ABSTRACT
Protein misfolding and aggregation into highly ordered fibrillar structures have been traditionally associated with pathological processes. Nevertheless, nature has taken advantage of the particular properties of amyloids for functional purposes, like in the protection of organisms against environmental changing conditions. Over the last decades, these fibrillar structures have inspired the design of new nanomaterials with intriguing applications in biomedicine and nanotechnology such as tissue engineering, drug delivery, adhesive materials, biodegradable nanocomposites, nanowires or biosensors. Prion and prion-like proteins, which are considered a subclass of amyloids, are becoming ideal candidates for the design of new and tunable nanomaterials. In this review, we discuss the particular properties of this kind of proteins, and the current advances on the design of new materials based on prion sequences.
KEYWORDS: Prions, amyloids, prion forming domains, prion-like proteins, self-assembly, nanotechnology
Introduction
Protein-based nanofibers are emerging nanomaterials with are finding an increasing number applications [1,2]. Amyloid proteins have the ability to organize into highly ordered fibrillar structures, thus emerging as interesting building blocks to generate new classes of supramolecular structures. These fibrillar assemblies are characterized by a cross-β structure in which β-strands are aligned perpendicularly to the fibril axis and are stabilized by a dense network of hydrogen bonds [3]. The abundant non-covalent interactions between polypeptide chains result in remarkably stable fibrils, with high elastic moduli, comparable to that of collagen, keratin and silk. Moreover, amyloid fibrils stand out by their high aspect ratio, being on the micrometer scale for length, but on the nanometer scale for diameter [4]. The self-templating properties of fibrils provide them the ability to self-propagate upon fragmentation, with the successive growing of the resulting fragments; a phenomenon known as seeding, which spontaneously generate new nanostructures, without any energy requirement [5]. Additionally, they display high resistance to proteolytic digestion, heat or chemical denaturation and insolubility in non-ionic detergents.
Although the aggregation of amyloid proteins has been traditionally associated with pathology [6], in recent years the presence of functional amyloids implicated in relevant biological processes have also been evidenced. It is not yet well completely understood which are the molecular mechanisms which differentiate toxic amyloids from beneficial functional amyloids. However, because there are increasing evidences pointing towards small soluble oligomeric species as the species responsible of cellular toxicity [7,8], it is suggested that a specific assembly machinery prevents from the accumulation of toxic oligomeric intermediates during the buildup of functional amyloids.
The rigidity, strength and the high level of resistance to degradation make amyloid fibrils ideal structural materials and, not surprisingly, these functionalities are exploited by some organisms. Notably, biofilms secreted by some bacteria include significant amounts of amyloid fibrils. Biofilms play a functional role in the defense of microorganisms against chemical and mechanical attacks. For instance, the biofilm from Escherichia coli contains an amyloid fibrillar component named curli, derived from the assembly of the protein CsgA. In this particular case the assembly process is known to be tightly controlled by specific cofactors [9]. Likewise, amyloid fibrils have also been found in biofilms secreted by other bacteria like Salmonella and Pseudomonas [10,11]. In this last case, the amyloid fibrils contribute to increase by 20-fold the stiffness of the biofilm, the amyloid component being the major contributor to the mechanical robustness of the biofilm. Moreover, insects like silk-moths also exploit the properties of amyloids in the eggshells, where they provide mechanical and chemical protection against environmental hazards, while displaying a porous structure that allows gases exchange [12]. Functional amyloids are not restricted to lower eukaryotic organisms; for instance, the polymerization of the protein pmel17 into fibrils provides a scaffold for the maturation of melanin in humans [13] and amyloid formation by the CPEB protein family facilitates memory consolidation in Aplysia [14], Drosophila [15] and mouse [16].
The stunning properties of functional amyloids have inspired the use of this kind of assemblies to build up new nanomaterials. Their biocompatibility is intended to complement, or replace, metallic and organic polymers which frequently turn out to be more toxic, or less eco-friendly. In addition, the twenty amino acid code provide protein and peptide-based assemblies with a multifunctionality that cannot be attained by any other means. Literature reports many examples of amyloids applied in biomedicine, being used as vehicles for drug delivery [17], as cell culture scaffolds able to sustain cell growth and differentiation [18], and to produce biomimetic tissues, such as a biomimetic bone [19] among others. Furthermore, there are other novel biotechnological applications emerging such as underwater adhesive materials [20]; biodegradable nanocomposites with shape memory, tunable fluorescence, conductivity or sensing properties [21,22], nanowires [23,24], biosensors [25,26] and light-emitting diodes [27,28].
Prion and prion-like proteins are considered as a subset of amyloid proteins with the distinctive ability to self-template and transmit between cells. Originally, prions were discovered as the pathogenic transmissible proteinaceous agents [29] responsible for several mammalian neurodegenerative diseases such as Bovine spongiform encephalopathy and scrapie, as well as Creutzfeldt Jakob disease, Kuru and Fatal familiar insomnia in humans [30]. Nevertheless, as in the case of amyloids, nature has taken advantage of the prion self-propagating features for functional purposes. Some of the best characterized functional prions are the yeast Sup35, Ure2, Swi1 and Mot3 proteins, which act as epigenetic elements in the adaptation to environmental fluctuations [31–35]. Functional prion proteins are, however, not restricted to yeast, and they have been also described to act in bacteria [36,37] or in plants [38].
In contrast to the extensive effort devoted to the design of new amyloid-based nanomaterials, examples of prion-based nanomaterials are still scarce. Here we review the use of prion proteins as nanostructures and how their special properties make them unique building blocks for certain nanotechnological applications.
General properties of prion proteins
Functional prion proteins fulfill the extraordinary ability to interconvert between a soluble state and an aggregated β-amyloid state under certain stimulus or conditions, keeping their globular domains folded and still functional [39]. This ability usually relies on a disordered and low-complexity region known as the Prion Domain (PrD). PrDs are well studied in yeast, were they are characterized by a biased composition enriched in Asn, Gln, Tyr, Ser and Gly, which are essentially polar residues, being depleted in hydrophobic amino acids [40] (Figure 1). This is somehow surprising, because it is well-established that classical amyloid proteins present at least one region enriched in hydrophobic residues with a high intrinsic aggregation propensity which has been defined as the amyloid core [41], and it is the formation of hydrophobic contacts between amyloid cores that usually triggers the amyloid state transition. This kind of sequences are absent in prion proteins, probably because their high amyloid potency would shift the equilibrium soluble and aggregated forms towards the aggregated state, even in the absence of triggering environmental signals. Instead, the amyloid nucleating sequences of PrDs are dominated by N/Q/S/Y/G residues [42–44] and, thus they are necessarily weaker than those of pathogenic amyloids. Upon an initial, relatively weak, intermolecular interaction, it is the accumulation of numerous weak contacts along the usually long PrD that is considered critical to sustain the aggregated state [45–48].
Figure 1.

Representation of the natural amino acid frequency in prion sequences. The size of the letter represents the frequency of each particular amino acid.
The amyloid core of the yeast prion Sup35 protein (GNNQQNY) has been deeply characterized, and the interactions mediating the formation of the amyloid assembly elucidated by x-ray diffraction crystallography [49]. The structure revealed which are the relevant residue contacts in the assembly of the Sup35 cross-β spine, highlighting the importance of hydrogen bonding, van der Waals interactions and π-π stacking between intermolecular Tyr residues [50]. Additional data on the properties of PrDs nucleating sequences come from the study of human prion-like proteins, as the case of the fused in sarcoma protein (FUS). FUS is considered a prion-like protein because of its similarity in composition to that of classical yeast prions and its prion-like behavior, although no infective capacity has been described for its assembly. Two recent studies reported the atomic interactions of the two [S/G]-Y-[S/G] tandem repeats from FUS LC domain in the aggregated state. Both structures highlight the important role of Tyr π-π stacking in PrDs aggregation, supported by the evidence that substitution of Tyr by Ala completely abolished the assembly [51,52].
Nanomaterials based on prions
Compared to classical amyloids, prion and prion-like proteins possess exclusive properties, like a generally slow and tunable aggregation kinetics [1]. This characteristic is considered optimal for the design of materials where the aggregation rate becomes a relevant factor to control during the assembly. The aggregation of classical amyloids is much faster, less controllable, and, in the case of globular proteins, harsh conditions are usually required to induce aggregation. Notably, many functional prion proteins are considered to be dynamic and reversible amyloids, due to their ability to break down the previously described weak interactions, in response to different environmental factors such as temperature, ionic strength or dilution [53].
The potential advantages of prion assemblages have been barely capitalized. However, a number of recent studies already begin to reveal their large potentiality in nanotechnology. The PrD domains of Sup35, Ure2, HET-s and FUS have been used as building blocks to obtain nanowires [54], bifunctional nanomaterials for antigenic detection [55,56], for enzyme immobilization [57,58], as redox biofilms [59] or in the generation of dynamic hydrogels [60]. Next, we review in more detail the different strategies exploited to obtain these remarkable functionalities.
Scheibel and collaborators reported the first application based on a yeast prion protein [54]. They used the protein Sup35, a translation terminator factor with the ability to switch from the soluble state to the aggregated form in response to certain environmental stresses [31,32]. Sup35 has two adjacent regions considered as responsible for its ability to switch between the soluble and the aggregated states: the N terminal and middle region (regions NM) [61]. The authors used NM assembled fibrils as self-templating molecules in the production of gold nanowires that could be used for the construction of nanodevices. First, they were able to produce fibers of different lengths with good insulator properties by modifying the assembly conditions. Additionally, by introducing a K184C mutation they produce gold-containing prion-based nanowires, with a width ranging 80–200 nm, high conductivity and low resistance (R = 86 Ω) [54]. In this pioneering work, the versatility of prion-sequence inspired nanomaterials was already demonstrated, since fibers of different lengths could be obtained, just by playing with the aggregation conditions.
In a second application, the same Sup35 PrD was used in the obtention of bifunctional protein nanowires able to increase the sensitivity of ELISA immunoassays [55]. Sup35 PrD was fused to two different proteins: protein G, with the ability to bind specifically to the Fc part of antibodies, and methyl-parathion hydrolase (MPH), an enzyme which transforms its initial substrate into a yellow-colored product. These hybrid fibers, decorated with both protein G and MPH, were employed, in an ELISA-like strategy, for the detection of the F1 protein from Yersinia pestis, which was previously immobilized in a microplate and incubated with its specific IgG antibody. The bifunctional fibrils were used in substitution of the classical enzyme conjugated secondary antibody. As a result, a remarkable increase in sensitivity, near 100-fold, was achieved, compared to the traditional HRP-secondary antibody detection method. An additional improvement of this strategy was developed by the same researchers through the fusion of Sup35 PrD with the biotin acceptor peptide (BAP), which is auto-biotinylated in the presence of biotin holoenzyme synthetase (BirA) [56]. This modification allowed increasing up to 2000 to 4000-fold the sensitivity of the assay in comparison to conventional ELISA. This approach takes profit of the aggregation under mild conditions of Sup35 PrD, without impairing the folding of the fused globular proteins, and exemplifies the potentiality of bifunctional hybrid fibers.
Another yeast prion used as nanoscaffold was Ure2p. This protein is involved in nitrogen metabolism regulation in Saccharomyces cerevisiae [62]. The PrD of Ure2p, corresponding to the N terminus (residues 1 to 93) [63], has been used to generate stable nanostructures intended of the immobilization of active retrievable enzymes: alkaline phosphatase (AP) and horse radish peroxidase (HRP). The enzymes were completely functional in the assembled amyloid-like assembly and their activity before and after the polymerization exhibited indistinguishable kinetic parameters [57]. In a subsequent work, the authors were able to produce enzymatically active microgels, with a porous architecture, that allowed the diffusion of substrates and products using enzymatic flow-chemistry [58]. In this case, they pre-selected the final shape of the gel by encapsulating Ure2-AP solutions into small droplets (around 20 μm diameter) by microfluidic techniques; in such a way that, when the prion domain driven polymerization took place into the droplet, the final shape of the microgel was spherical. Thus, they attained the desired gel shape while maintaining intact the enzymatic activity. Again, this was only possible due to both the slow aggregation kinetics of Ure2 and its ability to polymerize under mild conditions. This strategy can be extended to control the size and shape of a variety of functionalized microgels.
Altamura and collaborators generated a protein only redox assembly, inspired in bacterial biofilms and based on the use of the HET-s prion protein from Podospora anserine and rubredoxin (Rd) [59]. Rubredoxins are a class of low-molecular-weight iron-containing proteins that perform one-electron transfer processes. They demonstrated that the artificial biofilm can act as a bionanowire, with different thick and electrochemical properties. The Rd-HET-s nanowire can transport electrons allowing on site oxygen reduction.
Fused in sarcoma (FUS) low-complexity (LC) domain is the only human prion-like protein which has been used so far to obtain a fibrillar self-assembled nanomaterial. FUS is an RNA-binding protein with an important role in RNA transcription, processing and transport, which contains a 200 residues-long region at its N terminus, known as low complexity (LC) domain, responsible for the transition towards hydrogel and amyloid states [64]. The amino acid composition of this LC domain resembles that of yeast PrDs. In this approach, the slow assembly kinetics of the LC domain and the weak nature of the intermolecular contacts were exploited to obtain reversible self-assembled nanoscaffolds [60]. To this aim, the FUS LC domain was fused to either eGFP, mCherry, mMAPLE3 and PAtagRFP. Two of the globular proteins, FUS LC-EGFP and FUS LC-mCherry did not significantly impact the solubility of the LC domain alone and slowly assembled into functional fluorescence fibers. In contrast, the fusions of FUS LC with the hydrophobic tags, mMAPLE3 and PAtagRFP, aggregated faster, yielded significantly fewer fibers and precipitated at high concentration, stressing again the importance of a kinetic control of the assembly. Notably, FUS LC-eGFP and FUS LC-mCherry proteins assembled in hydrogels which could be depolymerized by increasing the temperature. Once more, hybrid hydrogels could be obtained just by combining different fusion of FUS LC proteins. The fact that varied functional domains or moieties can be easily assembled into the same nanostructure opens the door for a wide range of applications, like space confined multicatalytic reactions.
Extensive work has been devoted to understand the compositional determinants of prion domains [46,47,65] and the relevant interactions driving their amyloid formation [50–52]. Nevertheless, the de novo design of prion-inspired self-assembling peptides for nanotechnology applications remained essentially unexplored. To fill this gap we rationally designed four minimalist polar binary patterned peptides bioinspired on prion sequences and exploited them for the obtention of biomaterials with unique properties [66]. The design combined the five most enriched residues in PrDs, Asn, Gln, Ser, Gly and Tyr in a binary pattern, with only seven residues in length, resulting in four heptapeptides: NYNYNYN, QYQYQYQ, SYSYSYS and GYGYGYG. The central Tyr residue was intended to establish the characteristic π-π interactions, where the rest of residues provided the polar context and the rest of weak interactions. Thus, this minimal sequences mimic in different aspects the much larger PrDs used in the works described above. Effectively, the peptides showed the ability to slowly self-assemble into highly ordered amyloid fibrils under physiological conditions, none of them displaying cellular toxicity. It is worth to remark here that the high density of Tyr residues in these peptides allowed us to exploit the Di-Tyr covalent cross-link, which play an important role in overstabilization of protein scaffolds in nature [67] and also in pathologies such as Alzheimer [68] or Parkinson [69]. The controlled formation of intermolecular Di-Tyr cross-links, after the self-assembly reaction, resulted in fibrillar structures that were exceptionally stable in front of chemical denaturation. We also took advantage of the bioreductive properties of Tyr residues to induce the decoration of the fibrillar material with silver nanoparticles, resulting in bio-metallic nanowires with potential applications in electronics.
One potential application of a Tyr containing fibril is to act as a redox-active scaffold for developing an enzyme mimetic catalyst [70]. Monomeric tyrosyl radicals are easily quenched in aqueous solution. However, the chemical environment in prion-inspired amyloid assemblies might endorse their Tyr radicals with reduction potentials high enough to catalyze chemical reactions. We assessed the electrocatalytic ability of our four self-assembled nanostructures by exposing them to copper (II) as an oxidizing agent, in the presence of distilled pyrrole vapor. The fibrils catalyzed efficiently the polymerization of individual pyrrol moieties into a macromolecular polypyrrole (PPy) material (Figure 2) [66]. Importantly, not all the peptides exhibited the same electrocatalytic activity, which indicated that this property was sequence dependent and, accordingly, that it can be potentially tuned to develop synthetic prion-inspired nanodevices with different catalytic properties.
Figure 2.
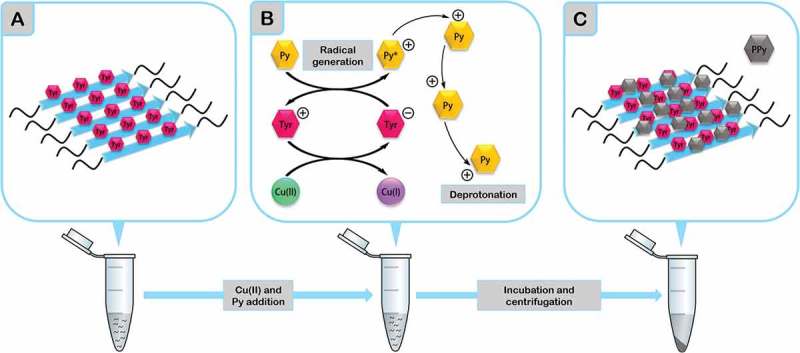
Schematic representation of the catalytic ability of synthetic prion-inspired peptides. A) Self-assembled peptides are centrifuged to recover just the fibrillar material after incubation. B) Copper (II) (in green) and distilled pyrrole vapor (yellow hexagons tagged as Py) are added to the fibrils and incubated for 24 hours. During this incubation, copper (II) is reduced to copper (I) subtracting electrons to Tyr residues, which at the same time receive electrons from Py. This reaction generates Py+ radicals, triggering Py deprotonation and C) inducing its polymerization into polypyrrole (PPy, grey hexagon). PPy becomes a grey insoluble precipitate which deposits specifically over the fibrils.
Why prion-based instead of other protein-based nanomaterials?
In this review we illustrated with a number of examples why the use of either natural or artificially designed prion-sequences for the obtention of new nanomaterials is becoming increasingly attractive. In the first place, prion proteins are essentially polar and soluble in many instances, but at the same time, they have the ability to convert into to the fibrillar state under specific conditions. In second place, the aggregated state is reversible under certain conditions, offering the possibility to design tunable materials that can be polymerized or depolymerized on demand. This feature can be exploited, for instance, in the obtention of nanomaterials which functionality can be controlled by switching the temperature, the ionic strength or the pH of the solution, Nevertheless, if a more stable fibrillar structure is required, this reversibility can be easily tuned off introducing post-assembly covalent interactions, either by including Cys residues in the sequence, which allows disulfide bridge formation [70] or by artificially inducing post-translational modifications such as Di-Tyr cross-links [66]. It is also important to highlight that, in contrast to the harsh conditions usually employed to induce the partial unfolding of classical amyloid proteins, the transition from the soluble to the fibrillar state in natural, but also in designed prionic sequences occurs under mild conditions. This property, together with the modularity of functional prion protein provides a unique context for the creation of materials in which these domains are fused to other proteins with multiple and specific functionalities. Last, but not least, the slower aggregation kinetics of prion sequences, when compared with classical amyloids, allows to sustain the correct fold of globular domains upon aggregation, to control the formation of nanofibrillar structures, ranging from single fibers and fiber networks to bulky hydrogels, and the possibility to model their final shape.
An important hallmark of prion domains is their high content of Tyr residues, that provide their assemblies with an intrinsic catalytic activity which can be modulated by changes in its chemical environment. This is exactly what happens in the catalytic sites of many enzymes, which suggests that prion-based assemblies might find applications in the development of novel enzymatic activities.
As a general conclusion, we are convinced that prion sequence-based biocompatible nanomaterials constitute worth to explore alternatives to classical amyloid-based ones. However, before we can translate these nanostructures into tangible technological applications we have to ensure that they can be fabricated in a large-scale, while maintaining their amazing properties.
Funding Statement
This work was funded by the Spanish Ministry of Economy and Competitiveness BIO2016-783-78310-R to S.V and by ICREA, ICREA-Academia 2015 to S.V. M. D.-C. was supported by the Spanish Ministry of Science and Innovation via a doctoral grant (FPU14/05786).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- [1].Knowles TPJ, Mezzenga R.. Amyloid fibrils as building blocks for natural and artificial functional materials. Adv Mater. 2016;28:6546–6561. [DOI] [PubMed] [Google Scholar]
- [2].Wei G, Su Z, Reynolds NP, et al. Self-assembling peptide and protein amyloids: from structure to tailored function in nanotechnology. Chem Soc Rev. 2017;46:4661–4708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Jahn TR, Makin OS, Morris KL, et al. The common architecture of cross-β amyloid. J Mol Biol. 2010;395:717–727. [DOI] [PubMed] [Google Scholar]
- [4].Knowles TPJ, Buehler MJ. Nanomechanics of functional and pathological amyloid materials. Nat Nanotechnol. 2011;6:469–479. [DOI] [PubMed] [Google Scholar]
- [5].Tanaka M, Collins SR, Toyama BH, et al. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. [DOI] [PubMed] [Google Scholar]
- [6].Bucciantini M, Giannoni E, Chiti F, et al. Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature. 2002;416:507–511. [DOI] [PubMed] [Google Scholar]
- [7].Walsh DM, Klyubin I, Fadeeva JV, et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416:535–539. [DOI] [PubMed] [Google Scholar]
- [8].Lashuel HA, Hartley D, Petre BM, et al. Neurodegenerative disease: amyloid pores from pathogenic mutations. Nature. 2002;418:291. [DOI] [PubMed] [Google Scholar]
- [9].Chapman MR, Robinson LS, Pinkner JS, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Collinson SK, Clouthier SC, Doran JL, et al. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zeng G, Vad BS, Dueholm MS, et al. Functional bacterial amyloid increases Pseudomonas biofilm hydrophobicity and stiffness. Front Microbiol. 2015;6:1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Iconomidou VA, Vriend G, Hamodrakas SJ. Amyloids protect the silkmoth oocyte and embryo. FEBS Lett. 2000;479:141–145. [DOI] [PubMed] [Google Scholar]
- [13].Fowler DM, Koulov AV, Alory-Jost C, et al. Functional amyloid formation within mammalian tissue. PLoS Biol. 2006;4:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Si K, Lindquist S, Kandel ER. A neuronal isoform of the aplysia CPEB has prion-like properties. Cell. 2003;115:879–891. [DOI] [PubMed] [Google Scholar]
- [15].Majumdar A, Cesario WC, White-Grindley E, et al. Critical role of amyloid-like oligomers of Drosophila Orb2 in the persistence of memory. Cell. 2012;148:515–529. [DOI] [PubMed] [Google Scholar]
- [16].Fioriti L, Myers C, Huang -Y-Y, et al. The persistence of hippocampal-based memory requires protein synthesis mediated by the prion-like protein CPEB3. Neuron. 2015;86:1433–1448. [DOI] [PubMed] [Google Scholar]
- [17].Silva RF, Araújo DR, Silva ER, et al. L-Diphenylalanine microtubes as a potential drug-delivery system: characterization, release kinetics, and cytotoxicity. Langmuir. 2013;29:10205–10212. [DOI] [PubMed] [Google Scholar]
- [18].Ruan H, Xiao R, Jiang X, et al. Biofunctionalized self-assembly of peptide amphiphile induces the differentiation of bone marrow mesenchymal stem cells into neural cells. Mol Cell Biochem. 2018;1–9. doi: 10.1007/s11010-018-3386-9. [DOI] [PubMed] [Google Scholar]
- [19].Li C, Born AK, Schweizer T, et al. Amyloid-hydroxyapatite bone biomimetic composites. Adv Mater. 2014;26:3207–3212. [DOI] [PubMed] [Google Scholar]
- [20].Zhong C, Gurry T, Cheng AA, et al. Strong underwater adhesives made by self-assembling multi-protein nanofibres. Nat Nanotechnol. 2014;9:858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Li C, Adamcik J, Mezzenga R. Biodegradable nanocomposites of amyloid fibrils and graphene with shape-memory and enzyme-sensing properties. Nat Nanotechnol. 2012;7:421–427. [DOI] [PubMed] [Google Scholar]
- [22].Li C, Bolisetty S, Mezzenga R. Hybrid nanocomposites of gold single-crystal platelets and amyloid fibrils with tunable fluorescence, conductivity, and sensing properties. Adv Mater. 2013;25:3694–3700. [DOI] [PubMed] [Google Scholar]
- [23].Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300:625–627. [DOI] [PubMed] [Google Scholar]
- [24].Sakai H, Watanabe K, Kudoh F, et al. Patterning nanofibrils through the templated growth of multiple modified amyloid peptides. Sci Rep. 2016;6:31993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Sasso L, Suei S, Domigan L, et al. Versatile multi-functionalization of protein nanofibrils for biosensor applications. Nanoscale. 2014;6:1629–1634. [DOI] [PubMed] [Google Scholar]
- [26].Wu X, Li M, Li Z, et al. Amyloid-graphene oxide as immobilization platform of Au nanocatalysts and enzymes for improved glucose-sensing activity. J Colloid Interface Sci. 2017;490:336–342. [DOI] [PubMed] [Google Scholar]
- [27].Rizzo A, Solin N, Lindgren LJ, et al. White light with phosphorescent protein fibrils in OLEDs. Nano Lett. 2010;10:2225–2230. [DOI] [PubMed] [Google Scholar]
- [28].Onur T, Yuca E, Olmez TT, et al. Self-assembly of bacterial amyloid protein nanomaterials on solid surfaces. J Colloid Interface Sci. 2018;520:145–154. [DOI] [PubMed] [Google Scholar]
- [29].Prusiner SB. Molecular biology of prion diseases. Science. 1991;252:1515–1522. [DOI] [PubMed] [Google Scholar]
- [30].Prions: SK. What are they good for? Annu Rev Cell Dev Biol. 2015;31:149–169. [DOI] [PubMed] [Google Scholar]
- [31].Serio TR, Lindquist SL. The yeast prion [PSI+]: molecular insights and functional consequences. Adv Protein Chem. 2001;59:391–412. [DOI] [PubMed] [Google Scholar]
- [32].Franzmann TM, Jahnel M, Pozniakovsky A, et al. Phase separation of a yeast prion protein promotes cellular fitness. Science. 2018;359:eaao5654. [DOI] [PubMed] [Google Scholar]
- [33].Wickner RB. [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. [DOI] [PubMed] [Google Scholar]
- [34].Du Z, Park K-W, Yu H, et al. Newly identified prion linked to the chromatin-remodeling factor Swi1 in Saccharomyces cerevisiae. Nat Genet. 2008;40:460–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Holmes DL, Lancaster AK, Lindquist S, et al. Heritable remodeling of yeast multicellularity by an environmentally responsive prion. Cell. 2013;153:153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pallarès I, Ventura S. The transcription terminator Rho: A first bacterial prion. Trends Microbiol. 2017;25:434–437. [DOI] [PubMed] [Google Scholar]
- [37].Shahnawaz M, Park K-W, Mukherjee A, et al. Prion-like characteristics of the bacterial protein Microcin E492. Sci Rep. 2017;7:45720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chakrabortee S, Kayatekin C, Newby GA, et al. Luminidependens (LD) is an Arabidopsis protein with prion behavior. Proc Natl Acad Sci U S A. 2016;113:201604478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Wickner RB, Shewmaker FP, Bateman DA, et al. Yeast prions: structure, biology, and prion-handling systems. Microbiol Mol Biol Rev. 2015;79:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Uptain SM, Lindquist S. Prions as protein-based genetic elements. Annu Rev Microbiol. 2002;56:703–741. [DOI] [PubMed] [Google Scholar]
- [41].Pastor MT, Esteras-Chopo A, Serrano L. Hacking the code of amyloid formation: the amyloid stretch hypothesis. Prion. 2007;1:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Schwartz JC, Wang X, Podell ER, et al. RNA seeds higher-order assembly of FUS protein. Cell Rep. 2013;5:918–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2014;130:130–133. [DOI] [PubMed] [Google Scholar]
- [44].Sabate R, Rousseau F, Schymkowitz J, et al. Amyloids or prions? That is the question. Prion. 2015;9:200–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Ross ED, Edskes HK, Terry MJ, et al. Primary sequence independence for prion formation. Proc Natl Acad Sci. 2005;102:12825–12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Toombs JA, McCarty BR, Ross ED. Compositional determinants of prion formation in yeast. Mol Cell Biol. 2010;30:319–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Alberti S, Halfmann R, King O, et al. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137:146–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Toombs JA, Petri M, Paul KR, et al. De novo design of synthetic prion domains. Proc Natl Acad Sci. 2012;109:6519–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nelson R, Sawaya MR, Balbirnie M, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Cuesta IG, Sánchez De Merás AMJ. Energy interactions in amyloid-like fibrils from NNQQNY. Phys Chem Chem Phys. 2014;16:4369–4377. [DOI] [PubMed] [Google Scholar]
- [51].Luo F, Gui X, Zhou H, et al. Atomic structures of FUS LC domain segments reveal bases for reversible amyloid fibril formation. Nat Struct Mol Biol. 2018;25:341–346. [DOI] [PubMed] [Google Scholar]
- [52].Hughes MP, Sawaya MR, Boyer DR, et al. Atomic structures of low-complexity protein segments reveal kinked b sheets that assemble networks. Science. 2018;701:698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kato M, Han TW, Xie S, et al. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 2012;149:753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Scheibel T, Parthasarathy R, Sawicki G, et al. Conducting nanowires built by controlled self-assembly of amyloid fibers and selective metal deposition. Proc Natl Acad Sci. 2003;100:4527–4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Men D, Guo YC, Zhang ZP, et al. Seeding-induced self-assembling protein nanowires dramatically increase the sensitivity of immunoassays. Nano Lett. 2009;9:2246–2250. [DOI] [PubMed] [Google Scholar]
- [56].Men D, Zhang ZP, Guo YC, et al. An auto-biotinylated bifunctional protein nanowire for ultra-sensitive molecular biosensing. Biosens Bioelectron. 2010;26:1137–1141. [DOI] [PubMed] [Google Scholar]
- [57].Zhou XM, Entwistle A, Zhang H, et al. Self-assembly of amyloid fibrils that display active enzymes. ChemCatChem. 2014;6:1961–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zhou X, Shimanovich U, Herling TW, et al. Enzymatically active microgels from self-assembling protein nanofibrils for microflow chemistry. ACS Nano. 2015;9:5772–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Altamura L, Horvath C, Rengaraj S, et al. A synthetic redox biofilm made from metalloprotein-prion domain chimera nanowires. Nat Chem. 2017;9:157–163. [DOI] [PubMed] [Google Scholar]
- [60].An B, Wang X, Cui M, et al. Diverse supramolecular nanofiber networks assembled by functional low-complexity domains. ACS Nano. 2017;11:6985–6995. [DOI] [PubMed] [Google Scholar]
- [61].Kushnirov V, Ter-Avanesyan M, Didichenko S, et al. Divergence and conservation of SUP2 (SUP35) gene of yeast Pichia pinus and Saccharomyces cerevisiae. Yeast. 1990;6:461–472. [DOI] [PubMed] [Google Scholar]
- [62].Bai M, Zhou JM, Perrett S. The yeast prion protein Ure2 shows glutathione peroxidase activity in both native and fibrillar forms. J Biol Chem. 2004;279:50025–50030. [DOI] [PubMed] [Google Scholar]
- [63].Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. [DOI] [PubMed] [Google Scholar]
- [64].Sun Z, Diaz Z, Fang X, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Sabate R, Rousseau F, Schymkowitz J, et al. What makes a protein sequence a prion?. PLoS Comput Biol. 2015;11:e1004013. doi: 10.1371/journal.pcbi.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Diaz Caballero M, Navarro S, Fuentes I, et al. Minimalist prion-inspired polar self-assembling peptides. ACS Nano. 2018;12:5394–5407. [DOI] [PubMed] [Google Scholar]
- [67].Partlow BP, Applegate MB, Omenetto FG, et al. Dityrosine cross-linking in designing biomaterials. ACS Biomater Sci Eng. 2016;2:2108–2121. [DOI] [PubMed] [Google Scholar]
- [68].Yang J, Yang J, Liang SH, et al. Imaging hydrogen peroxide in Alzheimer’s disease via cascade signal amplification. Sci Rep. 2016;6:35613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wördehoff MM, Shaykhalishahi H, Groß L, et al. Opposed effects of dityrosine formation in soluble and aggregated α-synuclein on fibril growth. J Mol Biol. 2017;429:3018–3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Jang HS, Lee JHJ, Park YS, et al. Tyrosine-mediated two-dimensional peptide assembly and its role as a bio-inspired catalytic scaffold. Nat Commun. 2014;5:1–11. [DOI] [PubMed] [Google Scholar]


