software (version 7.0) and the nucleic acid sequences of the PRNT gene from different species were obtained from the GenBank database (https://www.ncbi.nlm.nih.gov/nuccore/NC_019470.2). Moreover, the genotype distributions were analyzed by Hardy–Weinberg equilibrium using the χ2 test. Polymorphism information content (PIC) was calculated by Nei's method implemented in the GDIcall Online Calculator (http://www.msrcall.com/Gdicall.aspx) [20]. Distribution differences for genotypic and allelic frequencies among/between different breeds were calculated with the χ2 test implemented in SPSS (Version 18.0, IBM Corp., Armonk, NY, USA) [21]. Additionally, linkage equilibrium on the population of the seven pairs of alleles and haplotypes of seven polymorphic loci were also analyzed (http://analysis.bio-x.cn) [22–23]. Furthermore, independent-samples T test and analysis of variance (ANOVA), which are available in SPSS (version 18.0) [24, were used to explore associations of the SNPs in PRNT with several growth traits [e.g. body height (cm)] in different breeds. When necessary, the Bonferroni correction for multiple comparisons was performed. Particularly, data that did not follow a normal distribution and homogeneity of variances were analyzed by the non-parametric (Kruskal–Wallis) test in SPSS (version 18.0) [21]. P < 0.05 was considered to indicate statistical significance and all statistical tests were two-sided. Furthermore, correlation analysis of different growth traits was performed when those traits were significantly correlated with tested polymorphic loci.
3. Results
3.1. Novel SNPs loci of sheep PRNT and the calculation of genetic parameters
The sequence of the PCR product of PRNT consisted of three parts: a coding region upstream sequence (121 bp), a coding region sequence (159 bp), and a downstream control region sequence (81 bp).
Results of DNA Sanger sequencing revealed three novel SNP loci: g.93G>A (synonymous), g.162G>T, and g.190A>G, as well as four previously reported loci: g.17C>T (p.Ser6Phe), g.112G>C (p.Gly38Arg), g.129C>T, and g.144A>G (Fig. 1). Among these loci, g.17C>T, g.129C>T, and g.190A>G were present in Portuguese and the five tested sheep breeds. The novel SNP g.93G>A was only found in STHS and TS, whereas g.162G>T were detected in LFTS and STHS. Among the 285 sheep that were tested, the mutation frequencies of g.190A>G in LFTS and g.129C>T in other varieties were high (F>0.5), while the frequency of the g.93G>A mutation was extremely low in all tested breeds (0≤F≤0.02) (Table 1). Additionally, the g.93G>A locus was in Hardy–Weinberg equilibrium in STHS and TS, and g.162G>T was also in HWE in LFTS and STHS (Table 1).
Figure 1.
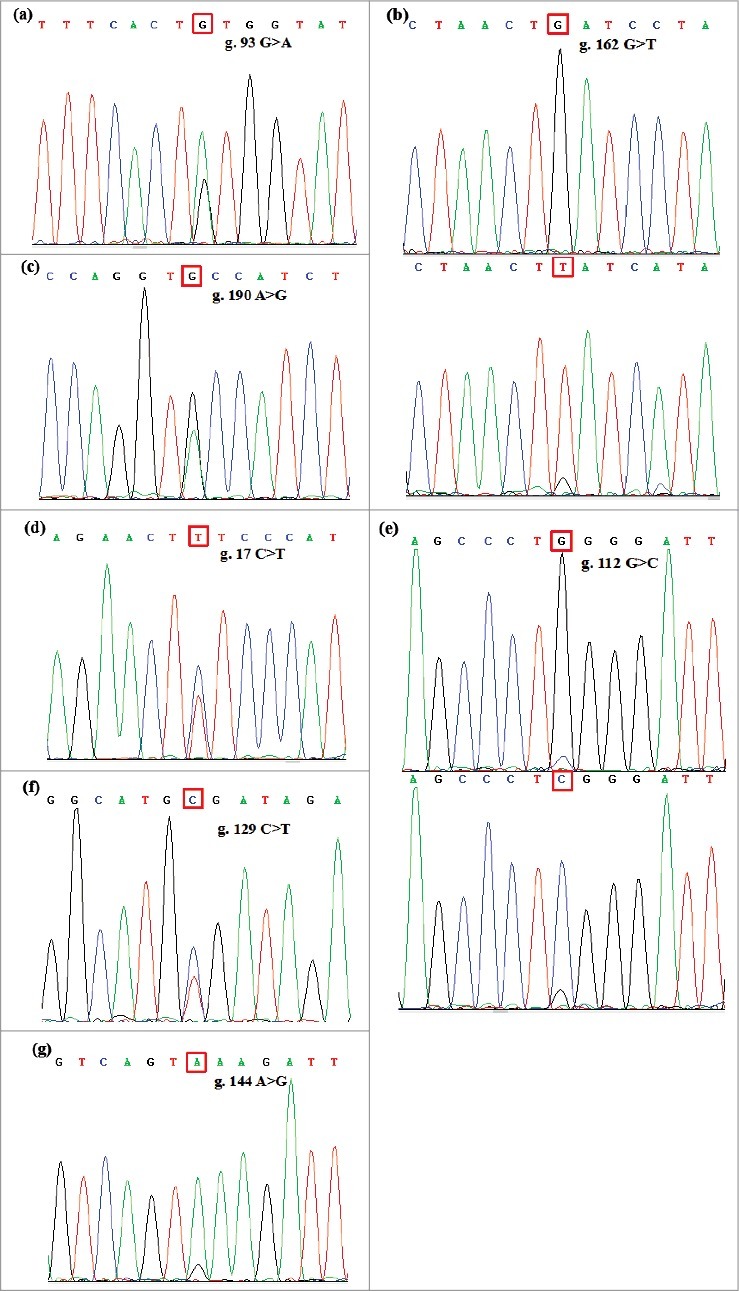
Sequence diagram of the three novel SNP variants loci of sheep PRNT gene: (a) g.93G>A, (b) g.162G>T, (c) g.190A>G; and the reported SNP loci: (d) g.17 C>T, (e) g.112G>C,(f) g.129 C>T, (g) g.144 A>G.
Table 1.
Diversity parameters for SNP loci of sheep PRNT gene.
| Breeds (N) | Loci | HoR | He | HoV | HWE(P) | MAF | PIC |
|---|---|---|---|---|---|---|---|
| LFTS (N = 38) | g.17C>T | 0.92 | 0.05 | 0.03 | P = 0.004 | 0.05 | 0.09 |
| g.93G>A | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.112G>C | 0.89 | 0.11 | 0.00 | P = 0.732 | 0.05 | 0.09 | |
| g.129C>T | 0.71 | 0.16 | 0.13 | P = 0.001 | 0.21 | 0.28 | |
| g.144A>G | 0.89 | 0.08 | 0.03 | P = 0.027 | 0.07 | 0.12 | |
| g.162G>T | 0.95 | 0.05 | 0 | P = 0.868 | 0.03 | 0.05 | |
| g.190A>G | 0.45 | 0.45 | 0.10 | P = 0.934 | 0.33 | 0.34 | |
| TS (N = 61) | g.17C>T | 0.93 | 0.07 | 0.00 | P = 0.791 | 0.03 | 0.06 |
| g.93G>A | 0.98 | 0.02 | 0.00 | P = 0.948 | 0.01 | 0.02 | |
| g.112G>C | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.129C>T | 0.25 | 0.72 | 0.03 | P = 0.001 | 0.39 | 0.36 | |
| g.144A>G | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.162G>T | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.190A>G | 0.67 | 0.28 | 0.05 | P = 0.486 | 0.19 | 0.26 | |
| STHS (N = 65) | g.17C>T | 0.85 | 0.15 | 0 | P = 0.481 | 0.14 | 0.13 |
| g.93G>A | 0.98 | 0.02 | 0 | P = 0.950 | 0.02 | 0.02 | |
| g.112G>C | 0.98 | 0.02 | 0 | P = 0.950 | 0.02 | 0.02 | |
| g.129C>T | 0.48 | 0.29 | 0.23 | P = 0.002 | 0.47 | 0.36 | |
| g.144A>G | 0.98 | 0.02 | 0 | P = 0.950 | 0.02 | 0.02 | |
| g.162G>T | 0.98 | 0.02 | 0 | P = 0.950 | 0.02 | 0.02 | |
| g.190A>G | 0.72 | 0.26 | 0.02 | P = 0.699 | 0.25 | 0.22 | |
| HS (N = 60) | g.17C>T | 0.90 | 0.10 | 0 | P = 0.683 | 0.05 | 0.09 |
| g.93G>A | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.112G>C | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.129C>T | 0.43 | 0.40 | 0.17 | P = 0.282 | 0.37 | 0.36 | |
| g.144A>G | 0.98 | 0.02 | 0.00 | P = 0.948 | 0.01 | 0.02 | |
| g.162G>T | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.190A>G | 0.77 | 0.23 | 0 | P = 0.394 | 0.12 | 0.18 | |
| SS (N = 61) | g.17C>T | 0.87 | 0.11 | 0.02 | P = 0.211 | 0.07 | 0.13 |
| g.93G>A | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.112G>C | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.129C>T | 0.44 | 0.49 | 0.07 | P = 0.252 | 0.31 | 0.34 | |
| g.144A>G | 0.98 | 0.02 | 0 | P = 0.949 | 0.01 | 0.02 | |
| g.162G>T | 1.00 | 0 | 0 | — | 0 | 0 | |
| g.190A>G | 0.65 | 0.33 | 0.02 | P = 0.394 | 0.18 | 0.25 |
Note: HoR, Homozygote reference; He, Heterozygote; HoV, Homozygote variant; HWE, Hardy-Weinberg equilibrium; MAF, minimum frequency of alleles; PIC, polymorphic information content. LFTS, Lanzhou Fat-Tail sheep; STHS, Small Tail Han sheep; TS, Tong sheep; HS, Hu sheep; SS, Sartuul sheep.
3.2. Sequence alignment
As shown by the multiple sequence alignment (Fig. 2), there were five SNP loci in the sheep PRNT coding region. The homology of the PRNT coding region among different species, such as Hereford, San Clemente goat, and Texel sheep, was high.
Figure 2.
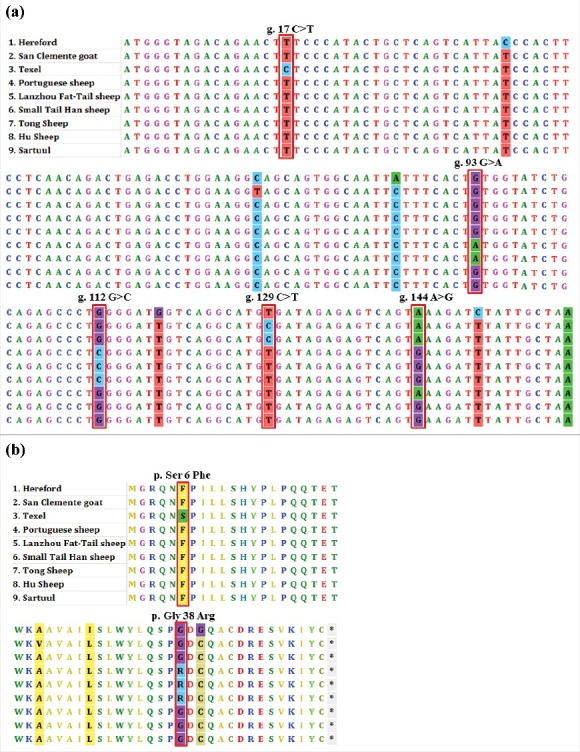
Coding region sequence (a) and amino acid sequence (b) alignment of PRNT gene in bovine, goat and different sheep breeds.
Based on the CDS of the PRNT gene, a Neighbor-Joining tree with bovine, goat, and different sheep breeds was constructed (Fig. 3). Interestingly, this analysis showed that the evolutionary relationship between San Clemente goat and Texel sheep was closer than that of the Texel and Tong sheep.
Figure 3.
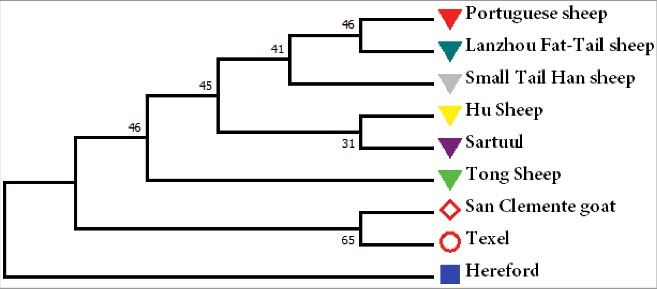
The Neighbor-Joining tree of bovine, goat and different sheep breeds basing the coding region sequence of PRNT gene.
3.3. Linkage equilibrium and haplotype analysis
Based on the D' test and r2 test for the seven pairs of alleles in the sheep PRNT gene, g.112G>C showed strong LD with g.144A>G in LFTS and STHS. Furthermore, in STHS, g.112G>C and g.190A>G were also in LD, and g.144A>G and g.162G>T also showed strong LD (Fig. 4).
Figure 4.
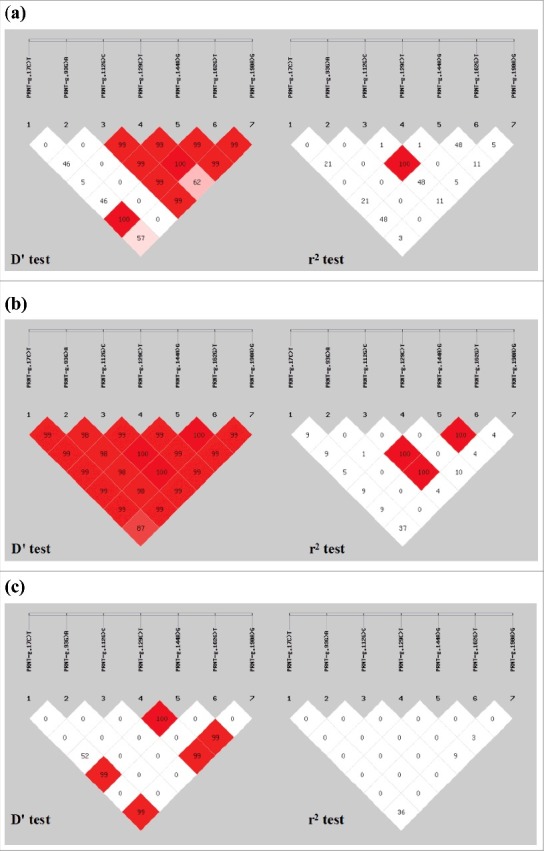
Genetic analysis of linkage disequilibrium on different populations of seven pairs alleles in sheep PRNT gene: (a) LFTS, (b) STHS, (c) TS, (d) HS, (e) SS. Loci chosen for analysis: loci1, g.17C>T; loci2, g.93G>A; loci3, g.112G>C; loci4, g.129C>T; loci5, g.144A>G; loci6, g.162G>T; loci7, g.190A>G.
Figure 4.
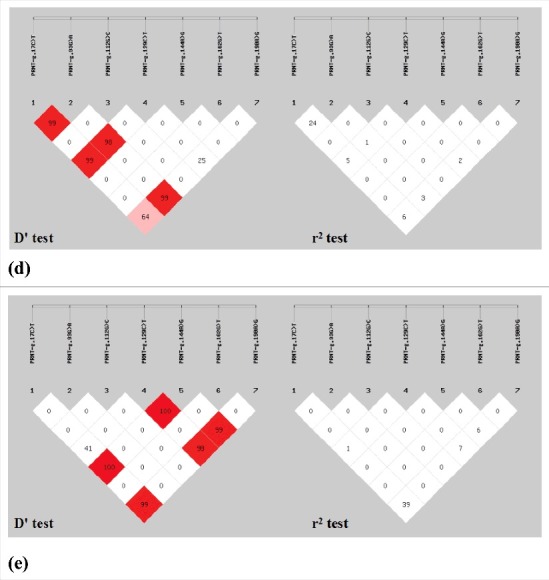
(Continued).
Moreover, 13 different haplotypes were found in 285 individuals. Among these sheep, ‘C-G-G-C-A-G-A’ showed the highest frequency and was detected in all of the groups that were tested (Fig. 5).
Figure 5.
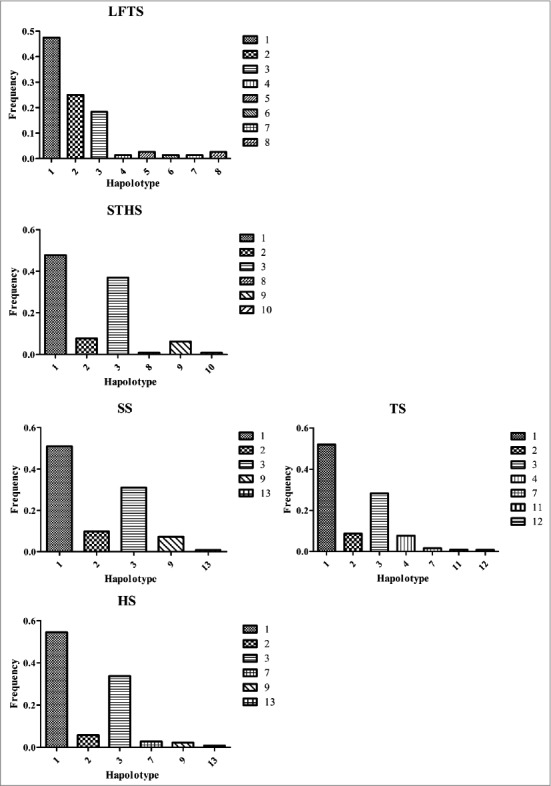
Haplotype frequency of seven mutation loci within the sheep PRNT gene.
Note: Loci chosen for hap-analysis: g.17C>T, g.93G>A, g.112G>C, g.129C>T, g.144A>G, g.162G>T, g.190A>G (All those frequency < 0 will be ignored in analysis.). Hap1, C G G C A G A; Hap2, C G G C A G G; Hap3, C G G T A G A; Hap4, C G G T A G G; Hap5, C G C C G G G; Hap6, T G G C A G A; Hap7, T G G T A G G; Hap8, T G C C G T G; Hap9, T G G C A G G; Hap10, T A G T A G A; Hap11, T G G T A G A; Hap12, T A G T A G G; Hap13, C G G C G G G.
3.4. Associations between PRNT polymorphisms and sheep growth performance
In view of the relationship between the linked homologous genes (PRNP and PRND) and sheep growth, a correlation analysis of the seven PRNT variant loci and sheep growth traits was conducted. All tested loci, except for g.93G>A, were significantly associated with ten different growth traits (P<0.05) (Table 2). For example, the g.190A>G genotypes significantly correlated with the body length index, head length, head depth, and teat numbers of Tong sheep (ewe). Based on the independent-samples T test findings, polymorphisms of g.112G>C and g.144A>G had the same effect on chest width of LFTS, which was consistent with the LD analysis results (Fig. 4). Furthermore, correlation analyses of different growth traits yielded results that were often consistent with the relevance of traits and loci. For example, in LFTS, ewes with a larger chest width tended to have larger values of body height, hip height, and chest width index; those traits were significantly correlated the g.162G>T genotype (Table 2). Meanwhile, chest width of LFTS showed a significant association with body height, hip height, and chest width index (Table 3).
Table 2.
Relationship between the different SNP locus of PRNT gene and growth related traits in different breed (LSMa±SE).
| Observed genotypes (LSMa±SE) |
|||||||
|---|---|---|---|---|---|---|---|
| Loci | Breeds | Size | Growth traits | Homozygote reference (N) | Heterozygote (N) | Homozygote variant (N) | P values |
| g.17C>T | HS(ewe) | 60 | HH | 106.037 ± 2.374 (54) | 90.667 ± 4.432 (6) | — | 0.040 |
| g.112G>C | LFTS(ewe) | 27 | ChW | 23.796 ± 0.720 (23) | 18.625 ± 0.851 (4) | — | 0.008 |
| ChWI | 68.585 ± 1.796 (23) | 52.045 ± 3.051 (4) | — | 0.003 | |||
| g.129C>T | STHS(ram) | 27 | BH | b61.382 ± 1.212 (11) | a,b63.722 ± 1.140 (9) | a66.000 ± 1.633 (7) | 0.022 |
| HS(ewe) | 60 | HH | b103.077 ± 3.556 (26) | b101.042 ± 3.324 (24) | a116.500 ± 4.590 (10) | 0.037 | |
| ChCI | b124.435 ± 1.463 (26) | b127.987 ± 1.422 (24) | a130.597 ± 2.229 (10) | 0.025 | |||
| g.144A>G | LFTS | 27 | ChW | 23.796 ± 0.720 (23) | 18.625 ± 0.851 (4) | — | 0.008 |
| (ewe) | ChWI | 68.585 ± 1.796 (23) | 52.045 ± 3.051(4) | — | 0.003 | ||
| g.162G>T | LFTS | 27 | ChW | 23.472 ± 0.702 (25) | 17.500 ± 0.500 (2) | — | 0.026 |
| (ewe) | BH | 76.008 ± 1.300 (25) | 62.500 ± 0.500 (2) | — | 0.008 | ||
| HH | 76.784 ± 1.317 (25) | 65.000 ± 5.000 (2) | — | 0.023 | |||
| ChWI | 67.837 ± 1.872 (25) | 52.373 ± 5.254 (2) | — | 0.028 | |||
| g.190A>G | STHS | 27 | BH | 64.238 ± 0.723 (21) | 60.283 ± 2.362 (6) | — | 0.040 |
| (ram) | ChC | 73.562 ± 1.187 (21) | 67.833 ± 1.493 (6) | — | 0.024 | ||
| HH | 65.000 ± 0.613 (21) | 60.950 ± 1.933 (6) | — | 0.013 | |||
| TS(ewe) | 41 | BLI | b110.705 ± 1.340 (30) | a115.644 ± 1.275 (9) | a,b112.452 ± 1.148 (2) | 0.038 | |
| HL | a20.017 ± 0.156 (30) | b19.056 ± 0.452 (9) | a20.500 ± 0.500 (2) | 0.014 | |||
| HD | a14.733 ± 0.151 (30) | b14.000 ± 0.220 (9) | a,b14.250 ± 0.250 (2) | 0.019 | |||
| TN | B2.867 ± 0.184 (30) | B2.222 ± 0.222 (9) | A4.000 ± 0.000 (2) | 1.26E-04 | |||
Note: HH, hip height; ChW, chest width; ChWI, chest width index; BH, body height; ChCI, chest circumference index; ChC, chest circumference; BLI, body length index; HL, head length; HD, head depth; TN, teat numbers. The values with different letters (a and b, or A and B) within the same row significantly at P < 0.05 and P < 0.01, respectively.
Table 3.
Correlation analysis of different growth traits in tested sheep breeds (pearson correlation/bilateral significance).
| Breed | Traits\Traits | ChW | ChWI | BH | HH |
|---|---|---|---|---|---|
| LFTS-ewe (n = 27) | ChW | 1/- | 0.718**/5.370E-5 | 0.509**/0.007 | 0.560**/0.002 |
| ChWI | 1/- | 0.401*/0.047 | 0.478*/0.016 | ||
| BH | 1/- | 0.819**/1.774E-7 | |||
| HH | 1/- | ||||
| BH | ChC | HH | |||
| STHS-ram (n = 27) | BH | 1/- | 0.445*/0.002 | 0.796**/6.782E-7 | |
| ChC | 1/- | 0.707**/3.745E-5 | |||
| HH | 1/- | ||||
| — | |||||
| BLI | HL | HD | TN | ||
| TS-ewe (n = 41) | BLI | 1/- | −0.216/0.174 | −0.127/0.430 | −0.086/0.593 |
| HL | 1/- | 0.104/0.519 | −0.037/0.818 | ||
| HD | 1/- | 0.075/0.643 | |||
| TN | 1/- | ||||
| HH | ChWI | ||||
| HS-ewe (n = 60) | HH | 1/- | — | ||
| ChWI | 1/- |
Note: ChW, chest width; ChWI, chest width index; BH, body height; HH, hip height; ChC, chest circumference; BLI, body length index; HL, head length; HD, head depth; TN, teat numbers. ChCI. The values with different asterisk (* and **) within significantly at P < 0.05 and P < 0.01, respectively.
Considering the reported linkage between PRND and PRNT or PRNP along with the findings of our previous study, we analyzed the relationship between the seven variant SNPs and reported indels mutation of sheep PRND (one indel) or PRNP (five indels); however, no significant association was found.
4. Discussion
In 2016, polymorphisms in the sheep PRNT gene were first detected in 567 sheep from eight Portuguese breeds [5]. In that study, two SNPs loci that resulted in missense mutations (p.Ser6Phe and p.Gly38Arg) and two synonymous mutations were identified. In this present study, g.17C>T and g.144A>G, which exhibited a low variant frequency in all tested Asian breeds, were also detected. By contrast, g.129C>T was present at a high frequency in five groups. A central principle of molecular evolution holds that organisms evolved to reserve those codons that yielded an advantage in gene function or other factors favored by natural selection [25–26]. Hence, for the aforementioned SNP loci that are present in both European and East Asian sheep groups may have evolved early in ovine evolution, and they may be closely related to some PRNT gene functions or certain traits in sheep. Consequently, they have been retained over a long evolutionary period. Mesquita inferred that those two missense mutations are closely related to a phosphorylation-dependent signaling pathway [5], but to date this hypothesis has not yet been confirmed. Notably, most missense mutations that change amino acids in protein domains can cause dramatic changes the protein that can impair its biological function [24,27,28]. By contrast, when missense mutations occur at the junctions of protein domains, the effect of amino acid substitutions will be weak or minimal [29]. Therefore, the reported two missense mutations (p.Ser6Phe and p.Gly38Arg) that were not eliminated by natural selection may by located at protein domain junctions. Moreover, missense nucleotide variants that are predicted to alter amino acid sequences at a protein binding site can also contribute to abnormal interactions between proteins [30]. Therefore, in accord with previous publications [5], the protein structure for the p.Ser6Phe and p.Gly38Arg polymorphic variants should be studied in detail in the future.
Additionally, except for the four reported SNPs, a synonymous mutation of CDS was newly identified and the downstream regulatory sequences of sheep PRNT were firstly studied with two novel mutations. Variation in the novel SNP g.162G>T was only found in LFTS and STHS with low mutation frequency. Previous studies indicated that SNPs that occur within non-protein-coding sequence can interfere with mRNA, such as by affecting RNA splicing or altering the access of transcription factors to gene regulatory elements, thereby affecting its allelic expression [31–33]. Thus, combing the results of association analysis in LFTS, ewes that are heterozygous for g.162G>T have a lower body height, head height, and chest width compared with wild-type individuals. Considering these data together, it is possible to infer that the occurrence of the g.162G>T mutation might inhibit the growth of sheep, so this mutation may be gradually eliminated by artificial or natural selection.
Our previous work identified the significant relationship between ovine growth performance and polymorphisms in PRND [6] or PRNP [11]. Herein, polymorphisms of the homologous gene PRNT were also found be strongly correlated with sheep growth traits. The strong linkage of SNP loci for PRNT and mutated loci within other growth-related genes might be a potential reason for this phenotypic association. Moreover, although no significant association of SNPs loci for PRNT and reported PRND indel mutations were found, other variants of ovine PRND (such as c.78G>A) that correlated with PRNT haplotypes [5] may affect sheep phenotypic traits, and this possibility merits future study.
Interestingly, in our study, the evolutionary relationship between the San Clemente goat and Texel sheep was closer than that of Texel and Tong sheep. Additionally, a previous study based on whole-genome SNPs revealed that LFTS had a shorter genetic distance with TS sheep than LFTS and STHS, which differed from our present findings [34]. In this present study, the Neighbor–Joining tree was only based on polymorphic loci of the ovine PRNT gene and the long-term artificial selection of these variants is thought to be the main cause of those differences.
In summary, three novel polymorphic loci in PRNT coding and regulatory regions were found in Chinese and Mongolian sheep, and strong correlations between these SNPs and economically important sheep traits were established. We highlighted cases in which a significant effect was detected, although it may have been one of many comparisons within the analyses of any particular study. Therefore, our findings may enrich current knowledge of the genetic variability of ovine PRNT and identify potentially useful DNA markers for the selection of individuals with MAS in sheep breeding.
Funding Statement
This work was supported by the Natural Science Foundation of Gansu Province (No.1610RJZA103) National Natural Science Foundation of China (No.31660642) Central Special Funds for Basic Research in Universities Operating Expenses of “An Excellent and Three Special” Discipline Construction (No.31920170170).
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Financial disclosure statements
The study was funded by the National Natural Science Foundation of China (No.31660642), Natural Science Foundation of Gansu Province (No.1610RJZA103), and Central Special Funds for Basic Research in Universities Operating Expenses of “An Excellent and Three Special” Discipline Construction (No.31920170170). We greatly thanked the staffs of TS elite reservation farm (Baishui county, Shannxi Province), Ruilin Sci-Tech Cluture and Breeding Limit Company (Yongjing county, Gansu Province), shanshan agriculture and animal husbandry Sci-tech company (Mengjin county, Henan Province), and Zhawkhan tsum sureg farm, (Tsagann Khairkhan sum, Zhawkhan province, Mongolia) for collecting samples.
References
- [1].Makrinou E, Collinge J, Antoniou M. Genomic characterization of the human prion protein (PrP) gene locus. Mamm Genome. 2002;13(12):696–703. doi: 10.1007/s00335-002-3043-0. [DOI] [PubMed] [Google Scholar]
- [2].Kocer A, Gallozzi M, Renault L, et al.. Goat PRND expression pattern suggests its involvement in early sex differentiation. Dev Dyn. 2007;236(3):836–842. doi: 10.1002/dvdy.21066. [DOI] [PubMed] [Google Scholar]
- [3].Pimenta J, Domingos A, Santos P, et al.. Is prnt a pseudogene? Identification of ram Prt in testis and ejaculated spermatozoa. PLoS One. 2012;7(8):e42957. doi: 10.1371/journal.pone.0042957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Kim YC, Jeong BH. The first report of prion-related protein gene (PRNT) polymorphisms in goat. Acta Vet Hung. 2017;65(2):291–300. doi: 10.1556/004.2017.028. [DOI] [PubMed] [Google Scholar]
- [5].Mesquita P, Garcia V, Marques MR, et al.. The prion-related protein (testis-specific) gene (PRNT) is highly polymorphic in Portuguese sheep. Anim Genet. 2016;47(1):128–132. doi: 10.1111/age.12380. [DOI] [PubMed] [Google Scholar]
- [6].Li J, Zhu XC, Ma L, et al.. Detection of a new 20bp insertion/deletion (indel) within sheep PRND gene using mathematical expectation (ME) method. Prion. 2017;11(2):143–150. doi: 10.1080/19336896.2017.1300740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Pereira RM, Mesquita P, Batista M, et al.. Doppel gene polymorphisms in Portuguese sheep breeds: insights on ram fertility. Anim Reprod Sci. 2009;114(1–3):157–166. doi: 10.1016/j.anireprosci.2008.10.003. [DOI] [PubMed] [Google Scholar]
- [8].Mesquita P, Batista M, Marques MR, et al.. Prion-like Doppel gene polymorphisms and scrapie susceptibility in Portuguese sheep breeds. Anim Genet. 2010;41(3):311–314. doi: 10.1111/j.1365-2052.2009.01992.x. [DOI] [PubMed] [Google Scholar]
- [9].Yang Q, Zhang SH, Liu LL, et al.. Application of mathematical expectation (ME) strategy for detecting low frequency mutations: An example for evaluating 14 bp insertion/deletion (indel) within the bovine PRNP gene. Prion. 2016;10:409–419. doi: 10.1080/19336896.2016.1211593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Yang Q, Zhang SH, Liu LL, Lei CZ, Qi XL, Lin FP, Qu WD, Qi XS, Liu JM, Wang RM, et al.. The evaluation of 23-bp and 12-bp insertion /deletion within the PRNP gene and their effects on growth traits in healthy Chinese native cattle breeds. J Appl Animal Res. 2017;46(1):505–511. doi: 10.1080/09712119.2017.1348950 [DOI] [Google Scholar]
- [11].Li J, Erdenee S, Zhang SL, et al.. Genetic effects of PRNP gene insertion/deletion (indel) on phenotypic traits in sheep. Prion. 2018;12(1):42–53. doi: 10.1080/19336896.2017.1405886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Zhao HD, He S, Zhu YJ, et al.. A novel 29 bp insertion/deletion (indel) variant of the LHX3 gene and its influence on growth traits in four sheep breeds of various fecundity. Archives Animal Breeding. 2017;60:79–85. doi: 10.5194/aab-60-79-2017. [DOI] [Google Scholar]
- [13].Lan XY, Pan CY, Chen H, et al.. An AluI PCR-RFLP detecting a silent allele at the goat POU1F1 locus and its association with production traits. Small Ruminant Res. 2007;73(1):8–12. doi: 10.1016/j.smallrumres.2006.10.009. [DOI] [Google Scholar]
- [14].Yang Q, Yan HL, Li J, et al.. A novel 14‐bp duplicated deletion within goat GHR gene is significantly associated with growth traits and litter size. Anim Genet. 2017;48:499–500. doi: 10.1111/age.12551. [DOI] [PubMed] [Google Scholar]
- [15].Zhang SH, Sun K, Bian YN, et al.. Developmental validation of an X-Insertion/Deletion polymorphism panel and application in HAN population of China. Sci Rep. 2015;5:18336. doi: 10.1038/srep18336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang XY, Wu XF, Jia WC, et al.. Novel nucleotide variations, haplotypes structure and associations with growth related traits of goat at motif-binding factor (ATBF1) gene. Asian-Australas J Anim Sci. 2015;28(10):1394–1406. doi: 10.5713/ajas.14.0860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Zhang SH, Xu H, Liu XF, et al.. The muscle development transcriptome landscape of ovariectomized goat. R Soc Open Sci. 2017;4:171415. doi: 10.1098/rsos.171415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang XY, Yang Q, Wang K, et al.. A novel 12-bp indel polymorphism within the GDF9 gene is significantly associated with litter size and growth traits in goats. Anim Genet. 2017;48(6):735–736. doi: 10.1111/age.12617. [DOI] [PubMed] [Google Scholar]
- [19].Cui Y, Zhang YH, Wei ZY, et al.. Pig KDM5B: mRNA expression profiles of different tissues and testicular cells and association analyses with testicular morphology traits. Gene. 2018;650:27–33. doi: 10.1016/j.gene.2018.01.092. [DOI] [PubMed] [Google Scholar]
- [20].Czarnik U, Grzybowski G, Zabolewicz T, et al.. Deletion/insertion polymorphism of the prion protein gene (PRNP) in Polish red cattle, Polish White-backed cattle and European bison (Bison bonasus L., 1758). Genetika. 2009;45(4):519–525. [PubMed] [Google Scholar]
- [21].Cui Y, Yan H, Wang K, et al.. Insertion/Deletion within the KDM6A gene is significantly associated with litter size in goat. Front Genet. 2018;9:91. doi: 10.3389/fgene.2018.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15(2):97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- [23].Li Z, Zhang Z, He Z, et al.. A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res. 2009;19(4):519–523. doi: 10.1038/cr.2009.33. [DOI] [PubMed] [Google Scholar]
- [24].Li Y, Wang K, Jiang YZ, et al.. 2:3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) inhibits human ovarian cancer cell proliferation. Cell Oncol (Dordr). 2014;37(6):429–437. doi: 10.1007/s13402-014-0206-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Adam Siepel, Leonardo Arbiza. Cis-regulatory Elements and Human Evolution. Curr Opin Genet Dev. 2014;0:81–89. doi: 10.1016/j.gde.2014.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Galtier N, Roux C, Rousselle M, Romiguier J, Figuet E, Glemin S, Bierne N, Duret L.. Codon usage bias in animals: disentangling the effects of natural selection, effective population size and GC-biased gene conversion. Mol Biol Evol. 2018;35(5):1092–1103. doi: 10.1093/molbev/msy015. [DOI] [PubMed] [Google Scholar]
- [27].William M. Shafer, Jacqueline T. Balthazar, Kayla E. Hagman, et al.. Missense mutations that alter the DNA-binding domain of the MtrR protein occur frequently in rectal isolates of Neisseria gonorrhoeae that are resistant to faecal lipids. Microbiology. 1995;141(4):907–911. doi: 10.1099/13500872-141-4-907. [DOI] [PubMed] [Google Scholar]
- [28].Mackeh R, Marr AK, Dargham SR, et al.. Single-Nucleotide Variations of the Human Nuclear Hormone Receptor Genes in 60,000 Individuals. J Endocr Soc. 2017;2(1):77–90. doi: 10.1210/js.2017-00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Tuz K, Hsiao YC, Juárez O, et al.. The Joubert syndrome-associated missense mutation (V443D) in the Abelson-helper integration site 1 (AHI1) protein alters its localization and protein-protein interactions. J Biol Chem. 2013;288(19):13676–13694. doi: 10.1074/jbc.M112.420786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Morris LE, Bloom GS, Frierson HF Jr, et al.. Nucleotide variants within the IQGAP1 gene in diffuse-type gastric cancers. Genes Chromosomes Cancer. 2005;42(3):280–286. doi: 10.1002/gcc.20150. [DOI] [PubMed] [Google Scholar]
- [31].Krawczak M, Reiss J, Cooper DN. The mutational spectrum of single base-pair substitutions in mRNA splice junctions of human genes: Causes and consequences. Hum Genet. 1992;90(1–2):41–54. doi: 10.1007/BF00210743. [DOI] [PubMed] [Google Scholar]
- [32].Li G, Pan T, Guo D, et al.. Regulatory variants and disease: the E-cadherin–160C/A SNP as an example. Mol Biol Int. 2014;2014:967565. doi: 10.1155/2014/967565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tak YG, Farnham PJ. Making sense of GWAS: using epigenomics and genome engineering to understand the functional relevance of SNPs in non-coding regions of the human genome. Epigenetics Chromatin. 2015;8(1):57. doi: 10.1186/s13072-015-0050-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhao YX, Yang J, Lv FH, et al.. Genomic Reconstruction of the History of Native Sheep Reveals the Peopling Patterns of Nomads and the Expansion of Early Pastoralism in East Asia. Mol Biol Evol. 2017;34(9):2380–2395. doi: 10.1093/molbev/msx181. [DOI] [PMC free article] [PubMed] [Google Scholar]


