Abstract
Exosomes can deliver therapeutic RNAs to neurons. The composition and the safety profile of exosomes depend on the type of the exosome-producing cell. Mesenchymal stem cells are considered to be an attractive cell type for therapeutic exosome production. However, scalable methods to isolate and manufacture exosomes from mesenchymal stem cells are lacking, a limitation to the clinical translation of exosome technology. We evaluate mesenchymal stem cells from different sources and find that umbilical cord-derived mesenchymal stem cells produce the highest exosome yield. To optimize exosome production, we cultivate umbilical cord-derived mesenchymal stem cells in scalable microcarrier-based three-dimensional (3D) cultures. In combination with the conventional differential ultracentrifugation, 3D culture yields 20-fold more exosomes (3D-UC-exosomes) than two-dimensional cultures (2D-UC-exosomes). Tangential flow filtration (TFF) in combination with 3D mesenchymal stem cell cultures further improves the yield of exosomes (3D-TFF-exosomes) 7-fold over 3D-UC-exosomes. 3D-TFF-exosomes are seven times more potent in small interfering RNA (siRNA) transfer to neurons compared with 2D-UC-exosomes. Microcarrier-based 3D culture and TFF allow scalable production of biologically active exosomes from mesenchymal stem cells. These findings lift a major roadblock for the clinical utility of mesenchymal stem cell exosomes.
Keywords: exosomes, tangential flow filtration, mesenchymal stem cell
Haraszti et al. optimize scalable production of exosomes from mesenchymal stem cells. Microcarrier-based three-dimensional culture and tangential flow filtration yield an increased number of biologically active exosomes than two-dimensional culture and differential ultracentrifugation.
Introduction
Exosomes are nano-sized (40–150 nm) extracellular vesicles, are surrounded by a lipid bilayer, and are derived from internal cellular compartments.1 Exosomes are released by most cell types and are considered to be part of the intercellular communication system, carrying RNAs and proteins locally and systemically.2, 3, 4, 5, 6, 7, 8 Information transferred via exosomes influences the phenotype of recipient cells.9, 10, 11, 12, 13, 14 Stem cell-derived exosomes are believed to mediate cellular restorative function11, 15, 16 and to modulate the inflammatory state.17, 18, 19, 20, 21, 22, 23 Due to their unique trafficking characteristics, exosomes are being explored as therapeutic RNA delivery vehicles9, 13, 14, 24, 25
The preclinical and clinical development of exosome technology as a delivery platform requires large quantities of exosomes. The isolation method of exosomes is required to be easily expandable to support large-scale manufacturing (e.g., scalable).26, 27 Current methods generate low yields of exosomes and are not scalable, a situation that so far has impeded studies to evaluate preclinical efficacy of exosomes in animals. A dose of 109–1011 exosomes administered per mouse is typically used to achieve biological outcomes.9, 11, 13, 14 Isolation of this exosome quantity requires the processing of liters of conditioned media to treat one animal. Therefore, exosome production to support a well-powered animal study can take several months. Exosomes are usually purified by size exclusion28, 29 or affinity chromatography,30 or by density gradient31, 32 or differential ultracentrifugation (UC).33 The gold standard for exosome retrieval, differential centrifugation, requires four to five sequential centrifugation steps. None of these methods is scalable. Unlike immortal tumor cells lines, the expansion of mesenchymal stem cells (MSCs) is limited in culture. Low yields of exosomes impede the use of mesenchymal stem cells for exosome production.
We combined the strengths of two production strategies to develop a robust and scalable strategy compatible with good manufacturing practices (GMPs) for exosome production from mesenchymal stem cells: (1) microcarrier-based three-dimensional (3D) cell culture is commonly used to grow adherent cells in bioreactors;34 and (2) tangential flow filtration (TFF) is a method to concentrate proteins or viruses from large amounts of cell culture media.35, 36, 37 The physicochemical characteristics of exosomes derived from two-dimensional (2D) or 3D cultures, as well as produced by differential UC or TFF, were compared. We show that both 3D culture and TFF improve the yield of exosomes to a cumulative extent of 140-fold. These exosomes were 7-fold more active in their ability to transfer therapeutic small interfering RNAs (siRNAs) to primary neurons. Thus, the method reported here advances the yield of mesenchymal stem cell exosomes and enables their preclinical exploration.
Results
Wharton’s Jelly-Derived Mesenchymal Stem Cells Produce the Most Exosomes
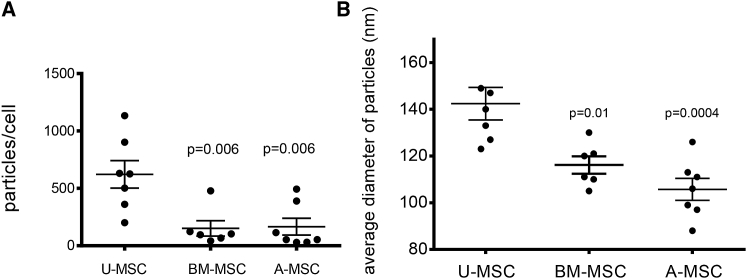
To develop a scalable method for exosome production suitable for manufacturing, we compared exosome yields and doubling times of mesenchymal stem cells derived from common sources: bone marrow, adipose tissue, and umbilical cord Wharton’s jelly (i.e., connective tissue of umbilical cord). Umbilical cord mesenchymal stem cells are distinct from cord blood hematopoietic stem cells. In traditional plastic flask-based cultures, umbilical cord mesenchymal stem cells grew faster (∼4-day doubling time) than mesenchymal stem cells from bone marrow or adipose tissue (∼7-day doubling time). Umbilical cord mesenchymal stem cells yielded four times as many exosomes per cell than did mesenchymal stem cells from bone marrow (p = 0.0063) or adipose tissue (p = 0.006) (Figure 1A). Exosomes derived from umbilical cord mesenchymal stem cells were also larger (140 ± 18 nm) than exosomes from bone marrow (116 ± 9 nm; p = 0.01) and adipose tissue (105 ± 12 nm; p = 0.0004) mesenchymal stem cells (Figure 1B). Based on their availability, favorable doubling time, and high yield of exosomes per cell, we used umbilical cord mesenchymal stem cells for the development of a scalable exosome isolation method.
Figure 1.
Umbilical Cord Mesenchymal Stem Cells Yield the Most Exosomes
(A) Yield of exosomes isolated by differential ultracentrifugation from mesenchymal stem cells derived from umbilical cord (U-MSC), bone marrow (BM-MSC), or adipose (A-MSC). Yield calculated as the number of exosomes in the isolated sample measured by Nanoparticle Tracking Analysis divided by the number of cells in the source cultures. Results of seven experiments are shown, with mean ± SD, one-way ANOVA. (B) Average sizes of U-MSC, BM-MSC, or A-MSC exosomes purified in (A).
3D Culture and Tangential Flow Filtration Enhances Exosome Yield
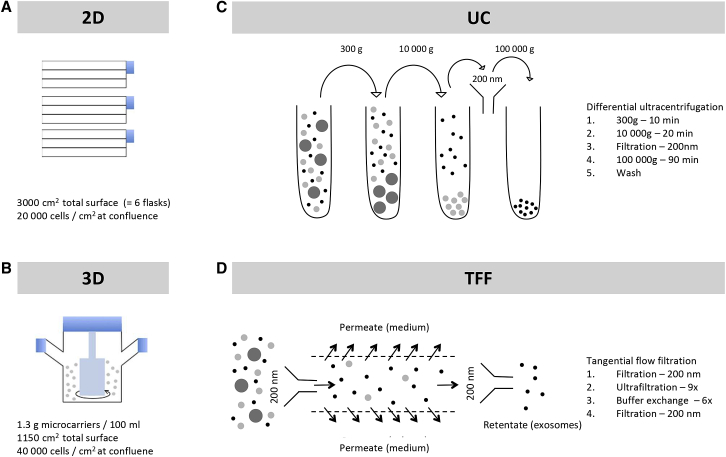
Conventional 2D cultures of adherent umbilical cord mesenchymal stem cells grown in three-layer plastic culture flasks show a density of 20,000 cells/cm2 at confluence (Figure 2A). To increase the expansion of umbilical cord mesenchymal stem cells, we used microcarrier-based 3D culture (a strategy commonly used for large-scale culture of adherent cells34) (Figure 2B), where cell density reached 40,000 cells/cm2, double the density obtained in 2D cultures.
Figure 2.
Scheme of Mesenchymal Stem Cell Culturing Methods and Exosome Isolation Methods
(A) Schematic of flask-based (two-dimensional) mesenchymal stem cell culture. Cells are cultured in triple-layer flasks in mesenchymal basal medium to a density of 20,000 cells/cm2. (B) Schematic of microcarrier-based (three-dimensional) mesenchymal stem cell cultures. Cells are cultured on microcarriers in serum-free/GMP-compatible medium in 250-mL spinner flasks to ∼40,000 cells/cm2. (C) Isolation of exosomes by differential ultracentrifugation. Exosomes are enriched from culture supernatants by sequential ultracentrifugation, with filtration and wash steps, as indicated. (D) Isolation of exosomes by tangential flow filtration. Exosomes are enriched from culture supernatants by tangential flow filtration using a 500-kDa cutoff cartridge, as indicated.
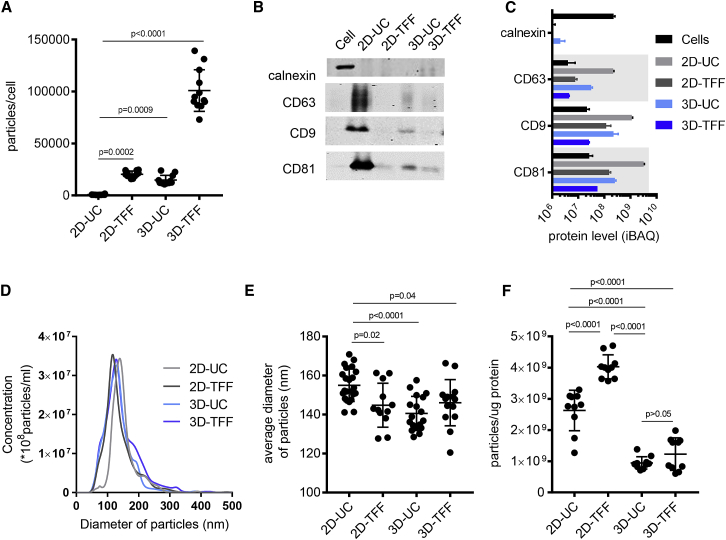
We then compared the gold standard method of exosome isolation, differential UC (Figure 2C), with TFF, a scalable concentration and buffer exchange strategy used during large-scale manufacturing of biologics35, 36 and viruses37 (Figure 2D). TFF isolates exosomes according to their size, whereas differential UC relies on both vesicle size and sedimentation properties. We combined both 2D and 3D cultures with either differential UC or TFF to evaluate the effect of cell culture method, as well as exosome isolation method, on exosome yield. Transmission electron microscopy confirmed the presence of lipid bilayer-surrounded vesicles in all exosome samples (Figure S1). Compared with the conventional 2D cell culture and differential UC (2D-UC-exosomes), TFF improved exosome yield 27-fold (2D-TFF-exosomes; p = 0.0002) and 3D culture 20-fold (3D-UC-exosomes; p = 0.0009) (Figure 3A). The cumulative effect of TFF and 3D culture (3D-TFF-exosomes) was a 140-fold increase of exosome yield compared with 2D-UC-exosomes (p < 0.0001) (Figure 3A). All exosomes were enriched in CD81 and CD9, and depleted of calnexin compared with parent cells (Figures 3B and 3C). CD63 was only enriched in 2D-UC- and 3D-UC-exosomes (Figures 3B and 3C). All exosomes showed a homogeneous size distribution (Figure 3D), and 2D-UC exosomes were slightly larger than other exosome variants (Figures 3D and 3E). 3D culture resulted in a 2- to 4-fold lower particle-to-protein ratio (0.9 × 109 ± 0.2 × 109 3D-UC-exosomes and 1.23 × 109 ± 0.5 × 109 3D-TFF-exosomes per μg protein versus 2.6 × 109 ± 0.6 × 109 2D-UC-exosomes and 4 × 109 ± 0.4 × 109 2D-TFF-exosomes per μg protein; p < 0.0001) than 2D culture (Figure 3F).
Figure 3.
Characterization of Exosomes
(A) Yield of exosomes isolated by 2D-UC, 2D-TFF, 3D-UC, or 3D-TFF (n = 12 measurements each). Yield calculated as the number of exosomes in an isolated sample measured by Nanoparticle Tracking Analysis divided by the number of cells in the source cultures. Plots show yield for each experiment, and the mean ± SD of all measurements, one-way ANOVA with Tukey’s multiple comparison test. (B) Western blot analyses of proteins present in exosomes (CD81, CD9, and CD63). Calnexin is a negative control, present in cells, but not in exosomes. (C) Protein levels detected via LC-MS/MS proteomics in exosome variants and cells. Protein content was determined by intensity-based absolute quantification (iBAQ) analysis.57 (D) Size distribution of exosomes isolated from two-dimensional (2D) or three-dimensional (3D) cultures by differential ultracentrifugation (UC) or tangential flow filtration (TFF). Concentration and size of exosomes were measured by nanoparticle tracking analysis. (E) Average size of 2D-UC- (n = 23), 2D-TFF- (n = 12), 3D-UC- (n = 18), and 3D-TFF-exosomes (n = 14) plotted, showing the mean ± SD of all measurements, one-way ANOVA with Tukey’s multiple comparison test. (F) Number of particles per microgram protein in exosome preparations isolated from 2D or 3D cultures by UC or TFF. Number of particles measured by Nanoparticle Tracking Analysis, and protein concentration measured by Bradford protein assay. Plots show result for each measurement, and the mean ± SD of all measurements, one-way ANOVA with Tukey’s multiple comparison test.
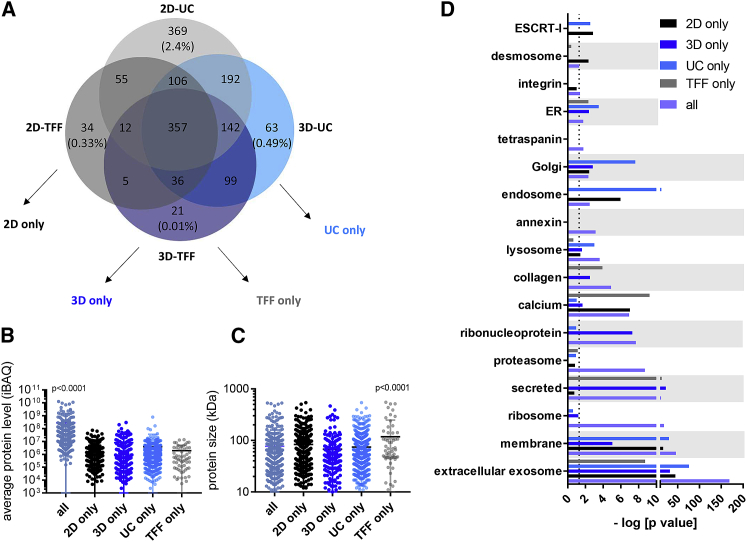
Proteomics analyses showed 357 high-abundance proteins detected in all exosome variants and 21–369 low-abundance proteins unique to an exosome variant (21 in 3D-TFF-exosomes only, 34 in 2D-TFF-exosomes only, 63 in 3D-UC exosomes only, and 369 in 2D-UC exosomes only, representing 0.01%, 0.33%, 0.49%, and 2.4% of all protein amount, respectively) (Figure 4A). Levels of proteins detected only in exosomes from 2D cultures (2D only), 3D cultures (3D only), or isolated by differential UC (UC only) or TFF (TFF only) were 35- to 146-fold lower than levels of proteins present in all exosome variants (p < 0.0001) (35-fold in 3D-only-exosomes, 96-fold in 2D-only-exosomes, 71-fold in UC-only-exosomes, and 146-fold in TFF-only-exosomes) (Figure 4B). Proteins unique to TFF exosomes (i.e., 2D-TFF-exosomes and 3D-TFF-exosomes) were larger (118 ± 130 kDa) than proteins present in other exosome variants (83 ± 70 kDa in 2D-only-exosomes, 67 ± 66 kDa in 3D-only-exosomes, and 74 ± 62 kDa in UC-only-exosomes) (p < 0.0001) (Figure 4C). However, the number of unique proteins detected in TFF-only-exosomes was low (60) compared with 424 unique proteins detected in 2D-only-exosomes, 183 in 3D-only-exosomes, and 624 in UC-only-exosomes. Gene ontology analysis revealed that proteins unique to 2D-only-exosomes were particularly enriched in desmosomal proteins and depleted of calcium-interacting proteins (Figure 4D). 3D-only-exosomes were enriched in ribonucleoproteins. 3D-only-exosomes and TFF-only-exosomes were both enriched in collagen and secreted proteins (e.g., albumin) (Figure 4D). 2D-only-exosomes and UC-only-exosomes were both enriched in “endosomal sorting complex required for transport” (ESCRT) proteins—a pathway involved in exosome formation38—as well as in endosomal proteins (Figure 4D). In addition, proteins present in all exosome variants were enriched in extracellular exosome, membrane, ribosomal, proteasomal, lysosomal, Golgi, and endoplasmic reticulum proteins, as well as in annexins, integrins, and tetraspanins (Figure 4D). Proteins found in all exosome variants are listed in Table S2. These findings suggest (1) that exosomes have a similar protein composition independent of culturing method of the parent cells or of isolation method, and (2) that high levels of secreted proteins are responsible for higher protein-to-vesicle ratio in TFF-exosome samples.
Figure 4.
Proteomic Content of Exosomes
(A) Venn diagram of proteins detected in 2D-UC-exosomes (light gray), 2D-TFF-exosomes (dark gray), 3D-UC exosomes (light blue), and 3D-TFF-exosomes (dark blue). Numbers represent the number of proteins detected in each group. Percentages represent the fraction of unique proteins of 2D-UC-only, 2D-TFF-only, 3D-UC-only, and 3D-TFF-only in the total protein amount of 2D-UC, 2D-TFF, 3D-UC, and 3D-TFF, respectively. Protein amount was determined by intensity-based absolute quantification (iBAQ) analysis.57 (B) Levels of proteins specific to two-dimensional culture (2D-UC and 2D-TFF), three-dimensional culture (3D-UC and 3D-TFF), differential ultracentrifugation (2D-UC and 3D-UC), and tangential flow filtration (2D-TFF and 3D-TFF), from (A), one-way ANOVA. Protein level was determined by intensity-based absolute quantification (iBAQ) analysis.57 (C) Size distribution of proteins specific to two-dimensional culture (2D-UC and 2D-TFF), three-dimensional culture (3D-UC and 3D-TFF), differential ultracentrifugation (2D-UC and 3D-UC), and tangential flow filtration (2D-TFF and 3D-TFF), from (A), one-way ANOVA. (D) Gene ontology analysis of proteins shared or unique to all exosome variants (lavender), 2D-only-exosomes (black), 3D-only-exosomes (dark blue), UC-only-exosomes (light blue), and TFF-only-exosomes (gray), from (A).
3D-TFF-Exosomes Deliver siRNA to Neurons Better Than 2D-UC-Exosomes
Exosome integrity is essential for biological activity and is therefore a major requirement for the development of large-scale isolation methods. We have previously shown that 2D-UC-exosomes can efficiently deliver therapeutic siRNAs to primary neurons.13 We therefore compared the ability of exosome variants to deliver Huntingtin siRNA to neurons, using Huntingtin silencing as a readout for efficient neuronal delivery.
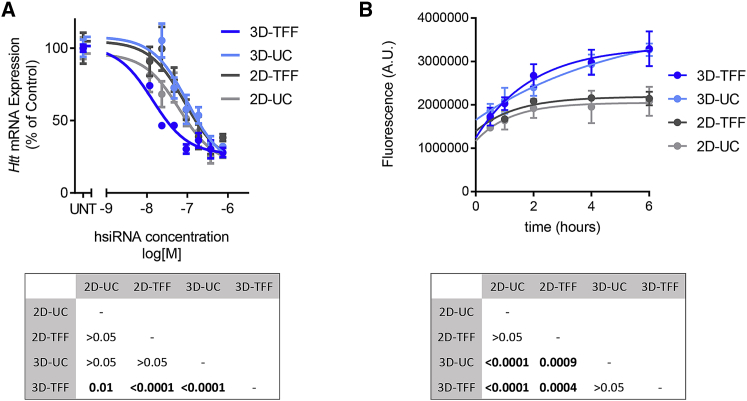
Seven days after treatment, we found that 3D-TFF-exosomes were ∼5-fold more efficient at siRNA transfer and Huntingtin silencing in neurons compared with 2D-UC-exosomes (3D-TFF half maximal inhibitory concentration [IC50], ∼13 nM; 2D-UC IC50 ∼63 nM; p < 0.0001) (Figure 5A). 3D-UC-exosomes and 2D-TFF-exosomes did not differ from conventional 2D-UC-exosomes in Huntingtin silencing (Figure 5A). To address whether the increased potency reflects better vesicle uptake or increased biological availability of internalized siRNAs, we treated neurons with all four types of exosomes loaded with equal amounts of fluorescently labeled siRNAs. Neurons internalized more 3D-TFF-exosomes and 3D-UC-exosomes containing fluorescent siRNA than 2D-UC-exosomes and 2D-TFF-exosomes (p < 0.0001) (Figure 5B). Thus, the culturing method of the parent cells predicted trafficking of exosomes into neurons, but not Huntingtin silencing activity. Nevertheless, enhanced trafficking of 3D-TFF-exosomes into neurons likely underlies their ability to support more efficient silencing.
Figure 5.
TFF-Exosomes Are More Efficient at Delivering siRNAs to Neurons
(A) Dose-response analysis showing Huntingtin (Htt) mRNA levels in mouse primary neurons treated with 2D-UC-, 2D-TFF-, 3D-UC-, and 3D-TFF-exosomes containing the indicated doses of siRNA. Each data point represents the mean ± SEM of n = 3 experiments, two-way ANOVA. (B) Time course of fluorescence in primary neurons treated with 2D-UC-, 2D-TFF-, 3D-UC-, and 3D-TFF-exosomes containing Cy3-labeled siRNA. Each data point represents the mean ± SEM of 62–143 cells per time point, two-way ANOVA. UNT, untreated.
Discussion
The development of exosomes as therapeutic delivery vehicles requires production and purification methods compatible with GMPs. 3D culture systems, xenofree medium, and TFF are suitable for GMP-grade biologics.35, 36, 37, 39 Here we show that 3D xenofree cultures of umbilical cord mesenchymal stem cells and TFF of the conditioned supernatants produced substantially higher yields of exosomes than standard 2D culture and UC. Whereas exosomes isolated from both 2D and 3D cultures via both UC and TFF have similar size distributions and protein contents, 3D-TFF-exosomes are more efficient at siRNA delivery to neurons and at inducing mRNA silencing.
The differences in protein-to-vesicle ratios, as well as in activity between exosome variants, probably reflect differences in their production and preparation methods. The high protein-to-vesicle ratio in 3D-exosome preparations might result from protein aggregates that form in GMP-compatible xenofree medium used for 3D cultures. Optimization of xenofree culture and purification conditions will undoubtedly be required to identify and generate exosomes with desired protein content and protein-to-vesicle ratios. Both cell culturing method and exosome isolation method contributed to improved siRNA transferring and gene silencing activity of 3D-TFF-exosomes. Differences in cellular uptake between exosomes from 3D and 2D cultures may be explained by differences in protein content: enrichment in some secreted proteins (such as AHSG and albumin) in 3D-exosomes may enhance endocytosis,40, 41, 42 and enrichment in ribonucleoproteins may lead to a more efficient intracellular path upon uptake.43 Centrifugal forces applied to exosomes may damage the integrity of exosomal membranes29 and compromise cellular uptake.44 Thus, lack of UC-related damaging effects may also contribute to better performance of 3D-TFF-exosomes compared with 2D-UC-exosomes.
Umbilical cord is an abundant source of stem cells. One human umbilical cord can yield an estimated number of 10 million Wharton’s jelly-derived mesenchymal stem cells.45, 46 By passage 6 this would translate into 6 × 1013 exosomes using 3D culture and TFF. Calculating with a range of 109–1011 exosomes per mouse for preclinical small-animal studies,9, 11, 13, 14 one umbilical cord may provide enough exosomes to treat 600–60,000 mice. Thus, large-scale animal studies may be powered by exosomes from low-passage (under 6) mesenchymal stem cells isolated from a single umbilical cord.
Cell culture conditions and exosome isolation methods should be developed together to advance exosome technology. Therapeutic virus production might serve as an example for exosome technology development.47 Although we use umbilical cord-derived mesenchymal stem cells, the cell culture and exosome isolation methods described here should work for other cell sources. Quality-control steps for large-scale exosome production need to be worked out in detail based on published recommendations for small-scale exosome isolation methods.48, 49 Protein-to-exosome ratio should be considered as part of the quality-control protocols.26
Genetic interference strategies are being developed into promising therapeutic drugs to treat genetically defined diseases. Here we used delivery of Huntingtin siRNAs and silencing as a readout for exosome activity produced at a large scale. We speculate that exosomes produced from microcarrier-based 3D xenofree cultures by TFF will prove useful as delivery vehicles for other therapeutic oligonucleotides that target numerous diseases: siRNAs, antisense oligonucleotides, and CRISPR guide RNAs. The effective, scalable exosome isolation method described here will facilitate the successful transition of the exosome technology to clinical applications.
Materials and Methods
All animal procedures were approved by the University of Massachusetts Medical School Institutional Animal Care and Use Committee (IACUC, protocol number A-2411) under “Preparation of primary cortical neurons”.
Oligonucleotides
Oligonucleotides were synthesized using standard phosphoramidite chemistry, as described previously.50, 51, 52 siRNA sequences and chemical modification patterns used in this study are described in Table S1.
2D Cell Culture
Mesenchymal stem cells from Wharton’s jelly of umbilical cord (PCS−500-010; ATCC, Manassas, VA, USA), adipose tissue (PCS-500-011; ATCC), or bone marrow (PT-2501; Lonza, Basel, Switzerland) were cultured in appropriate stem cell media (PCS-500-030 [ATCC] for umbilical cord- and adipose tissue-derived cells; or PT-3238 [Lonza] for bone marrow-derived cells) in the presence of 2% fetal bovine serum (FBS) and growth factors (PCS-500-040 [ATCC] for umbilical cord- and adipose tissue-derived cells; or PT-4105 [Lonza] for bone marrow-derived cells) at 37°C, 5% CO2. The final concentration of growth factors in umbilical cord-derived mesenchymal stem cells was 5 ng/mL recombinant human fibroblast growth factor (rhFGF), 5 ng/mL rhFGF, 5 ng/mL recombinant human epidermal growth factor (rhEGF). In addition, the media contained 2.4 mM L-alanyl-L-glutamine, 10 U/mL penicillin, 10 μg/mL streptomycin, and 25 ng/mL amphotericin B. Media were changed every 3 days, and cells were expanded until passage 6, to reach a total of 3,000-cm2 surface, equivalent to six T500 triple flasks (density at confluence 20,000 cells/cm2). We established that umbilical cord-derived mesenchymal stem cells can be frozen in the presence of 20% FBS and 10% DMSO in compete medium as described above. Freezing cell stocks early on enables re-starting mesenchymal stem cell cultures at lower passage numbers.
Isolation of Exosomes via Differential UC
Differential UC of exosomes relies on their vesicle size and sedimentation properties. Sequential centrifugation steps with increasing force of centrifugation deplete the conditioned medium from large particles and/or vesicles with high sedimentation rates. A final UC step sediments small vesicles or exosomes, leaving the smaller proteins in the supernatant.33
Medium was changed to exosome-depleted medium (i.e., stem cell medium was centrifuged at 100,000 × g for at least 17 hr), and cultures were incubated for 48 hr. Exosomes were then purified from the conditioned medium by differential UC, as described.13 In brief, cell debris were pelleted at 300 × g (10 min). Larger vesicles were pelleted at 10,000 × g (30 min), the supernatant filtered through a 0.2-μm membrane (Nalgene aPES; Thermo Fisher Scientific, Waltham, MA, USA), and exosomes pelleted at 100,000 Χ g (90 min) using 70-mL polycarbonate bottles (355622; Beckman Coulter, Brea, CA, USA) and Type 45 Ti rotor (339160; Beckman Coulter). Exosome pellets were washed once in 1 mL sterile PBS and centrifuged for 90 min at 100,000 × g in a tabletop ultracentrifuge using a TLA-110 rotor (366730; Beckman Coulter).
3D Cell Culture
Cells are grown on the surfaces of spherical support matrix beads and distributed in medium by stirring in a spinner flask.
In this method, a pump circulates the conditioned culture medium through membranes or filters with pores that are sized for a specific application. Particles that are smaller than the pore size pass through and are removed from the system (e.g., permeate). Larger particles than the pore size are withheld in the lumen of the fibers (e.g., retentate) and circulated back into the product. Multiple rounds of the ultrafiltration step lead to efficient particle concentration. In particular, we first passed the conditioned cell culture supernatant through a 200-nm pore size membrane to remove large vesicles and particles
The filtered conditioned medium was subjected to TFF using a hollow fiber filter with a 500-kDa molecular weight cutoff (MWCO) and concentrated 9-fold (volume reduced 9-fold). In the next step the cell culture medium was exchanged with PBS, by continuously feeding the system with PBS to replace the loss of permeate. The final product was sterile filtered using a 200-nm filter, resulting in a filtrate that contains exosomes in PBS (Figure 2B; see Materials and Methods).
Spinner flasks (250 mL) containing 3.2 g (1,150 cm2 total surface area) of Star-Plus Microcarriers (SoloHill; Pall Life Sciences, Port Washington, NY, USA) were autoclaved. Umbilical cord-derived mesenchymal stem cells were seeded to a density of 8,000 cells/cm2 in umbilical cord-derived stem cell medium, the impeller speed set to 36 rpm, and cells were cultured at 37°C. When cells were homogenously spread on microcarriers, medium was removed, microcarriers washed in PBS twice, and 250 mL of serum-free and xenofree StemPro medium was added (A1067501; Life Technologies, Carlsbad, CA, USA), and cells were cultured at 37°C and 36 rpm impeller speed.
Isolation of Exosomes via Tangential Flow Filtration
250 mL of conditioned medium (StemPro, A1067501; Life Technologies, Carlsbad, CA, USA) was collected after 48 hr. Collection was performed four times; conditioned medium was stored at 4°C and subsequently pooled together (final volume, 1 L). The conditioned medium was filtered through a 0.2-μm polyethersulfone (PES) membrane. Conditioned medium was then subjected to ultrafiltration in a TFF system using a 500-kDa cutoff TFF cartridge (MidiKros mPES 115 cm2, D02-E500-05-S; Spectrum Labs, Rancho Dominguez, CA, USA). A feed flow rate of 120 mL/min, transmembrane pressure <3.5 psi, and a crossflow rate >10:1 were maintained throughout the filtration operation. The conditioned medium was concentrated 9-fold and then buffer exchanged with 6× volume of PBS. The exosomes were 0.2 μm filtered (PES membrane) and stored in 0.1 M sucrose in a polyethylene terephthalate glycol (PETG) bottle at −80°C.
Characterization of Exosomes
Nanoparticle Tracking Analysis (NanoSight NS300; Malvern, Malvern, UK) was used to measure concentration and size distribution of exosomes. Samples were diluted 1:1,000 in PBS, manually injected into the instrument, and videos were acquired at ambient temperature at camera level 9 for 1 min per sample (n = 3). Exosomes were then frozen at −80°C in 0.1 M sucrose containing protease inhibitor cocktail (P8340; Sigma-Aldrich, St. Louis, MO, USA) until further use. Exosomes were visualized by transmission electron microscopy using a JEOL 1100 transmission electron microscope (JEOL, Peabody, MA, USA; Mass General Hospital) at 60 kV.53 For western blot analyses, exosome or cell pellets were suspended in radioimmunoprecipitation assay (RIPA) buffer (Pierce 899000; Thermo Fisher Scientific, Waltham, MA, USA) containing PMSF (36978; Thermo Fisher Scientific) and protease inhibitor cocktail (cOmplete Mini, 11836153001; Roche, Indianapolis, IN, USA), and samples were sonicated for 15 min. Insoluble material was pelleted by centrifugation for 15 min at 10,000 × g, 4°C. Proteins from 3 × 1010 vesicles or 6,000 cells were loaded onto NuPAGE 4%–12% Bis-Tris gels (Thermo Fisher Scientific, Waltham, MA, USA). After transfer to polyvinylidene fluoride (PVDF; Bio-Rad, Hercules, CA, USA), membranes were incubated with antibodies and washed, and images were captured using an Odyssey system (Li-Cor, Bad Homburg, Germany) according to manufacturer’s instructions. Primary antibodies used were Calnexin (C5C9; Cell Signaling, Danvers, MA, USA), CD63 (H5C6; BD Biosciences, San Jose, CA, USA), CD81 (B11; Santa Cruz Biotechnology, Dallas, TX, USA), and CD9 (C4; Santa Cruz Biotechnology). Liquid chromatography-mass spectrometry/mass spectrometry (LC-MS/MS) of exosomes was performed as described previously.53 Label-free quantification of proteins was performed via the iBAQ (intensity-based absolute quantification54) method in Scaffold Viewer (Proteome Software). This method accounts for the different lengths of proteins.
Loading siRNAs into Exosomes
4.5 × 1010 exosomes were co-incubated with 1 nmol of siRNA at 37°C for 1 hr in 500 μL of PBS (i.e., loading mixture). The exosome-siRNA mixture was centrifuged for 90 min at 100,000 × g, and supernatants containing unloaded siRNAs were removed. Pellets were suspended in 500 μL of PBS to measure fluorescence or in 300 μL of Neural Q medium to treat primary neurons.
To quantify loading of Cy3-labeled siRNA, a 200-μL aliquot was taken from the suspended exosome pellet or from the supernatant. Fluorescence was measured at 550 nm excitation, 570 nm emission on a TECAN instrument. The percent loaded siRNA was calculated as pellet/(pellet + supernatant). The siRNA copy number per exosome was estimated as: (percent of loaded siRNA) × (amount of siRNA initially mixed in with exosomes [mol]) × (Avogadro number)/(number of exosomes initially mixed in).
Preparation of Primary Cortical Neurons
Primary cortical neurons were isolated from wild-type FVBNj mouse embryos (embryonic day [E] 15.5). Pregnant females were euthanized by intraperitoneal injection with a solution of ketamine (100 mg/kg; KETASET; Zoetis, Kalamazoo, MI, USA) and xylazine (10 mg/kg; AnaSed; NDC59399-111-50; AKORN, Laker Forest, IL, USA), or with isoflurane (Isoflurane, USP; NDC66794-013-010; Piramal Critical Care, Bethlehem, PA, USA), followed by cervical dislocation. Embryos were removed and transferred to ice-cold DMEM/F12 medium (11320; Invitrogen). Brains were removed from DMEM, and meninges were carefully detached under a dissecting microscope. Cortices were isolated and dissolved in pre-warmed (37°C) papain-DNase solution for 30 min at 37°C, 5% CO2. Papain (54N15251; Worthington, Lakewood, NJ, USA) was dissolved in 2 mL Hibernate E (#HE; Brainbits, Springfield, IL, USA) and supplemented with 0.25 mL of 10 mg/mL DNase I (54M15168; Worthington) in Hibernate E. After 30-min incubation, the papain solution was removed and 1 mL of NeuralQ (N3100; Sigma-Aldrich) supplemented with 2.5% FBS was added to the tissue. Tissues were dissociated by trituration through a fire-polished, glass Pasteur pipet. Neurons were counted in a Neubauer chamber and diluted to 106 cells/mL. Cells were plated at a density of 105 neurons per well in 96-well plates pre-coated with poly-l-lysine (356515; BD BIOCOAT, Corning, NY, USA). Cells were incubated overnight at 37°C, 5% CO2, and an equal volume of NeuralQ supplemented with anti-mitotics, 0.484 μL/mL 5′ uridine 5′-triphosphate (UtP) (U6625; Sigma-Aldrich), and 0.2402 μL/mL 5′ 5-fluoro-2′-deoxyuridine (FdU) (F3503; Sigma-Aldrich) was added to prevent the growth of non-neuronal cells. Half the volume of media was replaced with fresh NeuralQ containing anti-mitotics every 48 hr until the experiments were performed.
Measurement of siRNA Silencing Activity in Neurons
Neurons were treated with siRNA-loaded exosomes (suspended in NeuralQ medium) and then incubated for 7 days at 37°C, 5% CO2. Treated neurons were lysed and mRNAs were quantified using QuantiGene 2.0 assay kit (Affymetrix, QS0011; Thermo Fisher Scientific),55 and probes for mouse Htt (Affymetrix, SB-14150; Thermo Fisher Scientific) and mouse Hprt (Affymetrix, SB-15463; Thermo Fisher Scientific). Datasets were normalized to housekeeping gene Hprt. Each measurement was run in duplicate and repeated three times independently.
Measurement of Live siRNA Uptake in Neurons
For the analysis of siRNA uptake in vitro, primary neuron nuclei plated in 35-mm glass-bottom dishes (P35G-1.5-10-C; MatTek, Ashland, MA, USA) were stained with NucBlue live cell stain (R37605; Thermo Fisher Scientific, Waltham, MA, USA), and neurons were treated with fluorescently labeled siRNA targeting Ppib gene (Table S1). Images were acquired with a Leica DM IRE2 (Leica Microsystems, Buffalo Grove, IL, USA) confocal microscope using a 40× oil-immersion objective and DAPI channel (exposure time, 50 ms), as well as mCherry channel (exposure time, 200 ms). Images were processed using ImageJ software56 (NIH, Bethesda, MD, USA). The relative uptake of siRNA, loaded in UC-exosomes or TFF-exosomes, was estimated based on pixel integrated density of five images for each time point.
Statistical Analysis
Data were analyzed using GraphPad Prism 7, version 7.04 (GraphPad Software, La Jolla, CA, USA).
In in vitro silencing experiments, IC50 values were determined by fitting a dose-response curve using “log(inhibitor) vs. response − variable slope (four parameters)” equation. Curves were compared using two-way ANOVA.
Differences in all comparisons were considered significant at p values <0.05.
Author Contributions
Conceptualization: R.A.H., Y.W., N.A., and A.K.; Methodology: R.A.H., M.S., Y.Y.S., M.-C.D., and R.W.; Validation: R.A.H., M.S., S.A.S., and A.K.; Formal Analysis: R.A.H.; Investigation: R.A.H., R.M., M.S., Y.Y.S., E.S., M. Dubuke, X.L., and A.C.; Resources: Y.W., M. DiFiglia, N.A., and A.K.; Writing – Original Draft: R.A.H.; Writing – Editing and Reviewing: R.A.H., M.S., Y.W., N.A., and A.K.; Visualization: R.A.H.; Supervision: M. DiFiglia, Y.W., N.A., and A.K.; Project Administration: R.A.H.; Funding Acquisition: Y.W., N.A., and A.K.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
We thank Darryl Conte for his assistance with text editing. This work was supported by NIH UH3 grant TR 000888 05 to N.A. and A.K.; UMass CCTS grant UL1 TR000161 to N.A., A.K., and Y.W.; NIH grants RO1GM10880304, RO1NS10402201, and S10 OD020012 to A.K.; and the CHDI Foundation (Research Agreement A-6119, JSC A6367) to N.A. M.-C.D. was supported by a Huntington’s Disease Society of America Postdoctoral Fellowship.
Footnotes
Supplemental Information includes one figure and two tables and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.09.015.
Contributor Information
Neil Aronin, Email: neil.aronin@umassmed.edu.
Anastasia Khvorova, Email: anastasia.khvorova@umassmed.edu.
Supplemental Information
References
- 1.Heijnen H.F., Schiel A.E., Fijnheer R., Geuze H.J., Sixma J.J. Activated platelets release two types of membrane vesicles: microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood. 1999;94:3791–3799. [PubMed] [Google Scholar]
- 2.Valadi H., Ekström K., Bossios A., Sjöstrand M., Lee J.J., Lötvall J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 3.Skog J., Würdinger T., van Rijn S., Meijer D.H., Gainche L., Sena-Esteves M., Curry W.T., Jr., Carter B.S., Krichevsky A.M., Breakefield X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frühbeis C., Fröhlich D., Kuo W.P., Amphornrat J., Thilemann S., Saab A.S., Kirchhoff F., Möbius W., Goebbels S., Nave K.A. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013;11:e1001604. doi: 10.1371/journal.pbio.1001604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korkut C., Li Y., Koles K., Brewer C., Ashley J., Yoshihara M., Budnik V. Regulation of postsynaptic retrograde signaling by presynaptic exosome release. Neuron. 2013;77:1039–1046. doi: 10.1016/j.neuron.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Abrami L., Brandi L., Moayeri M., Brown M.J., Krantz B.A., Leppla S.H., van der Goot F.G. Hijacking multivesicular bodies enables long-term and exosome-mediated long-distance action of anthrax toxin. Cell Rep. 2013;5:986–996. doi: 10.1016/j.celrep.2013.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zomer A., Maynard C., Verweij F.J., Kamermans A., Schäfer R., Beerling E., Schiffelers R.M., de Wit E., Berenguer J., Ellenbroek S.I.J. In vivo imaging reveals extracellular vesicle-mediated phenocopying of metastatic behavior. Cell. 2015;161:1046–1057. doi: 10.1016/j.cell.2015.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westergard T., Jensen B.K., Wen X., Cai J., Kropf E., Iacovitti L., Pasinelli P., Trotti D. Cell-to-cell transmission of dipeptide repeat proteins linked to C9orf72-ALS/FTD. Cell Rep. 2016;17:645–652. doi: 10.1016/j.celrep.2016.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beltrami C., Besnier M., Shantikumar S., Shearn A.I., Rajakaruna C., Laftah A., Sessa F., Spinetti G., Petretto E., Angelini G.D., Emanueli C. Human pericardial fluid contains exosomes enriched with cardiovascular-expressed microRNAs and promotes therapeutic angiogenesis. Mol. Ther. 2017;25:679–693. doi: 10.1016/j.ymthe.2016.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen S., Dooner M., Cheng Y., Papa E., Del Tatto M., Pereira M., Deng Y., Goldberg L., Aliotta J., Chatterjee D. Mesenchymal stromal cell-derived extracellular vesicles rescue radiation damage to murine marrow hematopoietic cells. Leukemia. 2016;30:2221–2231. doi: 10.1038/leu.2016.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saha B., Momen-Heravi F., Kodys K., Szabo G. MicroRNA cargo of extracellular vesicles from alcohol-exposed monocytes signals naive monocytes to differentiate into M2 macrophages. J. Biol. Chem. 2016;291:149–159. doi: 10.1074/jbc.M115.694133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didiot M.C., Hall L.M., Coles A.H., Haraszti R.A., Godinho B.M., Chase K., Sapp E., Ly S., Alterman J.F., Hassler M.R. Exosome-mediated delivery of hydrophobically modified siRNA for Huntingtin mRNA silencing. Mol. Ther. 2016;24:1836–1847. doi: 10.1038/mt.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., Li H., Haque F., Wang S., Croce C.M. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang S., Chuah S.J., Lai R.C., Hui J.H.P., Lim S.K., Toh W.S. MSC exosomes mediate cartilage repair by enhancing proliferation, attenuating apoptosis and modulating immune reactivity. Biomaterials. 2018;156:16–27. doi: 10.1016/j.biomaterials.2017.11.028. [DOI] [PubMed] [Google Scholar]
- 16.Willis G.R., Fernandez-Gonzalez A., Anastas J., Vitali S.H., Liu X., Ericsson M., Kwong A., Mitsialis S.A., Kourembanas S. Mesenchymal stromal cell exosomes ameliorate experimental bronchopulmonary dysplasia and restore lung function through macrophage immunomodulation. Am. J. Respir. Crit. Care Med. 2018;197:104–116. doi: 10.1164/rccm.201705-0925OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Fu L., Liang Y., Guo Z., Wang L., Ma C., Wang H. Exosomes originating from MSCs stimulated with TGF-β and IFN-γ promote Treg differentiation. J. Cell. Physiol. 2018;233:6832–6840. doi: 10.1002/jcp.26436. [DOI] [PubMed] [Google Scholar]
- 18.Monguió-Tortajada M., Roura S., Gálvez-Montón C., Pujal J.M., Aran G., Sanjurjo L., Franquesa M., Sarrias M.R., Bayes-Genis A., Borràs F.E. Nanosized UCMSC-derived extracellular vesicles but not conditioned medium exclusively inhibit the inflammatory response of stimulated T cells: implications for nanomedicine. Theranostics. 2017;7:270–284. doi: 10.7150/thno.16154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amarnath S., Foley J.E., Farthing D.E., Gress R.E., Laurence A., Eckhaus M.A., Métais J.Y., Rose J.J., Hakim F.T., Felizardo T.C. Bone marrow-derived mesenchymal stromal cells harness purinergenic signaling to tolerize human Th1 cells in vivo. Stem Cells. 2015;33:1200–1212. doi: 10.1002/stem.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skokos D., Botros H.G., Demeure C., Morin J., Peronet R., Birkenmeier G., Boudaly S., Mécheri S. Mast cell-derived exosomes induce phenotypic and functional maturation of dendritic cells and elicit specific immune responses in vivo. J. Immunol. 2003;170:3037–3045. doi: 10.4049/jimmunol.170.6.3037. [DOI] [PubMed] [Google Scholar]
- 21.Kim S.H., Bianco N.R., Shufesky W.J., Morelli A.E., Robbins P.D. Effective treatment of inflammatory disease models with exosomes derived from dendritic cells genetically modified to express IL-4. J. Immunol. 2007;179:2242–2249. doi: 10.4049/jimmunol.179.4.2242. [DOI] [PubMed] [Google Scholar]
- 22.Bai L., Shao H., Wang H., Zhang Z., Su C., Dong L., Yu B., Chen X., Li X., Zhang X. Effects of mesenchymal stem cell-derived exosomes on experimental autoimmune uveitis. Sci. Rep. 2017;7:4323. doi: 10.1038/s41598-017-04559-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Du Y.M., Zhuansun Y.X., Chen R., Lin L., Lin Y., Li J.G. Mesenchymal stem cell exosomes promote immunosuppression of regulatory T cells in asthma. Exp. Cell Res. 2018;363:114–120. doi: 10.1016/j.yexcr.2017.12.021. [DOI] [PubMed] [Google Scholar]
- 24.O’Loughlin A.J., Mäger I., de Jong O.G., Varela M.A., Schiffelers R.M., El Andaloussi S., Wood M.J.A., Vader P. Functional delivery of lipid-conjugated siRNA by extracellular vesicles. Mol. Ther. 2017;25:1580–1587. doi: 10.1016/j.ymthe.2017.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stremersch S., Vandenbroucke R.E., Van Wonterghem E., Hendrix A., De Smedt S.C., Raemdonck K. Comparing exosome-like vesicles with liposomes for the functional cellular delivery of small RNAs. J. Control. Release. 2016;232:51–61. doi: 10.1016/j.jconrel.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 26.Reiner A.T., Witwer K.W., van Balkom B.W.M., de Beer J., Brodie C., Corteling R.L., Gabrielsson S., Gimona M., Ibrahim A.G., de Kleijn D. Concise review: developing best-practice models for the therapeutic use of extracellular vesicles. Stem Cells Transl. Med. 2017;6:1730–1739. doi: 10.1002/sctm.17-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Colao I.L., Corteling R., Bracewell D., Wall I. Manufacturing exosomes: a promising therapeutic platform. Trends Mol. Med. 2018;24:242–256. doi: 10.1016/j.molmed.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Nordin J.Z., Lee Y., Vader P., Mäger I., Johansson H.J., Heusermann W., Wiklander O.P., Hällbrink M., Seow Y., Bultema J.J. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine (Lond.) 2015;11:879–883. doi: 10.1016/j.nano.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 29.Corso G., Mäger I., Lee Y., Görgens A., Bultema J., Giebel B., Wood M.J.A., Nordin J.Z., Andaloussi S.E. Reproducible and scalable purification of extracellular vesicles using combined bind-elute and size exclusion chromatography. Sci. Rep. 2017;7:11561. doi: 10.1038/s41598-017-10646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greening D.W., Xu R., Ji H., Tauro B.J., Simpson R.J. A protocol for exosome isolation and characterization: evaluation of ultracentrifugation, density-gradient separation, and immunoaffinity capture methods. Methods Mol. Biol. 2015;1295:179–209. doi: 10.1007/978-1-4939-2550-6_15. [DOI] [PubMed] [Google Scholar]
- 31.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wubbolts R., Leckie R.S., Veenhuizen P.T., Schwarzmann G., Möbius W., Hoernschemeyer J., Slot J.W., Geuze H.J., Stoorvogel W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003;278:10963–10972. doi: 10.1074/jbc.M207550200. [DOI] [PubMed] [Google Scholar]
- 33.Théry C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;Chapter 3 doi: 10.1002/0471143030.cb0322s30. Unit 3.22. [DOI] [PubMed] [Google Scholar]
- 34.van Wezel A.L. The large-scale cultivation of diploid cell strains in microcarrier culture. Improvement of microcarriers. Dev. Biol. Stand. 1976;37:143–147. [PubMed] [Google Scholar]
- 35.Grimm K.M., Trigona W.L., Heidecker G.J., Joyce J.G., Fu T.M., Shiver J.W., Keller P.M., Cook J.C. An enhanced and scalable process for the purification of SIV Gag-specific MHC tetramer. Protein Expr. Purif. 2001;23:270–281. doi: 10.1006/prep.2001.1507. [DOI] [PubMed] [Google Scholar]
- 36.Dizon-Maspat J., Bourret J., D’Agostini A., Li F. Single pass tangential flow filtration to debottleneck downstream processing for therapeutic antibody production. Biotechnol. Bioeng. 2012;109:962–970. doi: 10.1002/bit.24377. [DOI] [PubMed] [Google Scholar]
- 37.Potter M., Lins B., Mietzsch M., Heilbronn R., Van Vliet K., Chipman P., Agbandje-McKenna M., Cleaver B.D., Clément N., Byrne B.J., Zolotukhin S. A simplified purification protocol for recombinant adeno-associated virus vectors. Mol. Ther. Methods Clin. Dev. 2014;1:14034. doi: 10.1038/mtm.2014.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 39.Navé J.F., Taylor D., Tyms S., Kenny M., Eggenspiller A., Eschbach A., Dulworth J., Brennan T., Piriou F., Halazy S. Synthesis, antiviral activity and enzymatic phosphorylation of 9-phosphonopentenyl derivatives of guanine. Antiviral Res. 1995;27:301–316. doi: 10.1016/0166-3542(95)00023-f. [DOI] [PubMed] [Google Scholar]
- 40.Herrmann M., Schäfer C., Heiss A., Gräber S., Kinkeldey A., Büscher A., Schmitt M.M., Bornemann J., Nimmerjahn F., Herrmann M. Clearance of fetuin-A–containing calciprotein particles is mediated by scavenger receptor-A. Circ. Res. 2012;111:575–584. doi: 10.1161/CIRCRESAHA.111.261479. [DOI] [PubMed] [Google Scholar]
- 41.Jiang Y., Sverdlov M.S., Toth P.T., Huang L.S., Du G., Liu Y., Natarajan V., Minshall R.D. Phosphatidic acid produced by RalA-activated PLD2 stimulates caveolae-mediated endocytosis and trafficking in endothelial cells. J. Biol. Chem. 2016;291:20729–20738. doi: 10.1074/jbc.M116.752485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pang Z., Gao H., Chen J., Shen S., Zhang B., Ren J., Guo L., Qian Y., Jiang X., Mei H. Intracellular delivery mechanism and brain delivery kinetics of biodegradable cationic bovine serum albumin-conjugated polymersomes. Int. J. Nanomedicine. 2012;7:3421–3432. doi: 10.2147/IJN.S32514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Villarroya-Beltri C., Gutiérrez-Vázquez C., Sánchez-Cabo F., Pérez-Hernández D., Vázquez J., Martin-Cofreces N., Martinez-Herrera D.J., Pascual-Montano A., Mittelbrunn M., Sánchez-Madrid F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013;4:2980. doi: 10.1038/ncomms3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino A., Costa-Silva B., Shen T.L., Rodrigues G., Hashimoto A., Tesic Mark M., Molina H., Kohsaka S., Di Giannatale A., Ceder S. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527:329–335. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nazari-Shafti T.Z., Bruno I.G., Martinez R.F., Coleman M.E., Alt E.U., McClure S.R. High yield recovery of equine mesenchymal stem cells from umbilical cord matrix/Wharton’s jelly using a semi-automated process. Methods Mol. Biol. 2015;1235:131–146. doi: 10.1007/978-1-4939-1785-3_12. [DOI] [PubMed] [Google Scholar]
- 46.Mennan C., Brown S., McCarthy H., Mavrogonatou E., Kletsas D., Garcia J., Balain B., Richardson J., Roberts S. Mesenchymal stromal cells derived from whole human umbilical cord exhibit similar properties to those derived from Wharton’s jelly and bone marrow. FEBS Open Bio. 2016;6:1054–1066. doi: 10.1002/2211-5463.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lock M., Alvira M., Vandenberghe L.H., Samanta A., Toelen J., Debyser Z., Wilson J.M. Rapid, simple, and versatile manufacturing of recombinant adeno-associated viral vectors at scale. Hum. Gene Ther. 2010;21:1259–1271. doi: 10.1089/hum.2010.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lötvall J., Hill A.F., Hochberg F., Buzás E.I., Di Vizio D., Gardiner C., Gho Y.S., Kurochkin I.V., Mathivanan S., Quesenberry P. Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J. Extracell. Vesicles. 2014;3:26913. doi: 10.3402/jev.v3.26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lener T., Gimona M., Aigner L., Börger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F.A., Del Portillo H.A. Applying extracellular vesicles based therapeutics in clinical trials—an ISEV position paper. J. Extracell. Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. Hydrophobically modified siRNAs silence Huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Haraszti R.A., Roux L., Coles A.H., Turanov A.A., Alterman J.F., Echeverria D., Godinho B.M.D.C., Aronin N., Khvorova A. 5′-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017;45:7581–7592. doi: 10.1093/nar/gkx507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikan M., Osborn M.F., Coles A.H., Godinho B.M., Hall L.M., Haraszti R.A., Hassler M.R., Echeverria D., Aronin N., Khvorova A. Docosahexaenoic acid conjugation enhances distribution and safety of siRNA upon local administration in mouse brain. Mol. Ther. Nucleic Acids. 2016;5:e344. doi: 10.1038/mtna.2016.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Haraszti R.A., Didiot M.C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., Gao F., Narain N.R., DiFiglia M., Kiebish M.A. High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilhelm M., Schlegl J., Hahne H., Gholami A.M., Lieberenz M., Savitski M.M., Ziegler E., Butzmann L., Gessulat S., Marx H. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- 55.Coles A.H., Osborn M.F., Alterman J.F., Turanov A.A., Godinho B.M., Kennington L., Chase K., Aronin N., Khvorova A. A high-throughput method for direct detection of therapeutic oligonucleotide-induced gene silencing in vivo. Nucleic Acid Ther. 2016;26:86–92. doi: 10.1089/nat.2015.0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schneider C.A., Rasband W.S., Eliceiri K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haraszti R.A., Didiot M.C., Sapp E., Leszyk J., Shaffer S.A., Rockwell H.E., Gao Fei., Narain N.R., DiFiglia M., Kiebish High-resolution proteomic and lipidomic analysis of exosomes and microvesicles from different cell sources. J. Extracell. Vesicles. 2016;5:32570. doi: 10.3402/jev.v5.32570. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.