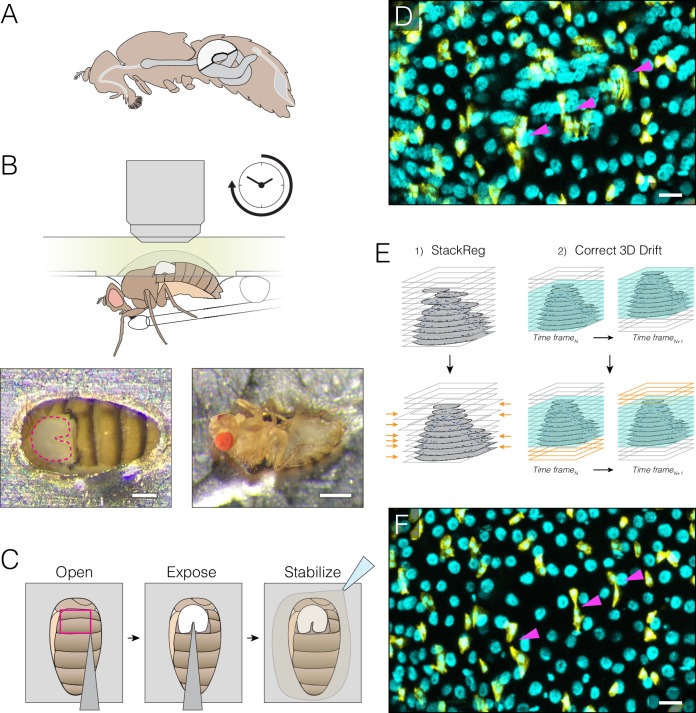
Figure 1. Extended imaging of the midgut in live Drosophila adults.
(A) Adult female midgut in situ, sagittal view. The white highlighted area indicates region R4a-b, also known as P1-2, (Buchon et al., 2013a; Marianes and Spradling, 2013)) of the midgut that will be exposed for imaging. (B–C) The midgut is accessed through a small cuticular window cut in the back of a live animal. (B) (Top) Schematic of the imaging apparatus. The animal is affixed to a modified petri dish ‘mount’. The chamber of the mount contains media. The underside of the mount supports a feeder tube. See and Fig. 1-fig. supplement 2. (Bottom) Dorsal (left) and ventral (right) views of an animal in the mount. In the left panel, the exposed midgut is outlined by the magenta dotted line. Scale bars: 0.25 mm (left), 0.5 mm (right). See Video 4. (C), Steps in preparing the midgut for imaging. See Video 1 tutorial. (D–F) Registration macros are applied post-acquisition to correct the blurring caused by tissue movements. (D), Before registration, blurring and duplications (arrowheads) are evident. This panel is a raw z-series projection of one movie time point. (E), During registration, two ImageJ plugins are applied in series. (1) 'StackReg' corrects for tissue movement during z-stack acquisition at a single time point. (2) 'Correct 3D Drift' corrects for global volume movements over multiple time points. (F), After registration, blurring and duplications are negligible. Cyan, all nuclei (ubi-his2av::mRFP); yellow, stem cells and enteroblasts (esg >LifeactGFP). Scale bars, 20 μm. See Video 6.
Figure 1—figure supplement 1. Mounts for upright, inverted and light-sheet microscopes.
Figure 1—figure supplement 2. Specifications for abdomen cutouts.
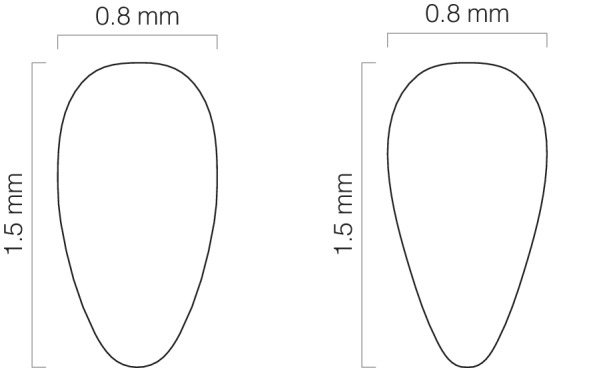
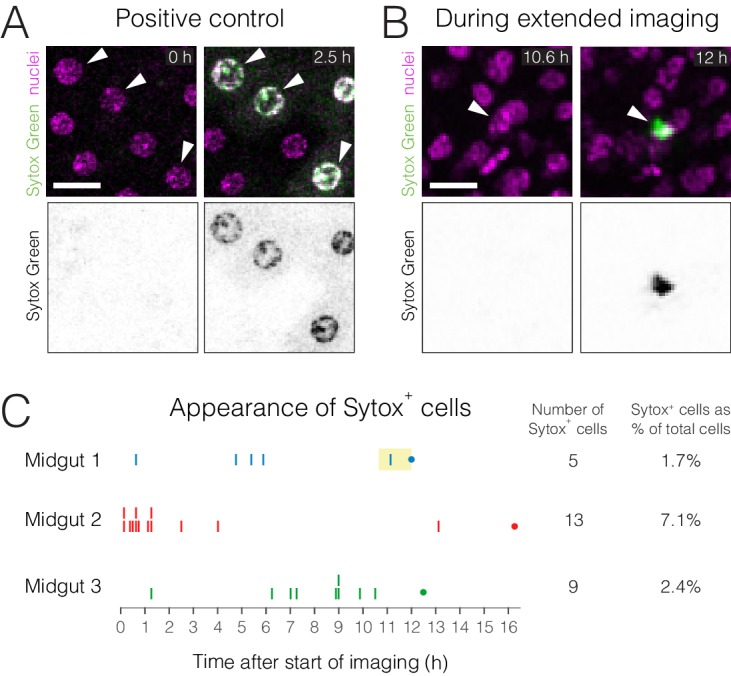
Figure 1—figure supplement 3. Cell viability during extended imaging.