Abstract
The tumor microenvironment (TME) represents a complex interplay between different cellular components, including tumor cells and cancer stem cells (CSCs), with the associated stroma; such interaction promotes tumor immune escape and sustains tumor growth. Several experimental approaches for cancer therapy are focused on TME remodeling, resulting in increased antitumor effects. We previously demonstrated that the hyaluronan synthesis inhibitor 4-methylumbelliferone (4Mu) decreases liver fibrosis and induces antitumor activity in hepatocellular carcinoma (HCC). In this work, 4Mu, in combination with an adenovirus encoding interleukin-12 genes (AdIL-12), elicited a potent antitumor effect and significantly prolonged animal survival (p < 0.05) in an orthotopic HCC model established in fibrotic livers. In assessing the presence of CSCs, we found reduced mRNA levels of CD133+, CD90+, EpCAM+, CD44+, and CD13+ CSC markers within HCC tumors (p < 0.01). Additionally, 4Mu downregulated the expression of the CSC marker CD47+ on HCC cells, promoted phagocytosis by antigen-presenting cells, and, combined with Ad-IL12, elicited a potent cytotoxic-specific T cell response. Finally, animal survival was increased when CD133low HCC cells, generated upon 4Mu treatment, were injected in a metastatic HCC model. In conclusion, the combined strategy ameliorates HCC aggressiveness by targeting CSCs and as a result of the induction of anticancer immunity.
Keywords: 4 methylumbiliferone, cancer stem cells, hepatocellular carcinoma, interleukin 12-based gene therapy, antitumoral immune response
The tumor microenvironment mediates resistance to different therapeutic approaches, including immunotherapy. In this issue of Molecular Therapy, Mazzolini et al. showed that tumor remodeling using 4-methylumbelliferone (4Mu) ameliorated hepatocellular carcinoma aggressiveness by targeting cancer stem cells and increasing antitumor immune response induced by gene therapy of interleukin-12.
Introduction
Hepatocellular carcinoma (HCC) is the third most common cause of cancer-related death worldwide,1 and its incidence and mortality are steadily increasing in Western countries.2, 3 Unfortunately, the number of new HCC cases being diagnosed each year is equal to the number of deaths from this cancer.4 Liver resection, transplantation, and tumor ablation are considered curative options, although they are applied in only 30%–40% of patients. HCC diagnosed at advanced stages has a dismal prognosis. The multikinase inhibitors sorafenib, in first line, and regorafenib, in second line, have been approved for advanced HCC,5, 6 although with modest impact on patient survival.
Immunotherapy of cancer is gaining interest and impact on patient overall survival in many solid tumors.7 In the field of HCC, nivolumab (anti-PD-1 monoclonal antibody) in second line appears to be the most promising strategy, as was observed by El-Khoueiry et al. in a phase 1/2 clinical trial.8 It is possible to increase the efficacy of checkpoint inhibitors through their combination with locoregional therapies, changing the tumor microenvironment (TME), which may have a synergistic effect or act upon mechanisms of primary or acquired resistance to checkpoint inhibitors.9 The TME contributes to cancer development in many solid tumors, including HCC.10 The HCC microenvironment constitutes a complex mixture of soluble factors, endothelial cells, hepatic stellate cells, tumor-associated macrophages (TAMs), regulatory T cells, cancer-associated fibroblasts, and cancer stem cells (CSCs) mixed within an excessive accumulation of extracellular matrix (ECM).11, 12 It has been demonstrated that TME components interact and may cooperate to prolong and sustain tumor dissemination.13
CSCs are defined as tumor-initiating cells with self-renewal and high proliferative capacity and with the ability to drive and maintain tumor growth.14 It has been proposed that HCC recurrence is favored, at least in part, by CSCs.15 CSCs have been also described as resistant to chemotherapy.16 CSCs are characterized by the presence of CSC-specific markers such as CD133, CD44, CD90, epithelial cell adhesion molecule (EpCAM), CD13, and CD47, which are involved in their capacity to initiate tumor growth, to form colonies, and to evade the immune system.17, 18, 19, 20 The TME acts as a fertile environment to grow the cancer seeds, and this is the reason that therapies directed to TME are gaining much attention by many scientists.21 We have previously reported that 4-methylumbelliferone (4Mu) selectively inhibits the synthesis of hyaluronan, a principal component of ECM, leading to the control of HCC growth in mice with advanced fibrosis,22 due in part to the inhibition of angiogenesis and reduction of IL-6 secretion by Kupffer cells.23 TME is also an interesting target for anticancer immunotherapy, and this is of particular importance considering the promising results obtained on HCC by immune checkpoint inhibitors.24, 25, 26 In addition to therapies that disrupt negative signaling mechanisms, immunotherapies destined to recruit immune effector cells into the tumors result in very attractive tools,27 since tumor-infiltrating T cells have been found to be functionally compromised in patients with HCC.28
The activation of effector immune cells is a key to mediate antitumor responses. Interleukin (IL)-12 induces the release of interferon gamma (IFNγ) and promotes antitumor activity.29 We have demonstrated that an adenovirus expressing IL-12 genes (AdIL-12) has an important antitumor effect.30 The efficacy of AdIL-12 therapy is, in part, due to its capability to modify different components of TME. IL-12 facilitates the recruitment of lymphocytes to the tumor, activates the specific cytotoxic T lymphocyte (CTL) response, and inhibits regulatory T cells and myeloid-suppressor-derived cells.31, 32
All these features make the HCC microenvironment an attractive target for cancer immunotherapy. Therefore, the aim of this work was to study whether 4Mu, by remodeling TME, could potentiate the immunotherapeutic effects generated by AdIL-12 in a murine model of HCC associated with fibrosis.
Results
4Mu+AdIl-12 Inhibits Tumor Growth in an Orthotopic Model of HCC Established in Fibrotic Livers
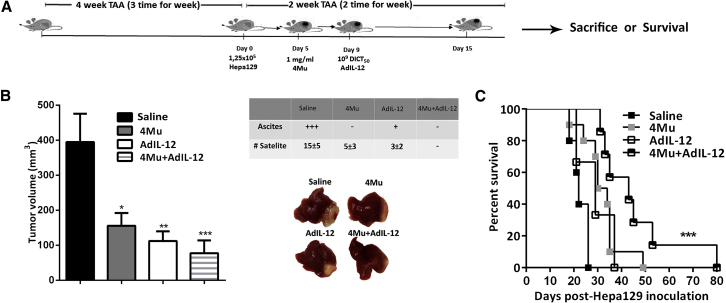
To assess the effects of 4Mu+AdIL-12 combined therapy on HCC tumor growth, we established an orthotopic tumor model associated with advanced fibrosis induced by thioacetamide (TAA) (Figure 1A). Tumor size from control (n = 8), 4Mu-treated (n = 8), AdIL-12-treated (n = 8), and 4Mu+AdIL-12-treated (n = 8) mice was analyzed after 2 weeks of Hepa 129 cell inoculation. 4Mu- or AdIL-12-treated mice showed a slight inhibition of HCC tumor growth in comparison with untreated mice (158 ± 36 mm3 and 112 ± 28 mm3 versus 395 ± 81 mm3; p < 0.05 and p < 0.01, respectively; Figure 1B). In addition, combined treatment induced a more potent decrease of HCC tumor volume (77 ± 37 mm3; p < 0.001) and significantly reduced the amount of carcinomatous ascites as well as the number of satellites nodules (Figure 1B, table) when compared with untreated mice. Survival of mice receiving 4Mu+AdIL-12 (n = 7) was significantly increased compared with mice receiving each single treatment (n = 6 per group) or saline (n = 5) (Figure 1C; p < 0.001).
Figure 1.
4Mu+AdIL-12 Inhibits Tumor Growth in an Orthotopic Model of HCC with Associated Liver Fibrosis
(A) Experimental model: 6- to 8-week-old male C3Hj/He mice were injected with thioacetamide (TAA; 200 mg/kg) for 4 weeks, 3 times per week, to induce fibrosis (injections i.p.). Mice received an intrahepatic inoculation of 1.25 × 105 syngeneic Hepa 129 cells (day 0). On day 5 after tumor implantation, mice were distributed in experimental groups (n = 8 per group) and received: (1) saline (control); (2) 4Mu, 200 mg/kg in drinking water; (3) AdIL-12, 1 × 109 DICT50 per milliliter, i.v., at day 7; (4) 4Mu+AdIL-12, 1 × 109 DICT50 per milliliter; and (5) 4Mu+Adβ-gal, 1 × 109 DICT50 per milliliter, i.v., at day 7. (B) Antitumoral efficacy of 4Mu+AdIL-12 on Hepa 129 tumors: mice were monitored, and tumor volumes were measured 15 days after Hepa 129 implantation. Tumor satellites were measured for each group (see table). *p < 0.05, saline versus 4Mu; **p < 0.01, saline versus AdIL-12; and ***p < 0.001, saline versus 4Mu+AdIL-12, one-way ANOVA and Tukey’s multiple comparisons test. Data from 3 independent experiments are expressed as the mean ± SEM. (C) Animal survival. ***p < 0.001, Kaplan-Meier, log-rank test. The experiment was carried out 2 times.
Combined Therapy Enhances Tumor-Infiltrating T Cells and Elicits a Potent Cytotoxic-Specific T Cell Response
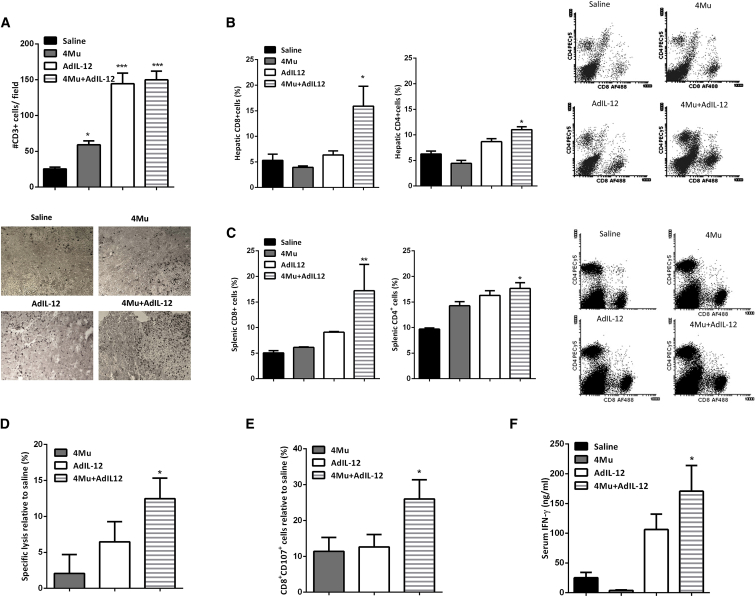
IL-12 antitumor activities are mediated by the activation of natural killer (NK) and T lymphocytes to secrete IFN-γ.33 First, we analyzed the presence of immune effector cells by immunohistochemistry in liver tumor sections. Only mice that received 4Mu and AdIL-12 in combination showed increased infiltrating CD3+ T cells within HCC tumors (p < 0.001; Figure 2A). In addition, the rate of CD8+ and CD4+ T cells was determined in liver tumor samples and spleen. The percentage of CD8+ T cells from total live cells in control mice was found to be about 5.3 ± 1.2% and 5.1 ± 0.4% (liver and spleen), respectively, while the percentage of CD8+ T cells in 4Mu+AdIL-12-treated mice was significantly higher: 16 ± 3.8% (hepatic) and 17 ± 5.1% (splenic), respectively; p < 0.05 versus saline (Figures 2B and 2C, left). Regarding CD4+ T cells, the percentage was 6.2 ± 0.6% and 9.7 ± 0.2% from total live cells within the liver and spleen, respectively, in control mice. However, the proportion of CD4+ T cells in the liver and spleen of 4Mu+AdIL-12-treated mice was significantly increased (11 ± 0.6% and 18 ± 1.1%, respectively; p < 0.05 versus saline) (Figures 2B and 2C, right).
Figure 2.
Combined Therapy Promotes Tumor-Infiltrating T Cells and Induces a Potent Cytotoxic-Specific T Cell Response
(A) Combined therapy showed higher levels of tumor-infiltrating CD3+ T cells. *p < 0.05, saline versus 4Mu; ***p < 0.001, saline versus 4Mu+AdIL-12; and ***p < 0.001, saline versus AdIL2, one-way ANOVA and Tukey’s multiple comparisons test. (B) Percentage of CD8+ and CD4+ T cells in liver tissue. *p < 0.05, saline versus 4Mu+AdIL12; one-way ANOVA and Tukey’s multiple comparisons test. (C) Levels of CD8+ and CD4+ T cells of total live cells in spleen of mice. *p < 0.05, saline versus 4Mu+AdIL-12; **p < 0.01, saline versus 4Mu+AdIL-12, one-way ANOVA and Tukey’s multiple comparisons test. (D) Splenocytes derived from 4Mu+AdIL12-treated mice exerted a potent CTL activity relative to that of the saline group against Hepa 129 cells, determined by LDH release. *p < 0.05, 4Mu+AdIL12, one-way ANOVA and Tukey’s multiple comparisons test. (E) Splenocytes derived from 4Mu+AdIL12-treated mice exerted a potent CTL activity relative to that of the saline group against Hepa 129 cells, determined by CD107 expression. *p < 0.05 for 4Mu+AdIL12, one-way ANOVA and Tukey’s multiple comparisons test. (F) Quantification of serum interferon gamma (IFN-γ) levels by ELISA; mice treated with combined therapy showed higher levels of IFN-γ; *p < 0.05 for saline versus 4Mu+AdIL12, one-way ANOVA and Tukey’s multiple comparisons test. Data are expressed as the mean ± SEM.
We next investigated whether the antitumor effect induced by 4Mu and AdIL-12 was mediated by the activation of CTLs. A slight CTL activity against Hepa 129 cells was elicited in mice from 4Mu or AdIL-12 treatment alone; however, combined treatment displayed a significantly higher CTL response against Hepa 129 (p < 0.05; Figures 2D and 2E). In addition, IFN-γ serum levels in 4Mu+AdIL-12-treated mice were significantly higher in comparison with those levels in the other groups (p < 0.001; Figure 2F).
4Mu Treatment Stimulates Phagocytosis of HCC Cells by Antigen-Presenting Cells and Potentiates the Antitumor Response Triggered by AdIL-12
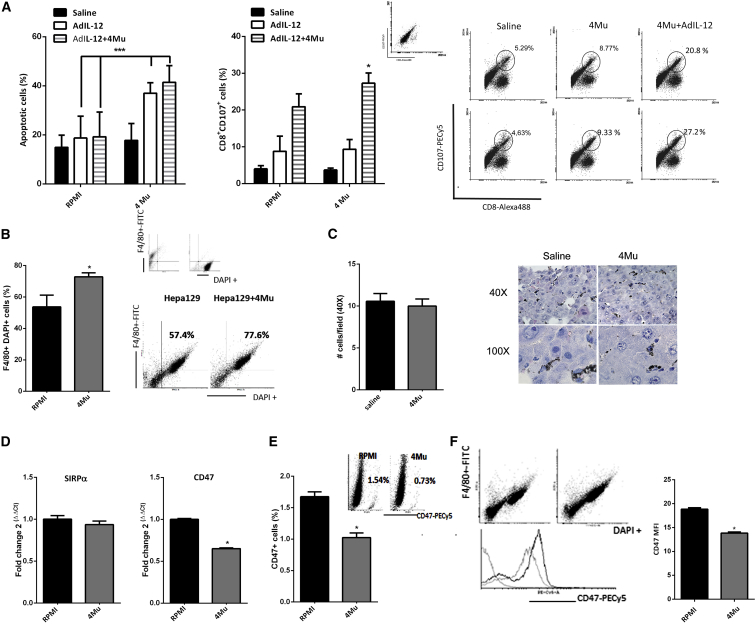
The antitumor immune response induced by 4Mu+AdIL-12 was superior to that of AdIL-12 alone. To investigate why 4Mu has the ability to boost CTL response, we first in vitro re-stimulated splenocytes from control, AdIL-12, or 4Mu+AdIL-12 groups with Hepa 129 cells previously exposed to 0.5 mM 4Mu for 72 hr. According to our previous data, 4Mu did not induce apoptosis in Hepa 129 cells at this dose.23 On day 5, splenocytes were harvested and added as effector cells, while Hepa 129 cells alone or pre-treated with 4Mu were used as target cells. When Hepa 129 cells were exposed to control, AdIL-12- or AdIL-12+4Mu-treated splenocytes, the percentages of apoptotic cells were 14 ± 2.0%, 18 ± 4.0%, and 19 ± 2.0%, respectively (Figure 3A, left). When Hepa 129 cells were pre-treated with 4Mu and exposed to splenocytes from control mice, the percentage of apoptotic cells was similar to that of Hepa 129 cells (17 ± 1.0%); however, when 4Mu-pretreated Hepa 129 cells were exposed to splenocytes derived from AdIL-12 or AdIL-12+4Mu groups, more apoptotic events were observed (37 ± 5.2% and 42 ± 3.5%, respectively; ***p ≥ 0.01, Kruskal-Wallis test). Similar results were obtained when we evaluated CTL activity (by CD107 expression on effector cells) against Hepa 129 or Hepa 129 pre-treated with 4Mu. When splenocytes from control mice were exposed to Hepa 129 cells, the percentage of degranulating T cells (CD8+CD107+) was similar to that for splenocytes from mice exposed to Hepa 129 cells pre-treated with 4Mu (13 ± 3.0% and 20 ± 3.5% respectively); however, when splenocytes derived from AdIL-12+4Mu groups were exposed to 4Mu-pre-treated Hepa 129 cells, the percentage of CD8+CD107+ cells was superior to that of Hepa 129 alone (47 ± 3.5% versus 24 ± 5.2%, respectively; Figure 3A, right; *p ≥ 0.05, Kruskal-Wallis test).
Figure 3.
4Mu Downregulates the Expression of CD47 on Hepa 129 Cells, Increases Phagocytosis by Macrophages, and Potentiates the Immune Response Induced by AdIL-12
(A) 4Mu-treated cells in vitro exposed to AdIL-12- or AdIL-12+4Mu-treated mouse splenocytes showed more apoptotic events. ***p < 0.01, Hepa 129 + 4Mu versus Hepa 129 (RPMI). Splenocytes from the AdIL-12+4Mu group show increased CD107 expression on CD8+ T cells. *p < 0.05, Kruskal-Wallis test. (B) Percentage of engulfed cells determined by flow cytometry (F4/80+DAPI+ cells). *p < 0.05, Hepa 129 + 4Mu versus Hepa 129, Mann-Whitney test. Small dot plot (above) corresponds to control Hepa 129 cells or macrophages alone. (C) Indian ink phagocytosis by liver macrophages. Quantification of phagocytosis showed no differences between 4Mu-treated and non-treated mice; ns (nonsignificant), saline versus 4Mu, Mann-Whitney test. (D) Left: peritoneal macrophages ex vivo treated with 4Mu exhibited mRNA levels of SIRP-α similar to that of non-treated cells; ns, Mann-Whitney test. Right: Hepa 129 + 4Mu showed a significant decrease of CD47 mRNA levels. *p < 0.05, Hepa 129 + 4Mu versus Hepa 129, Mann-Whitney test. (E) CD47 expression on Hepa 129 cells treated or non-treated with 4Mu. *p < 0.05, Mann-Whitney test. (F) CD47 median fluorescence intensity (MFI) on phagocytated cells F4/80+DAPI+ cells treated or non-treated with 4Mu. *p < 0.05, Mann-Whitney test. Data are expressed as the mean ± SEM.
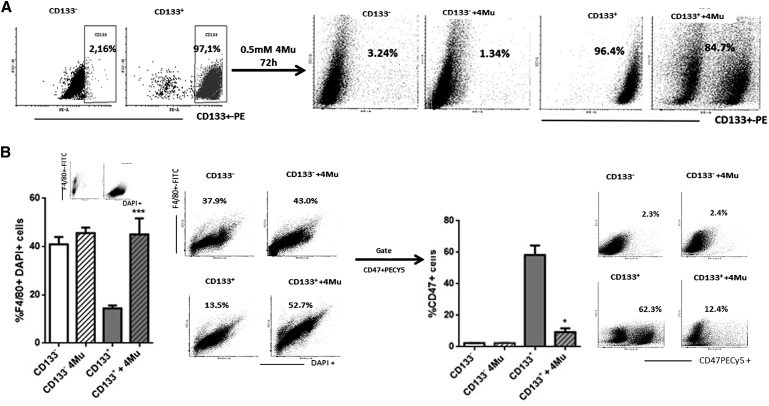
To evaluate whether 4Mu facilitates recognition and phagocytosis of Hepa 129 cells, we performed an in vitro phagocytosis assay using intraperitoneal macrophages (pMφ). To this end, Hepa 129 HCC cells were labeled with DAPI, co-cultured with pMφs for 2 hr, and incubated with fluorescein isothiocyanate (FITC)-labeled F4/80 antibody, and we quantified the presence of F4/80+DAPI+ cells, which represent macrophages that have phagocytosed Hepa 129 cells (upper right quadrant of scatterplots in Figure 3B, right). Interestingly, phagocytosis was significantly increased in Hepa 129 + 4Mu cells compared with Hepa 129 cells alone (RPMI; Figure 3B, left; *p < 0.05, Mann-Whitney test; for the phagocytosis assay at different time points, see Figure S1).To confirm whether 4Mu was able to modulate tumor cell “visibility” or macrophage phagocytic activity, we performed an in vivo Indian ink phagocytosis assay. Mice treated or not treated with 4Mu were intravenously injected with the black pigment and then sacrificed. Microphotography and quantification of H&E-stained livers from mice treated or not treated with 4Mu showed a similar profile of ink uptake by hepatic macrophages (Figure 3C).
It has been reported that CD47 interacts with the signal regulatory protein alpha (SIRPα) and suppresses the phagocytic activity of immune cells.34 CD47 expression on human solid tumors, and particularly on CSCs, is considered a mechanism for phagocytosis evasion, allowing tumor progression.35 First, we analyzed mRNA levels of CD47 on in-vitro-cultured Hepa 129 cells or Hepa 129+4Mu cells by real-time qPCR. Treatment with 4Mu significantly diminished the expression of CD47 (Figure 3D, right). Then, we confirmed a reduced expression of CD47+ cells induced by 4Mu by flow cytometry (Figure 3E). Phagocytated F4/80+DAPI+ Hepa 129 cells also showed reduced levels of CD47 when they were treated with 4Mu (Figure 3F). In contrast, mRNA levels of SIRPα were similar in both pMφ and pMφ + 4Mu (Figure 3D, left). These results suggest that 4Mu has the ability to decrease CD47 expression on HCC cells, while it has no effect on SIRPα expression levels; in addition, 4Mu did not change the innate phagocytic activity on macrophages. Then, phagocytosis of Hepa 129 cells by antigen-presenting cells (APCs) was improved, leading to a potent antitumor T cell response induced by IL-12 in mice with advanced HCC.
4Mu Reduces the Expression of CSC Markers on HCC
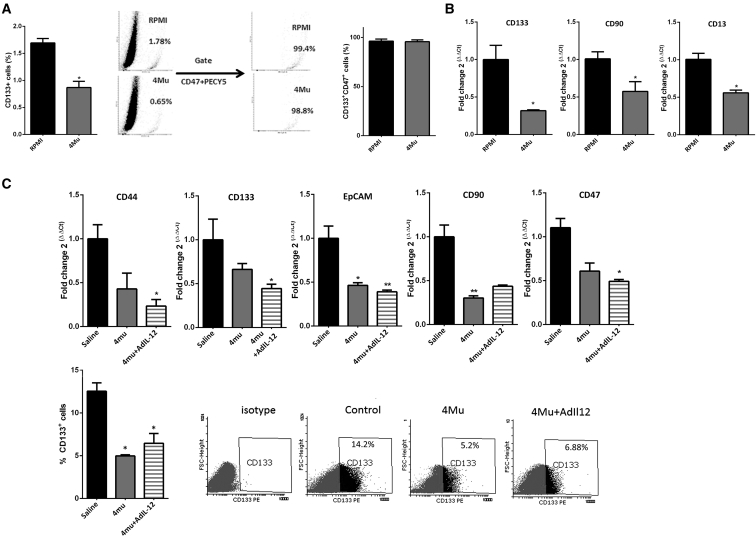
Considering the ability of 4Mu to modulate CD47 expression, we explored whether 4Mu could affect the expression of other molecules characteristic of CSCs. CD133 expression is a CSC marker for many tumor types, including HCC.36, 37, 38 We first analyzed CD133 on in vitro cultured HCC cells by flow cytometry. Although Hepa 129 cells showed low levels of CD133 (1.69 ± 0.16), the percentage of CD133+ was significatively reduced when Hepa 129 cells were treated with 0.5 mM 4Mu for 72 hr (0.80 ± 0.11; *p < 0.05, Mann-Whitney test; Figure 4A, left). Of note, we observed that the gate on CD133+ revealed that these cells were also CD47+ (Figure 4A, right). In addition, mRNA levels of the CSC markers CD133, CD90, and CD13 on Hepa 129 cells decreased after 4Mu incubation in vitro (Figure 4B). Similar results were observed in murine Hepa1.6 and BNL cells, as well as in human HepG2 and HuH7 HCC cells regarding CD133, CD13, CD90, and CD47 expression (Figure S2). Additionally, when 4Mu was withdrawn, the expression of CD133, CD90, EPCAM, CD47, and CD44 markers on Hepa 129 cells was restored (Figure S3). Then, we analyzed the expression of several CSC markers in HCC samples obtained from mice. 4Mu- and AdIL-12+4Mu-treated mice evidenced a 50% reduction of CD133, CD90, CD47, and EpCAM mRNA levels compared with that in control mice (Figure 4C, top). In addition, 4Mu and AdIL-12+4Mu groups showed a significant reduction in CD133+ expression within the tumors by flow cytometry (Figure 4C, bottom; *p < 0.05 versus control group, Kruskal-Wallis test).
Figure 4.
4Mu Reduces the Expression of CSC Markers on HCC In Vitro and In Vivo
(A) Flow cytometry of Hepa 129 cells in vitro exposed to 4Mu showed reduced levels of the CSC marker CD133. *p < 0.05, Hepa 129 + 4Mu versus Hepa 129 (RPMI), Mann-Whitney test. CD133+ cells were also CD47+. (B) mRNA levels of CD133, CD90, and CD13 CSC markers were determined in Hepa 129 or Hepa 129 + 4Mu cells. *p < 0.05, Hepa 129 + 4Mu versus Hepa 129, Mann-Whitney test. (C) mRNA expression levels of CD133, CD90, EpCAM, and CD47 markers were also determined in HCC tumor samples from mice (top). CD133: *p < 0.05. CD47: *p < 0.05. CD90: *p < 0.01, 4Mu-treated versus non-treated mice, or **p < 0.001, 4Mu+AdIL-12-treated versus non-treated mice, Kruskal-Wallis test. Levels of CD133 expression was determined by flow cytometry (bottom). *p < 0.05 for 4Mu-treated versus non-treated mice and 4Mu+AdIL-12-treated versus non-treated mice, Kruskal-Wallis test. Data are expressed as the mean ± SEM.
Changes Induced on CSCs by 4Mu Reduced the Aggressiveness of HCC In Vivo
We isolated CD133+ Hepa 129 cells by magnetic sorting to assess whether CSCs were the target of the effects of 4Mu. After isolation, CD133− Hepa 129 or CD133+ Hepa 129 cells were cultured in the presence of 0.5 mM 4Mu for 72 hr, and survival, cell cycle, duplication time, and apoptotic events were analyzed. No significant differences between groups on cell viability or apoptosis were observed; however, 4Mu induced a slightl arrest on the CD133+ cell cycle and enhanced the duplication time compared with CD133+/CD133− and CD133− plus 4Mu cells (Figure S4). Then, we confirmed that CD133− Hepa 129 cells treated or not treated with 4Mu did not acquire the CD133 marker during culture. On the other hand, CD133+ Hepa 129 cells preserved their marker (about 96%), while the expression of CD133+ decreased by around 10% when 4Mu was added to Hepa 129 cells (Figure 5A). Then, we performed a phagocytosis assay using FITC-F4/80-labeled pMφ and CD133−, CD133− plus 4Mu, CD133+, and CD133+ plus 4Mu Hepa 129 cells. HCC cells were also labeled with DAPI. Figure 5B shows that treatment of CD133+ with 4Mu induced higher percentages of F4/80+DAPI+ phagocytated cells (45.6 ± 3.8 versus 14.4 ± 1.25; ***p < 0.01, CD133+ plus 4Mu Hepa 129 cells versus CD133+ Hepa 129 cells; Kruskal-Wallis test). Then, we confirmed that only CD133+ Hepa 129 cells co-express CD47 (Figure 5C, left), and, importantly, CD133+ plus 4Mu Hepa 129 cells showed a 50% reduction in the expression of CD47 on F4/80+DAPI+ cells (Figure 5C, right); *p < 0.05, CD133+ plus 4Mu Hepa 129 cells versus CD133+ Hepa 129 cells; Kruskal-Wallis test). These results lead us to confirm that 4Mu interacts with liver CSCs, particularly with CD133+CD47+ cells, favoring tumor cell phagocytosis.
Figure 5.
CD133+ Hepa 129 Cells Treated with 4Mu Favors Phagocytosis by F4/80+ Macrophages and Showed Low Expression Levels of CD47
(A) CD133+ Hepa cells were isolated by magnetic sorting. After 72 hr of in vitro culture with 4Mu, CD133 expression was confirmed by flow cytometry. (B) Left: percentage of phagocytated cells (F4/80+DAPI+ cells); ***p < 0.001 for CD133+ Hepa 129 + 4Mu versus CD133+ Hepa 129, Kruskal-Wallis test. Right: CD133+ Hepa 129 cells + 4Mu showed a significant decrease in the expression of CD47 on phagocytated F4/80+DAPI cells, *p < 0.05 for CD133+ Hepa 129 + 4Mu versus CD133+ Hepa 129, Kruskal-Wallis test. Data are expressed as the mean ± SEM.
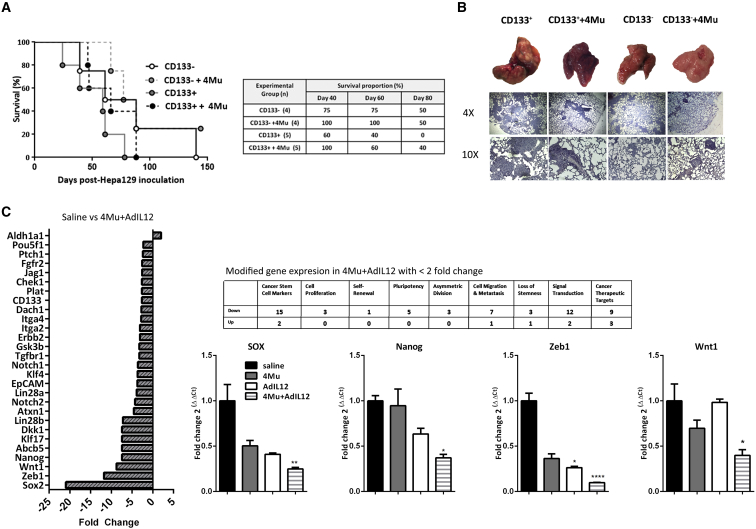
Next, we decided to evaluate whether 4Mu could modify the potential of HCC CSC cells to initiate and sustain tumor dissemination. Thus, male syngeneic C3Hj/He mice were injected with CD133−/CD133− plus 4Mu/CD133+/CD133+ plus 4Mu Hepa 129 cells. The percentages of survival of mice receiving CD133−, CD133− plus 4Mu, CD133+, and CD133+ plus 4Mu were 75%, 100%, 60%, and 100%, respectively, on day 40 after Hepa 129 inoculation (Figure 6A). In addition, the survival of mice receiving CD133+ plus 4Mu was superior to that of mice receiving CD133+ cells at days 60 and 80 (***p < 0.001, Kaplan-Meier, log-rank test). Histopathological examination of organs evidenced the presence of tumor metastases within the lungs. As shown in Figure 6B, mice receiving CD133+ cells showed more metastatic HCC nodules within the lungs in comparison with mice receiving CD133+ plus 4Mu or CD133− cells, indicating that 4Mu decreased CD133+ CSC capability to grow and disseminate. This effect was finally supported when we analyzed the gene expression profile from tumors of 4Mu+AdIL-12-treated mice by qPCR array. CSC markers, asymmetric division, cell migration, and metastasis, like in other CSC target genes, were downregulated in the combined therapy group. In addition, some CSC markers were validated by qPCR (Figure 6C).
Figure 6.
4Mu Decreased CD133+ CSC Capability to Growth and Disseminate
(A) C3Hj/He mice (n = 4–5 per group) were inoculated (i.v.) with (1) 1 × 105 CD133− cells; (2) 1 × 105 CD133− cells plus 4Mu; (3) 1 × 105 CD133+ cells; or (4) 1 × 105 CD133+ cells plus 4Mu. Animal survival was analyzed. ***p < 0.001, Kaplan-Meier, log-rank test. (B) Tissue sections of mice receiving CD133+ cells showed more metastatic HCC nodules within the lungs in comparison with mice receiving CD133+ plus 4Mu or CD133− cells. (C) CSC gene expression profiling by PCR array. (D) Real-time PCR confirmed downregulation of Nanog, Wnt, Sox, and Zeb1 expression in tumor samples of 4Mu+AdIL-12-treated mice compared to control mice. *p < 0.05; **p < 0.001; and ****p < 0.0001 versus saline (ANOVA and Tukey's post test). Data are expressed as the mean ± SEM.
Discussion
HCC develops in patients with underlying liver cirrhosis,39 a pre-neoplastic condition that results from the biological response to chronic damage and continuous hepatic remodeling.40 Cirrhosis is characterized by an alteration in the composition, metabolism, and deposition of ECM components, including hyaluronan (HA).41 In this context of cycles of tissue damage and regeneration, microenvironment factors, genetic and epigenetic changes in cells, and other molecular events promote hepatocarcinogenesis. Thus, hepatocarcinogenesis is a heterogeneous process in which the TME plays a key role.42 The liver TME is composed not only of cancer cells but also of immune cells, endothelial cells, hepatic stellate cells, tumor-associated fibroblasts, and CSCs within an ECM. Recent studies suggest that hepatocarcinogenesis may result from the hierarchical relationship of cancer cells with a subset of cells known as CSCs.43 Liver CSCs have been identified via several cell-surface antigens such as CD133, CD90, EpCAM, CD139, and CD24.44 Moreover, CSCs are able to resist chemotherapy and radiotherapy and establish a relapsed tumor or metastasis.45, 46 TME is typically an immunosuppressive microenvironment, and it is characterized by the presence of CD4+CD25+FoxP3+ T regulatory cells, type 2 macrophages, myeloid-derived suppressor cells, and Kupffer cells.47 A number of strategies destined to induce an effective immune response against cancer cells and to revert the immunosuppressive milieu have been carried out:48 (1) adoptive T cell therapy, (2) indirect immunological approaches (cytokines, immune checkpoint blockade monoclonal antibodies, and dendritic-cell-based vaccines), and (3) indirect non-immunological strategies (antigen-encoding mRNA, metronomic chemotherapy, and oncolytic viruses). Some of them are under evaluation in the clinic, particularly the use of immune checkpoint inhibitors.8, 9, 49 Although advances have been achieved, including objective responses and increased overall survival, several obstacles persist and sustain a hostile TME-generating tumor progression and treatment resistance.50
We and others have previously demonstrated that 4Mu has antitumor activity in many tumor types, including HCC;23 this effect is based on the inhibition of HA synthesis and other mechanisms of action not depending on the effects on HA. The capability of 4Mu to control tumor growth by inhibition of HA synthesis has been demonstrated in prostate, breast, pancreas, and liver cancer.22, 51, 52, 53 In addition, 4Mu affects several key steps of angiogenesis, including endothelial cell proliferation, adhesion, tube formation, and extracellular matrix remodeling.23, 54, 55 On the other hand, we have previously reported that AdIL-12-based immunotherapy reverts the immunosupression induced by CD4+CD25+FoxP3+ regulatory T cells, decreases the number of myeloid-derived suppressor cells (MDSCs), and induces IFNγ-secreting CD4 T lymphocytes with cytotoxic activity in a murine model of liver metastatic colorectal carcinoma.32 In the present work, we decided to investigate the effects of the modulation of TME by the specific HA synthesis inhibitor 4Mu and how this molecule synergizes with immune-gene therapy using AdIL-12 in an orthotopic HCC model established in fibrotic livers.
First, we demonstrated that the administration of 4Mu+AdIL-12 has an additive antitumor activity on HCC tumors and further prolonged animal survival. The combined therapy induced a potent antitumor effect that was superior to that of 4Mu or AdIL-12 therapy alone, leading to 80% of tumor growth inhibition and reduction of satellite nodules. Thus, combined therapy was an effective strategy to reduce tumor size and improved animal survival.
Activation of the adaptive immune arm is critical for generating a tumor-specific immune response.48 In this sense, IL-12 is a key cytokine for inducing a Th1-type response and a potent CTL activity mediated mainly by CD8+ T cells; IL-12 is able to induce IFNγ release from effector cells.29 Then, IFNγ strongly modifies the TME, and this event has been associated with good prognosis after transplantation or locoregional treatments of HCC.56 In this work, we demonstrated that the antitumoral effect achieved with the combined therapy was a consequence of the activation of CD4+ and CD8+ T cells within the liver, the production of higher serum levels of IFNγ, and, finally, a potent CTL response. We next investigated why 4Mu improves the specific CTL response by CD8+ T lymphocytes. It has been demonstrated that APCs are capable of presenting antigens from eaten cancer cells, promoting T cell activation.57 In this work, we observed that, by pre-conditioning HCC cells with 4Mu, it is possible to stimulate phagocytosis of cancer cells by macrophages through the modulation of CD47 expression. CD47 is highly expressed in many tumor types, particularly in CSCs, and it transduces inhibitory signals through SIRPα on macrophages and other APCs protecting CSCs from phagocytosis and favoring tumor cell proliferation.58, 59 The higher CD47 expression in tissue from HCC patients, compared to adjacent non-tumor liver, has been previously reported.60 In our work, we demonstrated that 4Mu was capable to reduce CD47 expression on HCC cells, including CSCs, promoting phagocytosis of cancer cells and stimulating a specific T cell response against HCC. A role of SIRP-expressing dendritic cells (DCs) in antitumor responses has been reported, including in HCC;61 it is possible that CD47 downregulation by 4Mu may affect the response mediated by both macrophages and DCs. Notably, the ability of 4Mu to modulate CD47 expression was also observed in other murine and human HCC cell lines (Figure S5). We also found that CD47+ HCC cells were also CD133+, a well-known liver CSC marker. Therefore, we decided to evaluate whether 4Mu, in addition to its capacity to decrease CD47, is able to modulate other CSC markers. Our results demonstrated that 4Mu ameliorated the expression of CD133, CD90, EpCAM, CD44, and CD13 in HCC cells in vitro and also in HCC tumors in vivo. The clinical role of CSCs in HCC growth is under study, particularly to elucidate the reasons for tumor recurrence or to predict therapeutic response.62 It has been demonstrated that high expression levels of CD133 and CD44 were related to poor prognosis in HCC patients and that expansion of CD44+ is correlated with high tumor recurrence after liver transplantation.63, 64 Therefore, strategies aimed at reducing the presence of CSCs are gaining much attention in the field. In our hands, 4Mu clearly modulates the presence of CSC markers and reduced in nearly 50% the expression of CD47 on isolated CD133+ cells; this change promotes phagocytosis of CSCs by macrophages, which contributes to overcoming the escape to the immune system.
CSCs are defined as a small subset of cells that selectively possesses the ability to self-renew and to initiate tumor growth. Differentiation capacity is verified by a decrease in the expression of CSC markers together with an increase in the expression of differentiation markers; differentiated cells lose their tumorigenic potential.65 We demonstrated that 4Mu was able to modify CSC markers and, at the same time, modulate their self-renewal ability. To assess whether our therapy has the capacity to inhibit HCC dissemination and metastases, we intravenously administered CD133+ cells, pre-treated or not with 4Mu, and evaluated their capability to generate metastases in mice. Animals that received CD133+ plus 4Mu or CD133− cells showed a reduced number of lung metastases in comparison with mice receiving CD133+ cells. Importantly, mouse survival was increased in mice injected with 4Mu-treated CD133low HCC cells in comparison with mice injected with CD133+.
All in all, our results showed that combined therapy demonstrated the capacity to recruit effector cells in the TME and to modulate the presence of CSCs and their immunogenicity. In conclusion, the combined strategy ameliorates HCC aggressiveness by targeting CSCs and as a result of the induction of anticancer immunity.
Materials and Methods
Animals
Six- to 8-week-old male C3Hj/He mice were purchased from Centro Atómico Ezeiza, Buenos Aires, Argentina. Animals were maintained at our Animal Resources Facilities in accordance with the experimental ethical committee and the NIH guidelines on the ethical use of animals. The Animal Care Committee from the School of Biomedical Sciences, Universidad Austral, approved the experimental protocol.
Cell Lines
Hepa 129 cells (syngeneic with C3H/He mice) were kindly provided by Dr. Volker Schmitz (Bonn University, Bonn, Germany). Hepa 129, BNL, and Hepa 1.6 HCC cells were grown in RPMI 1640 (GIBCO, Invitrogen Argentina, Buenos Aires, Argentina) with 10% fetal bovine serum (FBS). Hep3B and Huh7 human HCC cells (ATCC, Manassas, VA, USA) were kindly provided by Jesús Prieto, University of Navarra, Spain, and were grown in DMEM (GIBCO) with 5% FBS.
Drugs
For in vitro studies, murine or human HCC cells (5,000 per well) were incubated for 24 hr and then treated with 4Mu (Sigma-Aldrich, St. Louis, MO, USA) at different concentrations (0.25, 0.5, or 1 mM) or control vehicle solution (Hank’s balanced salt solution; HBSS) for an additional 72 hr.
Adenoviral Vector
Construction and purification of the non-replicative recombinant adenovirus, serotype 5, under the control of the cytomegalovirus (CMV) promoter and encoding for murine IL-12 genes (AdIL-12), was previously described.31 The purified virus was dialyzed and stored at −80°C. The virus titer was determined by plaque assay. The dose of vectors was expressed as 50% tissue culture infectious doses (TCID50) per milliliter.
In Vivo Experiments
Experimental Model of HCC Associated with Fibrosis
C3H/He mice were treated intraperitoneally (i.p.) with 200 mg/kg thioacetamide (TAA) (Sigma-Aldrich, St. Louis, MO, USA) 3 times a week for 4 weeks. On day 28, mice were anesthetized, and orthotopic tumors were established by sub-capsular inoculation of 1.25 × 105 Hepa 129 cells into the left liver lobe by laparotomy66 (at day 0). Five days after tumor implantation, mice were distributed in groups (n = 8 per group) and received: (1) saline (control); (2) 4Mu, 200 mg/kg drinking water; (3) AdIL-12, 1 × 109 DICT50 per milliliter, intravenously (i.v.), at day 7; (4) 4Mu+AdIL-12, 1 × 109 DICT50 per milliliter, i.v., at day 7; or (5) 4Mu+ Adβ-gal, 1 × 109 DICT50 per milliliter, i.v., at day 7. Fifteen days after tumor implantation, mice were sacrificed; ascites, tumor volume, and numbers of HCC satellites were quantified. This experiment was repeated 3 times.
i.v. HCC Metastatic Model
Isolated CD133+ or CD133− Hepa 129 cells were treated, or not treated, with 0.5 mM 4Mu for 72 hr. C3H mice were separated in 4 groups and received: (1) CD133− Hepa 129 cells (1 × 105; n = 4); (2) CD133+ Hepa 129 cells (1 × 105; n = 4); (3) 4Mu-treated CD133− Hepa 129 cells (1 × 105; n = 5), or (4) 4Mu-treated CD133+ Hepa 129 cells (1 × 105; n = 5). Animal survival was analyzed. This experiment was repeated 3 times.
Ex Vivo Experiments
Quantification of CD3+ Cells on HCC Tumors
5-μm paraffin-embedded liver sections were rehydrated and incubated with rabbit anti-mouse CD3 polyclonal antibody (1:100; ab5690, Abcam, Cambridge, UK) overnight. After washing, slides were incubated with peroxidase-linked biotinylated goat anti-rabbit secondary antibody (Jackson ImmunoResearch, West Grove, PA, USA) for 2 hr, washed twice with PBS, and washed twice with 0.1 M acetate buffer before incubation with a solution of diaminobenzidine (DAB; Sigma-Aldrich, St. Louis, MO, USA), ammonium nickel sulfate, and H2O2. CD3+ cells from liver sections were detected by taking 10 images per slide at 200× (Nikon Eclipse E800, Global Nikon, Melville, NY, USA), and the percentage of the positive area was calculated with ImageJ software (NIH, Bethesda, MD, USA).
Flow Cytometry
Tumor lysates from mice treated with 4Mu, AdIL-12, and 4Mu+AdIL-12 or saline were obtained after liver perfusion with collagenase (Sigma-Aldrich, St. Louis, MO, USA). Splenocytes from control or treated mice were also obtained through mechanic disruption of spleens. Cell suspensions were then treated with red blood cell (RBC) lysis buffer (0.15 mol/L NH4Cl, 1 mmol/L KHCO3, 0.1 mmol/L Na2-EDTA) and washed with PBS 1% BSA (Sigma-Aldrich, St. Louis, MO, USA). Then, 1 × 106 cells were suspended in a 1% PBS solution and stained with fluorochrome-conjugated antibodies: anti-CD4-PECy5 (553050; BD Biosciences, Franklin Lakes, NJ, USA), anti-CD8 Alexa Fluor 488 (557668; BD Biosciences, Franklin Lakes, NJ, USA), anti-CD133-APC (141208; BioLegend, San Diego, CA, USA), anti- F4/80 (ab105155; Abcam, Cambridge, UK); and anti-CD47-PECy5 (ab108415; Abcam, Cambridge, UK) for 45 min at 4°C. Hepa 129 cells were suspended in 1% BSA/PBS and stained with anti-CD133-APC and anti CD47-PECy5 antibodies. Samples were analyzed by flow cytometry (FACSAria, BD Biosciences, Franklin Lakes, NJ, USA), and data were analyzed using Cyflogic v1.2.1 software.
Cytotoxicity Assay
1 × 107 splenocytes from control or treated mice were stimulated in vitro with mitomycin-C-treated Hepa 129 cells (1 × 106 cells per well in 24-well plates). On day 5, cells were harvested, washed, adjusted to 2 × 106 cells per milliliter, and added to 96-well plates (effector cells). To determine specific CTL activity, Hepa 129 cells were used as the target at 2 × 105 per milliliter. After incubation for 4 hr at 37°C, plates were centrifuged and cell-free supernatants were obtained. Levels of released lactate dehydrogenase (LDH) were evaluated with the LDH Cytotoxicity Detection Kit (Sigma-Aldrich, St. Louis, MO, USA) and expressed as percentage of lysis.
Detection of T Cell Degranulation
1 × 107 splenocytes from control or treated mice were stimulated in vitro with mitomycin-C-treated Hepa 129 cells (1 × 106 cells per well in 24-well plates). On day 5, cells were harvested, washed, adjusted to 2 × 106 cells per milliliter, and added to 96-well plates (effector cells). To determine specific T cell degranulation, Hepa 129 cells were used as the target at 2 × 105 cells per milliliter. During incubation for 4 hr at 37°C in the presence of 5 μg/mL monensin, cells were stained with the fluorochrome-conjugated antibodies anti-CD8-Alexa Fluor 488 and anti-CD107-PECy5 (BioLegend, San Diego, CA, USA). Then, they were analyzed by flow cytometry (FACSAria, BD Biosciences, Franklin Lakes, NJ, USA) using Cyflogic v1.2.1 software.
ELISA
Serum samples were analyzed by ELISA to quantify IFNγ (BD Biosciences, Franklin Lakes, NJ, USA), following the manufacturer’s recommendations.
Apoptosis Assays
Splenocytes from control or treated mice (1 × 107) were stimulated in vitro with mitomycin-C-treated Hepa 129 cells treated or not with 4Mu (1 × 106 cells per well in 24-well plates). On day 5, cells were harvested, washed, adjusted to 2 × 106 per milliliter, and added to 96-well-plates (effector cells). 4Mu-treated or untreated Hepa 129 cells were used as the target at 2 × 105 per milliliter. After incubation for 18 hr at 37°C, apoptosis was analyzed by acridine orange (AO) and ethidium bromide (EB) staining and visualized by fluorescence microscopy (Nikon Eclipse E800 Global Nikon, Melville, NY, USA). Assays were performed in quadruplicate and repeated at least twice.
Phagocytosis Assays
Macrophages were isolated from the peritoneal cavity (pMφ), incubated in serum-free medium for 2 hr in a 24-well tissue-culture plate, and immunostained with anti-F4/80 (Abcam, Cambridge, UK). Then, Hepa 129 cells treated or no treated with 4Mu (5 × 105), labeled with DAPI and immunostained with anti-CD47-PECy5, were exposed to 5 × 104 pMφ and incubated for 8 min, 15 min, 30 min, 1 hr, 2 hr, and 3 hr at 37°C. Phagocytosis was determined by flow cytometry detection of F4/80+DAPI+ cells.
Indian Ink Phagocytosis Assay
Mice with fibrosis induced by TAA treated or not treated with 4Mu were injected i.v. with 100 μL Indian ink and then sacrificed. Liver samples were taken, fixed, and embedded in paraffin for H&E staining.
Pathological Analysis
5-μm paraffin-embedded liver and lung tissue samples were stained with H&E, following standard procedures.
RNA Isolation and qPCR Analysis
Total RNA samples were isolated using TRI Reagent (Sigma-Aldrich, St. Louis, MO, USA), and total RNA (2 mg) was reverse transcribed (through qRT-PCR) with 200 U SuperScript II Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) using 500 ng oligo(dT) primers. cDNAs were subjected to real-time qPCR (Stratagene Mx3005p, Stratagene, La Jolla, CA, USA). For qRT-PCR, the mRNA levels of CD133, CD44, CD90, EpCAM, CD13, CD47, and SIRP-α were quantified by SYBRGreen (Invitrogen, Carlsbad, CA, USA), using the following primers: human CD133, forward: 5′-AAACAGTTTGCCCCCAGGAA-3′ and reverse: 5′-ACAATCCATTCCCTGTGCGT-3′; human CD47, forward: 5′-CGCTGTGGTTGGACTGATCT-3′ and reverse: 5′-GGGGTTCCTCTACAGCTTTCCPCR-3′; mouse CD47, forward: 5′-ATGCTTCTGGACTTGGCCTC-3′ and reverse: 5′-CCG ACCAAAGCAAGGACGTA-3′; mouse SIRP-α, forward: 5′-AGAAAGCCAAGGGGTCAACATC-3′ and reverse: 5′-TCTCTTTGGGCAGATTCAGGTC-3′. Amplifications were carried out using a cycle of 95°C for 10 min and 40 cycles under the following parameters: 95°C for 30 s, 56°C for 30 s, and 72°C for 1 min. At the end of the PCR reaction, the temperature was increased from 60°C to 95°C at a rate of 2°C/min, and fluorescence was measured every 15 s to construct the melting curve. Values were normalized to levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH; used as housekeeping) transcript (forward: 5′- CATCTCTGCCCCCTCTGCTG-3′; reverse: 5′-GCCTGCTTCACCACCTTCTTG-3′). Data were processed by the ΔΔCt method. The relative amount of the PCR product amplified from untreated cells was set as 1. A non-template control (NTC) was run in every assay, and all determinations were performed as triplicates for each animal (n = 5 per group) in two separated experiments. The relative expression was calculated according to the following equation: relative expression (RE) = 2 − ΔΔCt.
PCR Array
Equal amounts of RNA from saline- or 4Mu+AdIL12-treated mouse tumor samples (n = 3 per group) were pooled, and an RT-PCR kit (QIAGEN, Hilden, Germany) was used to obtain cDNA. A PCR for GAPDH was assessed to examine the quality of cDNA. Expression levels of 84 genes related to CSC markers were evaluated (PAMM-176ZA; SABiosciences, Frederick, MD, USA) following the manufacturer’s instructions. The mRNA expression levels obtained for each gene were normalized to the expression of the GAPDH housekeeping gene. Finally, data results were analyzed by using the manufacturer’s website, and scores were expressed as fold change (ΔΔCt) relative to control (saline group).
In Vitro Assays
mRNA Expression on HCC Cell Lines
mRNA levels of CD133, CD13, CD90, EpCAM, CD44, and CD47 were determined in murine Hepa 129, Hepa1.6, and BNL cells, as well as in human HepG2 and HuH7 HCC cell lines by real-time qPCR when HCC cells were exposed to 0.5 mM 4Mu for 72 hr. For Hepa 129 cells, mRNA levels of CD133, CD90, EpCAM, CD44, and CD47 were also determined by qPCR at 24, 48, and 72 hr post-4Mu withdrawal.
Cell Isolation by MACS
Hepa 129 cells were labeled with primary CD133/1 antibody (rat IgG1130-092-564, Miltenyi Biotec, Bergisch Gladbach, Germany), and the CD133+ cells were subsequently magnetically isolated using magnetic-activated cell sorting (MACS) columns. CD133+ cells were enriched using LS columns according to the manufacturer’s instructions. The purity of sorted cells was evaluated by flow cytometry. Trypan blue staining was used to assess cell viability.
Proliferation Assay
Isolated CD133+ or CD133− Hepa cells were cultured in a flask and treated, or not treated, with 4Mu for 72 hr. Cell doubling times during logarithmic growth were calculated according to Hayflick’s formula: T = t × [ln2/ln(Nt/N0)] (T = population doubling time; t = appointed time after subculture; Nt = number of cells at the appointed time; N0 = number of cells at the beginning of subculture).
Viability Assay
Isolated CD133+ or CD133− Hepa cells (5 × 103) were seeded in 96-well cell-culture plates and treated, or not treated, with 4Mu for 72 hr. At the indicated time points, cells were incubated with 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT; Promega, Madison, WI, USA) for 4 hr at 37°C with 5% C2O2. Isopropanol/acid chloride was used to stop the reaction. Absorbance was determined at 560 nm using a microplate spectrophotometer (Thermo Scientific, Waltham, MA, USA). The experiment was carried out 2 times independently in 5 replicates.
Cell-Cycle Analysis
Isolated CD133+ or CD133− Hepa cells were cultured in a flask and were treated or not treated with 4Mu for 72 hr. Then, cells were fixed with 70% ethanol, 30% PBS, and kept at −20°C for at least 24 hr. Cells were centrifuged for 10 min at 2,000G, at 4°C, and washed twice with PBS before incubation with 5 mg propidium iodine in PBS together with 180 U/mL RNase for 30 min at room temperature; samples were analyzed by cytometry (FACSAria, BD Biosciences, Franklin Lakes, NJ, USA)
Statistical Analysis
All experiments were repeated at least 3 times. Values were expressed as mean ± SEM. Mann-Whitney, Tukey, or Kruskal-Wallis (ANOVA) multiple comparison tests were used to evaluate the statistical differences between groups. Mice survival was analyzed by a Kaplan-Meier curve. A p value of < 0.05 was considered as significant. Prism software (GraphPad, San Diego, CA, USA) was used for statistical analysis.
Author Contributions
Conception and design: M.M. and G.M.; Development of methodology: M.M. and G.M.; Acquisition of data: M.M.R., E.F., A.O., and M.M.; Analysis and interpretation of data: M.M.R., J.B., C.A., M.G., M.M., and G.M.; Writing, review, and/or revision of the manuscript: M.M.R., M.M., and G.M.; Administrative, technical, or material support: C.A., M.G., A.O., and L.D.; Study supervision: G.M.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We would like to thank Plácida Baz, Ariel Billordo, Paula Roselló, Franco Puebla, Santiago Cabrera, and Guillermo Gastón for their professional technical assistance. This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica (PICT-2012-1407 and PICT-2015-2036), Instituto Nacional del Cáncer (Asistencia Financiera a Proyectos de Investigación en Cáncer de Origen Nacional III), Fundación para el progreso de la Medicina (GF04), and Universidad Austral.
Footnotes
Supplemental information includes five figures and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.09.012.
Supplemental Information
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.El-Serag H.B. Hepatocellular carcinoma. N. Engl. J. Med. 2011;365:1118–1127. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 3.Choo S.P., Tan W.L., Goh B.K., Tai W.M., Zhu A.X. Comparison of hepatocellular carcinoma in Eastern versus Western populations. Cancer. 2016 doi: 10.1002/cncr.30237. Published September 13, 2016. [DOI] [PubMed] [Google Scholar]
- 4.Bray F., Ferlay J., Laversanne M., Brewster D.H., Gombe Mbalawa C., Kohler B., Piñeros M., Steliarova-Foucher E., Swaminathan R., Antoni S. Cancer incidence in five continents: inclusion criteria, highlights from Volume X and the global status of cancer registration. Int. J. Cancer. 2015;137:2060–2071. doi: 10.1002/ijc.29670. [DOI] [PubMed] [Google Scholar]
- 5.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., SHARP Investigators Study Group Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 6.Bruix J., Qin S., Merle P., Granito A., Huang Y.H., Bodoky G., Pracht M., Yokosuka O., Rosmorduc O., Breder V., RESORCE Investigators Regorafenib for patients with hepatocellular carcinoma who progressed on sorafenib treatment (RESORCE): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;389:56–66. doi: 10.1016/S0140-6736(16)32453-9. [DOI] [PubMed] [Google Scholar]
- 7.Berraondo P., Labiano S., Minute L., Etxeberria I., Vasquez M., Sanchez-Arraez A., Teijeira A., Melero I. Cellular immunotherapies for cancer. OncoImmunology. 2017;6:e1306619. doi: 10.1080/2162402X.2017.1306619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.El-Khoueiry A.B., Sangro B., Yau T., Crocenzi T.S., Kudo M., Hsu C., Kim T.Y., Choo S.P., Trojan J., Welling T.H. Nivolumab in patients with advanced hepatocellular carcinoma (CheckMate 040): an open-label, non-comparative, phase 1/2 dose escalation and expansion trial. Lancet. 2017;389:2492–2502. doi: 10.1016/S0140-6736(17)31046-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duffy A.G., Ulahannan S.V., Makorova-Rusher O., Rahma O., Wedemeyer H., Pratt D., Davis J.L., Hughes M.S., Heller T., ElGindi M. Tremelimumab in combination with ablation in patients with advanced hepatocellular carcinoma. J. Hepatol. 2017;66:545–551. doi: 10.1016/j.jhep.2016.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tlsty T.D., Coussens L.M. Tumor stroma and regulation of cancer development. Annu. Rev. Pathol. 2006;1:119–150. doi: 10.1146/annurev.pathol.1.110304.100224. [DOI] [PubMed] [Google Scholar]
- 11.Hernandez-Gea V., Toffanin S., Friedman S.L., Llovet J.M. Role of the microenvironment in the pathogenesis and treatment of hepatocellular carcinoma. Gastroenterology. 2013;144:512–527. doi: 10.1053/j.gastro.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rani B., Cao Y., Malfettone A., Tomuleasa C., Fabregat I., Giannelli G. Role of the tissue microenvironment as a therapeutic target in hepatocellular carcinoma. World J. Gastroenterol. 2014;20:4128–4140. doi: 10.3748/wjg.v20.i15.4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanahan D., Coussens L.M. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21:309–322. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Bao B., Ahmad A., Azmi A.S., Ali S., Sarkar F.H. Overview of cancer stem cells (CSCs) and mechanisms of their regulation: implications for cancer therapy. Curr. Protoc. Pharmacol. 2013;Chapter 14 doi: 10.1002/0471141755.ph1425s61. Unit 14.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mishra L., Banker T., Murray J., Byers S., Thenappan A., He A.R., Shetty K., Johnson L., Reddy E.P. Liver stem cells and hepatocellular carcinoma. Hepatology. 2009;49:318–329. doi: 10.1002/hep.22704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valent P., Bonnet D., De Maria R., Lapidot T., Copland M., Melo J.V., Chomienne C., Ishikawa F., Schuringa J.J., Stassi G. Cancer stem cell definitions and terminology: the devil is in the details. Nat. Rev. Cancer. 2012;12:767–775. doi: 10.1038/nrc3368. [DOI] [PubMed] [Google Scholar]
- 17.Yang Z.F., Ngai P., Ho D.W., Yu W.C., Ng M.N., Lau C.K., Li M.L., Tam K.H., Lam C.T., Poon R.T., Fan S.T. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 18.Terris B., Cavard C., Perret C. EpCAM, a new marker for cancer stem cells in hepatocellular carcinoma. J. Hepatol. 2010;52:280–281. doi: 10.1016/j.jhep.2009.10.026. [DOI] [PubMed] [Google Scholar]
- 19.Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 20.Lee T.K., Cheung V.C., Lu P., Lau E.Y., Ma S., Tang K.H., Tong M., Lo J., Ng I.O. Blockade of CD47-mediated cathepsin S/protease-activated receptor 2 signaling provides a therapeutic target for hepatocellular carcinoma. Hepatology. 2014;60:179–191. doi: 10.1002/hep.27070. [DOI] [PubMed] [Google Scholar]
- 21.Fidler I.J. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 22.Piccioni F., Malvicini M., Garcia M.G., Rodriguez A., Atorrasagasti C., Kippes N., Piedra Buena I.T., Rizzo M.M., Bayo J., Aquino J. Antitumor effects of hyaluronic acid inhibitor 4-methylumbelliferone in an orthotopic hepatocellular carcinoma model in mice. Glycobiology. 2012;22:400–410. doi: 10.1093/glycob/cwr158. [DOI] [PubMed] [Google Scholar]
- 23.Piccioni F., Fiore E., Bayo J., Atorrasagasti C., Peixoto E., Rizzo M., Malvicini M., Tirado-González I., García M.G., Alaniz L., Mazzolini G. 4-methylumbelliferone inhibits hepatocellular carcinoma growth by decreasing IL-6 production and angiogenesis. Glycobiology. 2015;25:825–835. doi: 10.1093/glycob/cwv023. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y., Ramjiawan R.R., Reiberger T., Ng M.R., Hato T., Huang Y., Ochiai H., Kitahara S., Unan E.C., Reddy T.P. CXCR4 inhibition in tumor microenvironment facilitates anti-programmed death receptor-1 immunotherapy in sorafenib-treated hepatocellular carcinoma in mice. Hepatology. 2015;61:1591–1602. doi: 10.1002/hep.27665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazzolini G.D., Malvicini M. Immunostimulatory monoclonal antibodies for hepatocellular carcinoma therapy. Trends and perspectives. Medicina (B. Aires) 2018;78:29–32. [PubMed] [Google Scholar]
- 26.Iñarrairaegui M., Melero I., Sangro B. Immunotherapy of hepatocellular carcinoma: facts and hopes. Clin. Cancer Res. 2018;24:1518–1524. doi: 10.1158/1078-0432.CCR-17-0289. [DOI] [PubMed] [Google Scholar]
- 27.Moreno-Cubero E., Larrubia J.R. Specific CD8(+) T cell response immunotherapy for hepatocellular carcinoma and viral hepatitis. World J. Gastroenterol. 2016;22:6469–6483. doi: 10.3748/wjg.v22.i28.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou G., Sprengers D., Boor P.P.C., Doukas M., Schutz H., Mancham S. Antibodies against immune checkpoint molecules restore functions of tumor-infiltrating T cells in hepatocellular carcinomas. Gastroenterology. 2017;153:1107–1119.e10. doi: 10.1053/j.gastro.2017.06.017. [DOI] [PubMed] [Google Scholar]
- 29.Berraondo P., Etxeberria I., Ponz-Sarvise M., Melero I. Revisiting interleukin-12 as a cancer immunotherapy agent. Clin. Cancer Res. 2018;24:2716–2718. doi: 10.1158/1078-0432.CCR-18-0381. [DOI] [PubMed] [Google Scholar]
- 30.Sangro B., Mazzolini G., Ruiz J., Herraiz M., Quiroga J., Herrero I., Benito A., Larrache J., Pueyo J., Subtil J.C. Phase I trial of intratumoral injection of an adenovirus encoding interleukin-12 for advanced digestive tumors. J. Clin. Oncol. 2004;22:1389–1397. doi: 10.1200/JCO.2004.04.059. [DOI] [PubMed] [Google Scholar]
- 31.Malvicini M., Rizzo M., Alaniz L., Piñero F., García M., Atorrasagasti C., Aquino J.B., Rozados V., Scharovsky O.G., Matar P., Mazzolini G. A novel synergistic combination of cyclophosphamide and gene transfer of interleukin-12 eradicates colorectal carcinoma in mice. Clin. Cancer Res. 2009;15:7256–7265. doi: 10.1158/1078-0432.CCR-09-1861. [DOI] [PubMed] [Google Scholar]
- 32.Malvicini M., Ingolotti M., Piccioni F., Garcia M., Bayo J., Atorrasagasti C., Alaniz L., Aquino J.B., Espinoza J.A., Gidekel M. Reversal of gastrointestinal carcinoma-induced immunosuppression and induction of antitumoural immunity by a combination of cyclophosphamide and gene transfer of IL-12. Mol. Oncol. 2011;5:242–255. doi: 10.1016/j.molonc.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lasek W., Zagożdżon R., Jakobisiak M. Interleukin 12: still a promising candidate for tumor immunotherapy? Cancer Immunol. Immunother. 2014;63:419–435. doi: 10.1007/s00262-014-1523-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willingham S.B., Volkmer J.P., Gentles A.J., Sahoo D., Dalerba P., Mitra S.S., Wang J., Contreras-Trujillo H., Martin R., Cohen J.D. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc. Natl. Acad. Sci. USA. 2012;109:6662–6667. doi: 10.1073/pnas.1121623109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCracken M.N., Cha A.C., Weissman I.L. Molecular pathways: activating T cells after cancer cell phagocytosis from blockade of CD47 “don’t eat me” signals. Clin. Cancer Res. 2015;21:3597–3601. doi: 10.1158/1078-0432.CCR-14-2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Curley M.D., Therrien V.A., Cummings C.L., Sergent P.A., Koulouris C.R., Friel A.M., Roberts D.J., Seiden M.V., Scadden D.T., Rueda B.R., Foster R. CD133 expression defines a tumor initiating cell population in primary human ovarian cancer. Stem Cells. 2009;27:2875–2883. doi: 10.1002/stem.236. [DOI] [PubMed] [Google Scholar]
- 37.Chen Y.L., Lin P.Y., Ming Y.Z., Huang W.C., Chen R.F., Chen P.M., Chu P.Y. The effects of the location of cancer stem cell marker CD133 on the prognosis of hepatocellular carcinoma patients. BMC Cancer. 2017;17:474. doi: 10.1186/s12885-017-3460-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miyata T., Oyama T., Yoshimatsu T., Higa H., Kawano D., Sekimura A., Yamashita N., So T., Gotoh A. The clinical significance of cancer stem cell markers ALDH1A1 and CD133 in lung adenocarcinoma. Anticancer Res. 2017;37:2541–2547. doi: 10.21873/anticanres.11597. [DOI] [PubMed] [Google Scholar]
- 39.European Association for the Study of the Liver. European Organisation for Research and Treatment of Cancer EASL-EORTC clinical practice guidelines: management of hepatocellular carcinoma. J. Hepatol. 2012;56:908–943. doi: 10.1016/j.jhep.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 40.Thorgeirsson S.S., Grisham J.W. Molecular pathogenesis of human hepatocellular carcinoma. Nat. Genet. 2002;31:339–346. doi: 10.1038/ng0802-339. [DOI] [PubMed] [Google Scholar]
- 41.Vigetti D., Karousou E., Viola M., Deleonibus S., De Luca G., Passi A. Hyaluronan: biosynthesis and signaling. Biochim. Biophys. Acta. 2014;1840:2452–2459. doi: 10.1016/j.bbagen.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 42.Yang J.D., Nakamura I., Roberts L.R. The tumor microenvironment in hepatocellular carcinoma: current status and therapeutic targets. Semin. Cancer Biol. 2011;21:35–43. doi: 10.1016/j.semcancer.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yoo J.E., Kim Y.J., Rhee H., Kim H., Ahn E.Y., Choi J.S., Roncalli M., Park Y.N. Progressive enrichment of stemness features and tumor stromal alterations in multistep hepatocarcinogenesis. PLoS ONE. 2017;12:e0170465. doi: 10.1371/journal.pone.0170465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu L., Li H., Fu S., Chen X., Lu L. Surface markers of liver cancer stem cells and innovative targeted-therapy strategies for HCC. Oncol. Lett. 2018;15:2039–2048. doi: 10.3892/ol.2017.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ma S., Lee T.K., Zheng B.J., Chan K.W., Guan X.Y. CD133+ HCC cancer stem cells confer chemoresistance by preferential expression of the Akt/PKB survival pathway. Oncogene. 2008;27:1749–1758. doi: 10.1038/sj.onc.1210811. [DOI] [PubMed] [Google Scholar]
- 46.Nishiyama M., Tsunedomi R., Yoshimura K., Hashimoto N., Matsukuma S., Ogihara H., Kanekiyo S., Iida M., Sakamoto K., Suzuki N. Metastatic ability and the epithelial-mesenchymal transition in induced cancer stem-like hepatoma cells. Cancer Sci. 2018;109:1101–1109. doi: 10.1111/cas.13527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Greten T.F., Duffy A.G., Korangy F. Hepatocellular carcinoma from an immunologic perspective. Clin. Cancer Res. 2013;19:6678–6685. doi: 10.1158/1078-0432.CCR-13-1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Longo V., Gnoni A., Casadei Gardini A., Pisconti S., Licchetta A., Scartozzi M., Memeo R., Palmieri V.O., Aprile G., Santini D. Immunotherapeutic approaches for hepatocellular carcinoma. Oncotarget. 2017;8:33897–33910. doi: 10.18632/oncotarget.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sangro B., Gomez-Martin C., de la Mata M., Iñarrairaegui M., Garralda E., Barrera P., Riezu-Boj J.I., Larrea E., Alfaro C., Sarobe P. A clinical trial of CTLA-4 blockade with tremelimumab in patients with hepatocellular carcinoma and chronic hepatitis C. J. Hepatol. 2013;59:81–88. doi: 10.1016/j.jhep.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 50.Wang X., Hassan W., Jabeen Q., Khan G.J., Iqbal F. Interdependent and independent multidimensional role of tumor microenvironment on hepatocellular carcinoma. Cytokine. 2018;103:150–159. doi: 10.1016/j.cyto.2017.09.026. [DOI] [PubMed] [Google Scholar]
- 51.Yoshida E., Kudo D., Nagase H., Shimoda H., Suto S., Negishi M., Kakizaki I., Endo M., Hakamada K. Antitumor effects of the hyaluronan inhibitor 4-methylumbelliferone on pancreatic cancer. Oncol. Lett. 2016;12:2337–2344. doi: 10.3892/ol.2016.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yates T.J., Lopez L.E., Lokeshwar S.D., Ortiz N., Kallifatidis G., Jordan A., Hoye K., Altman N., Lokeshwar V.B. Dietary supplement 4-methylumbelliferone: an effective chemopreventive and therapeutic agent for prostate cancer. J. Natl. Cancer Inst. 2015;107:107. doi: 10.1093/jnci/djv085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Urakawa H., Nishida Y., Wasa J., Arai E., Zhuo L., Kimata K., Kozawa E., Futamura N., Ishiguro N. Inhibition of hyaluronan synthesis in breast cancer cells by 4-methylumbelliferone suppresses tumorigenicity in vitro and metastatic lesions of bone in vivo. Int. J. Cancer. 2012;130:454–466. doi: 10.1002/ijc.26014. [DOI] [PubMed] [Google Scholar]
- 54.Malvicini M., Fiore E., Ghiaccio V., Piccioni F., Rizzo M., Olmedo Bonadeo L., García M., Rodríguez M., Bayo J., Peixoto E. Tumor microenvironment remodeling by 4-methylumbelliferone boosts the antitumor effect of combined immunotherapy in murine colorectal carcinoma. Mol. Ther. 2015;23:1444–1455. doi: 10.1038/mt.2015.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.García-Vilas J.A., Quesada A.R., Medina M.A. 4-methylumbelliferone inhibits angiogenesis in vitro and in vivo. J. Agric. Food Chem. 2013;61:4063–4071. doi: 10.1021/jf303062h. [DOI] [PubMed] [Google Scholar]
- 56.Budhu A., Forgues M., Ye Q.H., Jia H.L., He P., Zanetti K.A., Kammula U.S., Chen Y., Qin L.X., Tang Z.Y., Wang X.W. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111. doi: 10.1016/j.ccr.2006.06.016. [DOI] [PubMed] [Google Scholar]
- 57.Feng M., Chen J.Y., Weissman-Tsukamoto R., Volkmer J.P., Ho P.Y., McKenna K.M., Cheshier S., Zhang M., Guo N., Gip P. Macrophages eat cancer cells using their own calreticulin as a guide: roles of TLR and Btk. Proc. Natl. Acad. Sci. USA. 2015;112:2145–2150. doi: 10.1073/pnas.1424907112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu X., Pu Y., Cron K., Deng L., Kline J., Frazier W.A., Xu H., Peng H., Fu Y.X., Xu M.M. CD47 blockade triggers T cell-mediated destruction of immunogenic tumors. Nat. Med. 2015;21:1209–1215. doi: 10.1038/nm.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Murata Y., Kotani T., Ohnishi H., Matozaki T. The CD47-SIRPα signalling system: its physiological roles and therapeutic application. J. Biochem. 2014;155:335–344. doi: 10.1093/jb/mvu017. [DOI] [PubMed] [Google Scholar]
- 60.Xiao Z., Chung H., Banan B., Manning P.T., Ott K.C., Lin S., Capoccia B.J., Subramanian V., Hiebsch R.R., Upadhya G.A. Antibody mediated therapy targeting CD47 inhibits tumor progression of hepatocellular carcinoma. Cancer Lett. 2015;360:302–309. doi: 10.1016/j.canlet.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu Q., Wen W., Tang L., Qin C.J., Lin Y., Zhang H.L., Wu H., Ashton C., Wu H.P., Ding J. Inhibition of SIRPα in dendritic cells potentiates potent antitumor immunity. OncoImmunology. 2016;5:e1183850. doi: 10.1080/2162402X.2016.1183850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sia D., Villanueva A., Friedman S.L., Llovet J.M. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin B., Chen T., Zhang Q., Lu X., Zheng Z., Ding J., Liu J., Yang Z., Geng L., Wu L. FAM83D associates with high tumor recurrence after liver transplantation involving expansion of CD44+ carcinoma stem cells. Oncotarget. 2016;7:77495–77507. doi: 10.18632/oncotarget.12715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao Q., Zhou H., Liu Q., Cao Y., Wang G., Hu A., Ruan L., Wang S., Bo Q., Chen W. Prognostic value of the expression of cancer stem cell-related markers CD133 and CD44 in hepatocellular carcinoma: from patients to patient-derived tumor xenograft models. Oncotarget. 2016;7:47431–47443. doi: 10.18632/oncotarget.10164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen W., Dong J., Haiech J., Kilhoffer M.C., Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cells Int. 2016;2016:1740936. doi: 10.1155/2016/1740936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kornek M., Raskopf E., Tolba R., Becker U., Klöckner M., Sauerbruch T., Schmitz V. Accelerated orthotopic hepatocellular carcinomas growth is linked to increased expression of pro-angiogenic and prometastatic factors in murine liver fibrosis. Liver Int. 2008;28:509–518. doi: 10.1111/j.1478-3231.2008.01670.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.