Abstract
Background: Lung adenocarcinoma has a strong tendency to develop into bone metastases, especially spinal metastases (SM). Long noncoding RNAs (lncRNAs) play critical roles in regulating several biological processes in cancer cells. However, the mechanisms underlying the roles of lncRNAs in the development of SM have not been elucidated to date.
Methods: Clinical specimens were collected for analysis of differentially expressed lncRNAs. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to examine the effects of these genes on pathways. RNA pull-down was utilized to identify the targeting protein of lncRNAs. The effects of lncRNA on its target were detected in A549 and SPCA-1 cells via perturbation of the lncRNA expression. Oncological behavioral changes in transfected cells and phosphorylation of kinases in the relevant pathways, with or without inhibitors, were observed. Further, tumorigenicity was found to occur in experimental nude mice.
Results: LINC00852 and the mitogen-activated protein kinase (MAPK) pathway were found to be associated with SM. Moreover, the LINC00852 target S100A9 had a positive regulatory role in the progression, migration, invasion, and metastasis of lung adenocarcinoma cells, both in vitro and in vivo. Furthermore, S100A9 strongly activated the P38 and REK1/2 kinases, and slightly activated the phosphorylation of the JNK kinase in the MAPK pathway in A549 and SPCA-1 cells.
Conclusion: LINC00852 targets S100A9 to promote progression and oncogenic ability in lung adenocarcinoma SM through activation of the MAPK pathway. These findings suggest a potential novel target for early intervention against SM in lung cancer.
Keywords: lung adenocarcinoma, spinal metastasis, lncRNA, S100A9, mitogen-activated protein kinase
Introduction
Lung cancer remains the leading cause of cancer-related deaths worldwide, with lung adenocarcinoma being the main subtype1. Moreover, lung cancer exhibits a strong tendency to develop into bone metastases; an estimated 30 to 40% of lung cancer patients develop bone metastasis, among which spinal metastasis (SM) is the most common2,3. The development of SM represents a deleterious stage in the course of the disease that has a strong impact on the patient's quality of life and is associated with poor prognosis and high morbidity4,5. Therefore, the prevention of SM of malignant tumors is critical.
Non-coding RNAs are a type of RNA that is not involved in encoding proteins. Non-coding RNAs may be divided into two categories based on the number of nucleotides contained: small non-encoding RNAs, such as microRNAs and circRNAs; and long non-coding RNAs (lncRNAs), which are more than 200 nt in length. Recent studies showed that lncRNAs affect diverse physiological and pathological cellular processes at multiple levels, including gene expression, epigenetic modulation, and post-transcriptional control6,7. The roles of lncRNAs in the pathogenesis and development of bone tumors, as well as in bone metastases, are recognized8,9. However, the mechanisms underlying the roles of lncRNAs in SM remain to be elucidated.
Cancer metastasis requires evasion of immune surveillance and the involvement of numerous cell-lethal signals. The outcome is co-determined by the various immune-related molecules in the metastatic microenvironment10,11. S100A9 is a calcium-binding protein. Under physiological conditions, S100A9 and other S100 family proteins are intracellular proteins. However, tumor cells constantly secrete S100A9 to recruit myeloid-derived suppressor cells, thereby promoting cancer growth via inflammatory pathways and forming a special pre-metastatic immunosuppressive niche12. Therefore, the S100A9 protein plays a critical role in cancer metastasis 13-15.
In this study, we profiled lncRNA expression in lung adenocarcinoma SM and primary lung adenocarcinoma tissues. The differentially expressed lncRNAs were selected to lncRNA target prediction. The binding relationships between the predicted lncRNAs and their target effector molecules were further validated. Finally, the predicted lncRNAs and their target proteins were studied with respect to their roles in various tumorigenic processes in vitro and in vivo.
Materials and methods
Patients and treatments
Fifty-four clinical specimens from 2012 to 2017 were selected randomly from the biobank of the Department of Orthopaedic Surgery and Thoracic Surgery at Zhongshan Hospital, Fudan University. These included 18 cases of primary lung adenocarcinoma, 18 cases of lung adenocarcinoma SM, and 18 healthy vertebral samples (fracture cases). Tumor diagnoses were verified based on postoperative pathological reports. No patients received treatment before surgery. All tissue samples had been collected during surgery, snap-frozen in a liquid nitrogen tank, and then stored in a freezer at -80°C until use. Informed consent was provided by the patients before surgery. We obtained approval from the institutional review board of Zhongshan Hospital, Fudan University.
Genome-wide analysis of lncRNAs and lncRNA target prediction
The SBC human ceRNA microarray v1.0 (Shanghai Biotechnology Corporation, China) was used for analysis of differentially expressed lncRNAs in clinical specimens. Microarray experiments were performed following a protocol from Agilent Technologies Inc., Shanghai Biotechnology Corporation. Abnormally expressed genes were added to the Database for Annotation, Visualization and Integrated Discovery (DAVID; http://david.abcc.ncifcrf.gov/) v6.7. The Kyoto Encyclopedia of Genes and Genomes (KEGG) was used to analyze the potential effects of these genes in tumor pathways. The differentially expressed lncRNAs were then used for target prediction, using an algorithm described in a previous report16. First, we searched for target genes in cis using the genome browser of the University of California, Santa Cruz (UCSC) (http://genome.ucsc.edu/); these were defined as genes transcribed within a 10-kbp window upstream or downstream of lncRNAs. Second, the interaction of lncRNA with mRNA molecules were determined based on mRNA sequence complementarity and RNA double-chain energy prediction. BLAST software was used for first-round screening. Finally, we searched for trans-acting target genes using RNAplex software.
Biotinylated RNA pull-down assay
RNA pull-down experiments were performed using a Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo, Waltham, MA, USA) according to the manufacturer's instruction, with some modifications. In brief, the sequence of LINC00852 was amplified using PCR primers with the T7 promoter sequence. The forward (F) and reverse (R) primers for the sense RNA template were as follows: 5'-TAATACGACTCACTATAGGGCTGGGAAGGGGATTCTAAGC-3' and 5'-TGGTCTGTGGGAAGGGAATA-3', respectively, while those for the anti-sense template were 5'-TAATACGACTCACTATAGGGTGGTCTGTGGGAAGGGAATA-3' and 5'-C TGGGAAGGGGATTCTAAGC-3'. In vitro transcription was performed using the RiboMAX Large Scale RNA Production System (Promega, Madison, WI, USA), and the resulting RNA was purified using the TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA was then biotinylated using a Pierce RNA 3' End Desthiobiotinylation Kit (Thermo, Waltham, MA, USA) and purified again with TRIzol. Next, A549 cells were lysed in Pierce IP Lysis Buffer (Thermo, Waltham, MA, USA). RNA-protein complexes were also eluted for mass spectroscopy experiments and for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), followed by silver staining and western blotting.
Cell lines and transduction/transfection
Human lung adenocarcinoma cell lines A549 and SPCA-1 were purchased from Zhong Qiao Xin Zhou Biotechnology Co., Ltd. (Shanghai, China). Cells were cultured in F12K or RPMI1640 (GIBCO-BRL; Invitrogen, Carlsbad, CA) medium containing 10% fetal bovine serum (FBS) and 1X penicillin/ streptomycin (Invitrogen, Carlsbad, CA, USA). We constructed overexpression lentivirus vectors (GeneChem, Shanghai, China) and knockdown siRNA sequences (GenePharma, Shanghai, China) targeting LINC00852 and S100A9, respectively. The siRNA sequence targeting LINC00852 was 5'-GCCCAAGATTCTACATTTCTAAG-3', while that targeting S100A9 was 5'-UAGAAAUGUAGAAUCUUGGGC-3'. Lentivirus transductions were performed by seeding 3-4 × 104/ml A549 and SPCA-1 cells in six-well plates. Cells were transduced with LINC00852- and S100A9-overexpressing lentiviruses using polybrene at a final concentration of 6 μg/ml (Sigma, St. Louis, MO, USA). Two or three days after infection, the cell lines successfully transduced with the lentivirus-mediated vector were isolated using 5 μg/ml of puromycin. For siRNA transfection, A549 and SPCA-1 cells were seeded onto a six-well plate and transfected with 100 nM LINC00852 siRNA and S100A9 siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Six groups of cells were created for functional analysis: two overexpression groups with either LINC00852 or S100A9; two knockdown groups with either si-LINC00852 or si-S100A9; LINC00852+si-S100A9; and si-LINC00852+S100A9.
Western blotting
Total protein was extracted from clinical samples and A549 and SPCA-1 cells using a lysis buffer containing a phosphorylase inhibitor cocktail (Abcam, Cambridge, MA, USA) and phenylmethanesulfonyl fluoride (Beyotime, Shanghai, China). The protein concentration was detected using a Bio-Rad protein assay kit. Bromophenol blue 2× (Ameresco, Solon, OH, USA) was added as a loading buffer. An equal amount of each sample with 20 μg protein was electrophoresed on an 8-12% SDS polyacrylamide gel and transferred onto polyvinylidene fluoride membranes using an electric transfer system (BIO-RAD, Hercules, CA, USA). Subsequently, these membranes were incubated with primary antibody after blocking with 5% skimmed milk powder for 2 h at room temperature. The main primary antibodies were S100A9 (1:1000) (Abcam, Cambridge, MA, USA), ERK1/2 MAPK (1:2000), p-ERK1/2 MAPK (1:2000), P38 MAPK (1:1000), p-P38 MAPK (1:1000), SAKP/JNK (1:1000), p-SAKP/JNK (1:1000) (CST, Danvers, MA, USA), and GAPDH (1:5000) (Beyotime, Shanghai, China). These antibodies were added and incubated at 4°C overnight. Goat anti-rabbit (1:5000) or goat anti-mouse (1:5000) IgG-HRP (BBI Life Science, Shanghai, China) was then added and incubated at 37°C for 2 h. The color reaction was observed using an electro-chemiluminescence detection reagent (SAB, College Park, MD, USA). GAPDH was used as an internal control.
Quantitative real-time PCR analyses
A total of 2 μg of RNA from frozen clinical samples or cell lines was reverse-transcribed to obtain cDNA using a PrimeScript kit (Takara Bio, Otsu, Japan). Subsequently, real-time PCR analyses were conducted using GoTaq ® qPCR Master Mix (Takara Bio, Otsu, Japan), and qPCR data collection was performed using a thermocycler ABI 7500 instrument (Thermo, Waltham, MA, USA). The expression ratio was calculated according to the 2-ΔΔCt method, and the results were normalized to the expression of GAPDH. The primer sequences for LINC00852, S100A9, and GAPDH, respectively, were as follows. F: 5'-CGTTGCCTACAGTCAAGTCAGT-3'; R: 5'-GCCATGGTTCCCTTACTGATAC-3'. F: 5'-GCCATGGTTCCCTTACTGATAC-3'; R: 5'-CAGGTCCTCCATGATGTGTTCTA-3'. F: 5'-TGTTCGTCATGGGTGTGAAC-3'; R: 5'-ATGGCATGGACTGTGGTCAT-3'.
CCK-8 assays
A549 and SPCA-1 cell suspensions were added to a 96-well plate at a density of 1 × 104/ml. The next day, the cells were incubated with 10 μl of CCK-8 solution (Dojindo, Tokyo, Honshu, Japan) for 2 h. The absorbance was measured at 450 nm using a multifunctional microplate reader (Thermo, Waltham, MA, USA) on days 1, 2, and 3.
Cell apoptosis assay
Approximately 5 × 105 of A549 and SPCA-1 cells were harvested and suspended in 500 μl of binding buffer. Then, 5 μl of Annexin V-APC and 5 μl of 7-AAD staining solution (Invitrogen, Carlsbad, CA, USA) were added, and the cells were incubated at 37°C for 15 min in the dark. Flow cytometry analysis was used (BD-FACSCalibur, Franklin Lakes, NJ, USA) to detect red fluorescence of cells. Finally, the results were analyzed using FlowJo software, version 10.0 (FlowJo, Ashland, OR, USA).
Scratch-wound assay
A549 cells were seeded to six-well plates. After 2-3 days, when cells had spread over the entire bottom of the culture cluster in a monolayer, a 100-μl pipette tip was used to create a “scratch” in the wells. PBS was then used to wash the wells. Subsequently, 2 ml of F12K culture medium without FBS was added to maintain the culture. The change in the distance between the two edges of the wound was observed by optical microscopy (Olympus-IX51, Tokyo, Japan).
Transwell assays
Invasion assays were performed with 8-μm pore size inserts (Corning, Kennebunk, ME, USA) in a 24-well plate. The upper compartment of the polycarbonate filter was pre-coated with 100 μl of Matrigel (5 mg/ml; Sigma-Aldrich, St. Louis, MO, USA). After drying, the A549 cells (1 × 105) were seeded into the upper chamber containing 100 μl of FBS-free F12K culture medium, and 600 μl of F12K culture medium containing 10% FBS was added to the lower chamber. The whole culture plate was incubated at 24 h. Finally, the lower surface of the upper chamber was observed at six random fields of view after fixation with 4% paraformaldehyde and 0.1% crystal staining.
Xenograft mouse model of SM
Forty BALB/c nude mice (specific-pathogen-free grade, male and female in half) of 4-6 weeks of age were obtained from the Vital River Laboratory Animal Technology Co., Ltd. (Beijing, China). The animal studies were approved by the Animal Ethics Committee of Zhongshan Hospital, Fudan University. The mice were randomly divided into four equal groups and anesthetized using sodium pentobarbital (35-40 mg/kg). Approximately 1 × 106 A549 cells were transfected with LINC00852, si-LINC00852, S100A9, or si-S100A9 and suspended in 200 μl of serum-free media. Then, cells were injected into the left ventricles of mice in each respective group to establish SM animal models17. After 6-8 weeks, the mice underwent positron emission tomography (PET) scans (SuperNova, Kunshan, Jiangsu, China). If a suspected SM was found, the lesion was subject to a further micro computed tomography (micro-CT) scan (Quantum GX, PerkinElmer, Waltham, MA, USA) and pathological examination.
Statistical analyses
Differences in means between the groups were statistically evaluated using one-way analysis of variance or chi-squared tests. Comparison between the groups was performed using the least significant difference test with multiple comparisons. Bivariate correlations were statistically evaluated using Spearman's grade correlation. All analyses were performed using the SPSS statistical software version 16.0 (SPSS UK, Ltd., Woking, UK). P < 0.05 was considered to indicate statistical significance.
Results
LINC00852 and S100A9 in lung adenocarcinoma SM
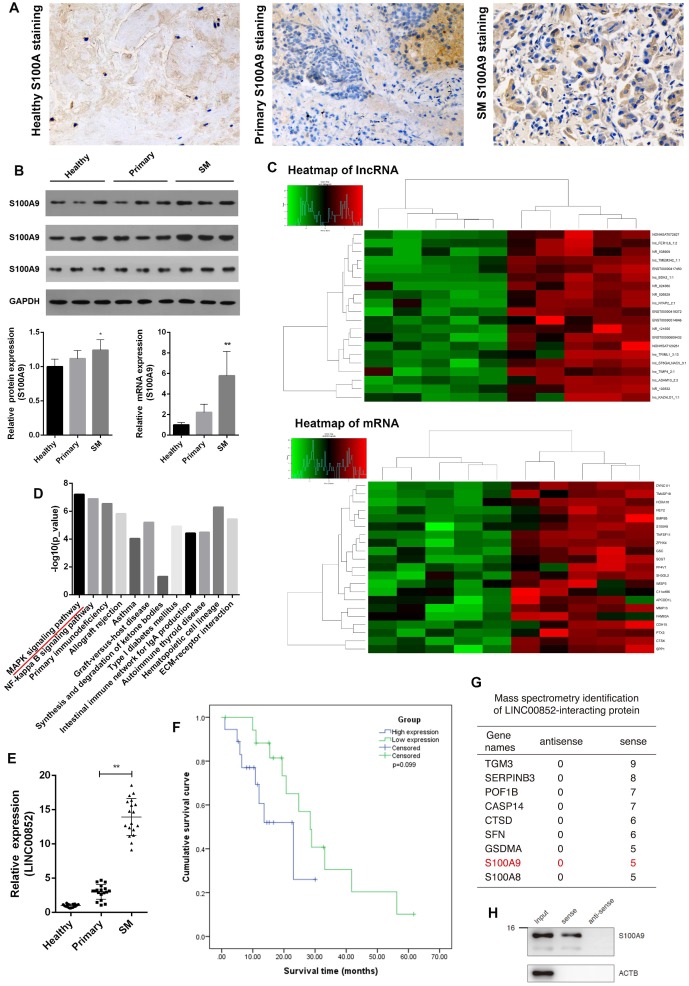
Immunohistochemistry of clinical specimens showed substantially higher levels of S100A9 proteins in lung adenocarcinoma SM lesions than in primary lung adenocarcinoma tissues. Similarly, western blotting and real-time PCR analyses showed that protein and mRNA levels of S100A9 were correspondingly elevated in lung adenocarcinoma SM compared with those in primary lung adenocarcinoma and healthy vertebrae. These results indicate a relationship between S100A9 and SM (Figure 1A,B). To further investigate the potential role of the lncRNA targeting S100A9 in SM, these clinical specimens were subjected to lncRNA target prediction. The results showed that LINC00852 was associated with lung adenocarcinoma SM. In addition, LINC00852 is considered to play a role in lung adenocarcinoma SM by targeting S100A9 (Figure 1C). To validate the results of the genome-wide analysis, we chose four differentially expressed lncRNAs at random for qPCR; the results were consistent with the microarray data (Figure S1 and Table S1). LINC00852 levels were significantly increased in lung adenocarcinoma SM compared with those in primary lung adenocarcinoma and healthy spinal osseous tissues (P < 0.05) (Figure 1E). Moreover, the level of LINC00852 was related to the involvement of spinal segments (Table 1). The patients were divided into two equal groups on the basis of LINC00852 expression (high or low). Survival analysis indicated that high LINC00852 expression also showed a trend towards poor survival (P > 0.05) (Figure 1F). RNA pull-down was used to identify the target effector through which the lncRNA exerts its oncogenic effect (Table S2). Mass spectrometry and western blotting indicated that S100A9 binds LINC00852 (Figure S2 and Figure 1G,H).
Figure 1.
LINC00852 and its target S100A9 are overexpressed in human lung adenocarcinoma spinal metastasis (SM). (A) Clinical samples were collected and analyzed by immunohistochemical staining with S100A9 antibody (1:200). Representative staining is shown (400×). (B) Results of western blotting and real-time PCR assays showing that S100A9 was overexpressed in SM; *P < 0.05, **P < 0.01. (C,D) Results of lncRNA target prediction and signal pathway prediction using high-throughput microarray screening and the Kyoto Encyclopedia of Genes and Genomes (KEGG), showing that LINC00852 expression and MAPK pathway were enriched in SM tissues. (E) Results of real-time PCR assay showing that LINC00852 is overexpressed in cancer tissue of SM; **P < 0.01. (F) Duration of survival in patients with high LINC00852 expression was shorter than that of patients with low LINC00852 expression; however, the difference was not statistically significant. (G,H) RNA pull-down verifying LINC00852 binding to S100A9.
Table 1.
Correlation expression of LINC00852 in spinal metastasis tissues and primary lung adenocarcinoma tissues
| Variable | SM (n=18) | Primary (n=18) | χ2 / F / r | p |
|---|---|---|---|---|
| Gender (Male/Female) | 14/4 | 11/7 | 1.178§ | 0.471 |
| Age (years, mean ± SD) | 62.72 ± 10.29 | 60.78 ± 8.58 | 0.379† | 0.542 |
| Clinical stage | ||||
| IA | 0 | 3 | 0.215¶ | 0.130 |
| IB | 0 | 4 | ||
| IIA | 0 | 5 | ||
| IIB | 0 | 5 | ||
| III | 0 | 1 | ||
| Involved segment | ||||
| I | 6 | 0 | 0.352¶ | 0.030* |
| II | 7 | 0 | ||
| III | 2 | 0 | ||
| More than III | 3 | 0 | ||
* indicates significant difference at p < 0.05. §, † and ¶ represent chi-squared tests (χ2), F-test (F) and correlation coefficient (r), respectively.
LINC00852 promotes cell proliferation and inhibits cell apoptosis
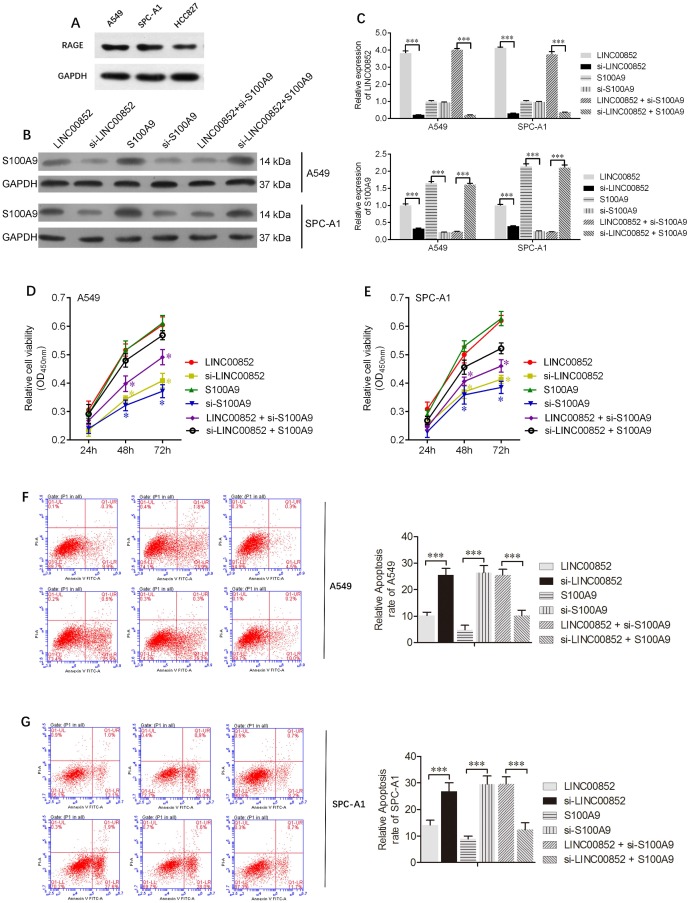
To further understand the effects of lncRNAs on lung adenocarcinoma cells, lentiviruses and/or siRNAs were used to upregulate or downregulate the expression of LINC00852 and/or S100A9 in A549 and SPCA-1 cells. The expression levels of S100A9 protein, as well those of LINC00852 and S100A9 mRNA, in A549 and SPCA-1 cells are shown in Figure 2 B,C. CCK8 assays showed that overexpression of LINC00852 or S100A9 promoted cell proliferation, while knockdown of LINC00852 or S100A9 reduced cell proliferation. In addition, we measured cell apoptosis by flow cytometry and found that overexpression of LINC00852 or S100A9 inhibited cell apoptosis, while repression of LINC00852 or S100A9 increased cell apoptosis (Figure 2F,G). These results suggest that the targeting of S100A9 by LINC00852 promotes cell proliferation and inhibited apoptosis.
Figure 2.
LINC00852 or S100A9 promotes cell proliferation and inhibits cell apoptosis. (A) Differential expression of the RAGE receptor in lung adenocarcinoma cell lines. (B,C) Stably overexpressing LINC00852 or S100A9 cell lines (A549 and SPC-A1) were constructed by lentiviral infection. Specific siRNA was used to inhibit the expression of LINC00852 or S100A9 in A549 and SPC-A1 cells. The expression levels of LINC00852 and S100A9 were determined by western blotting and qPCR. (D,E) CCK-8 assay was used to demonstrate A549 and SPC-A1 cell proliferation after transduction/ transfection. The experiment was repeated three times; *P < 0.05. (F,G) Annexin V-APC assay was used to detect apoptosis of A549 and SPC-A1 cells after lentivirus transduction and/or siRNA transfection. The experiment was repeated three times. ***P < 0.001
LINC00852 induces cell migration and invasion
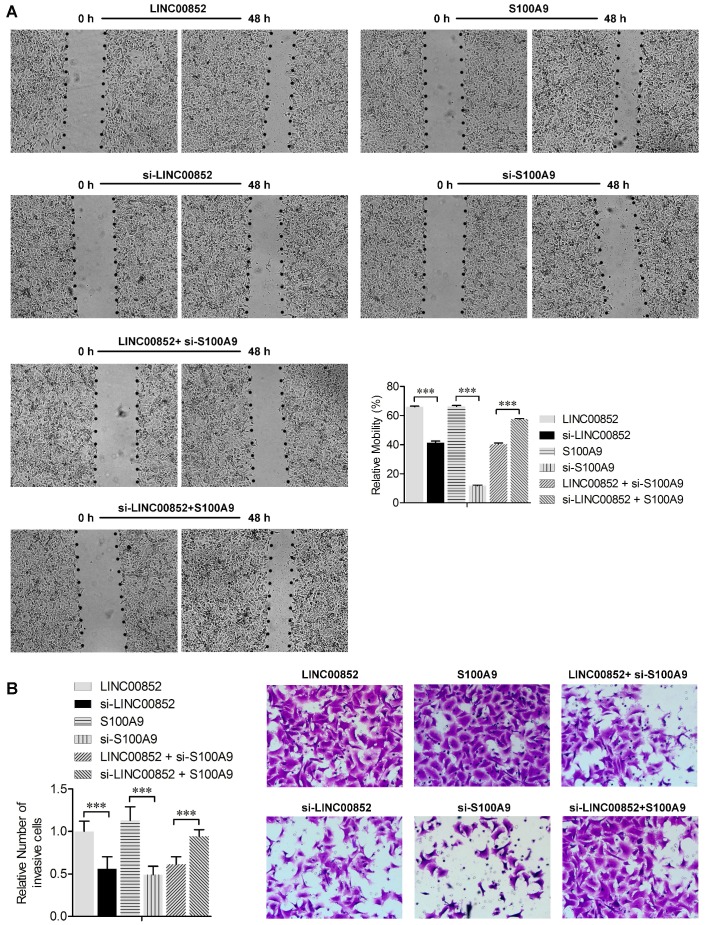
We further investigated the effects of LINC00852 and S100A9 on migration and invasion of cancer cells using wound healing and transwell assays, respectively. The wound healing assay showed that overexpression of LINC00852 or S100A9 in A549 cells increased cell migration rates, while knockdown of LINC00852 or S100A9 decreased the rates of cell migration (P < 0.01) (Figure 3A). Transwell assays showed that A549 cell invasion was positively regulated by LINC00852 or S100A9 expression. In LINC00852-overexpressing A549 cells, cell invasion increased by 44% compared with LINC00852-repressed A549 cells. In S100A9-overexpressing A549 cells, cell invasion increased by 57% compared with that in S100A9-repressed A549 cells (Figure 3B). These results indicate that LINC00852 promotes cell migration and invasion by targeting S100A9.
Figure 3.
LINC00852 or S100A9 induces cell migration and invasion. (A) Cell migration was measured by scratch-wound assay. Representative images are shown (40×). The results of three independent experiments are summarized; ***P < 0.001. (B) Cell invasion was determined by transwell assay. Representative images are shown (200×). The results of three independent experiments are shown. ***P < 0.001.
LINC00852 target S100A9 activates the MAPK pathway
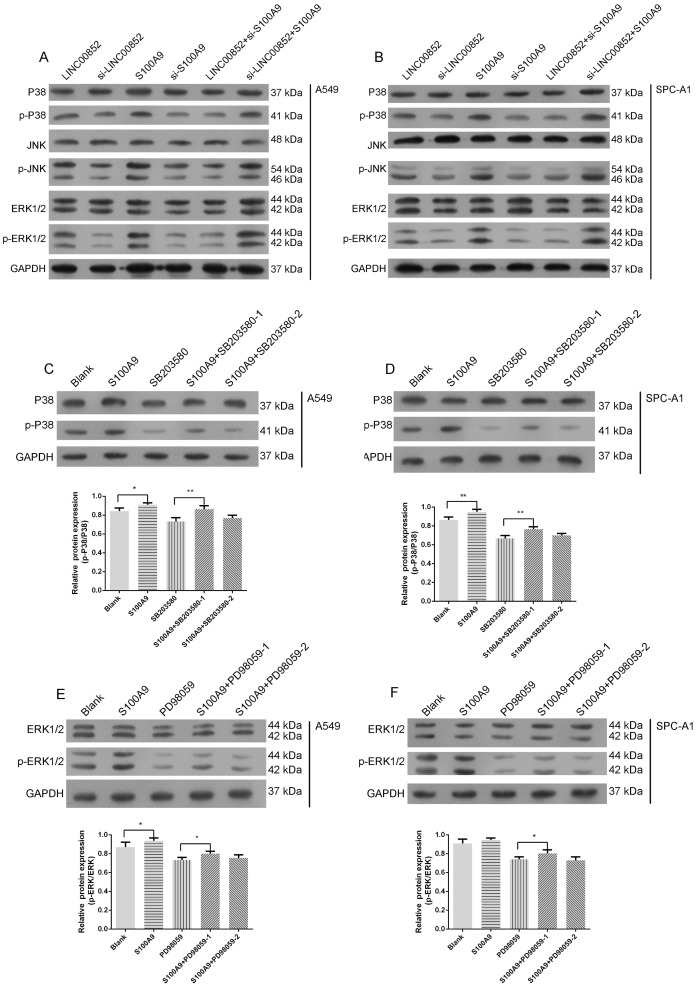
Signal pathway prediction using KEGG indicated a potential relationship between the MAPK pathway and lung adenocarcinoma SM (Figure 1D). Accordingly, we further examined the activity of this pathway by examining the expression levels of the relevant phosphorylated kinases. Expression levels of p-P38 and p-ERK were strongly decreased, while those of p-JNK were slightly decreased, in LINC00852- or S100A9-knockdown A549 and SPCA-1 cells, relative to those in cells in which LINC00852 or S100A9 were overexpressed (Figure 4A-B). When S100A9 was upregulated in A549 and SPCA-1 cells, the expression levels of phosphorylated P38 and ERK were strongly increased, but decreased again when SB203580 and PD98059 were used to block phosphorylated P38 and ERK, respectively. Furthermore, there was a stronger decrease in expression when high concentrations of inhibitor were used than that observed with low concentrations (Figure 4C-F).
Figure 4.
S100A9 activates the MAPK pathway. (A,B) P38, JNK, ERK, and their phosphorylation expression levels in transfected A549 and SPC-A1 cells were determined by western blotting. (C,D) A549 and SPC-A1 cells were transfected with or without LV-S100A9 and pretreated without or with SB203580 (0.5 μM or 5 μM for 24 hours), respectively. Expression levels of p-P38 and P38 were determined by western blotting; *P < 0.05; **P < 0.01. (E,F) A549 and SPC-A1 cells were transfected with or without LV-S100A9 and pretreated without or with PD98059 (1 μM or 10 μM for 24 hours), respectively; expression levels of p-ERK1/2 and ERK1/2 were determined by western blotting; *P < 0.05.
LINC00852 facilitates lung cancer SM in vivo
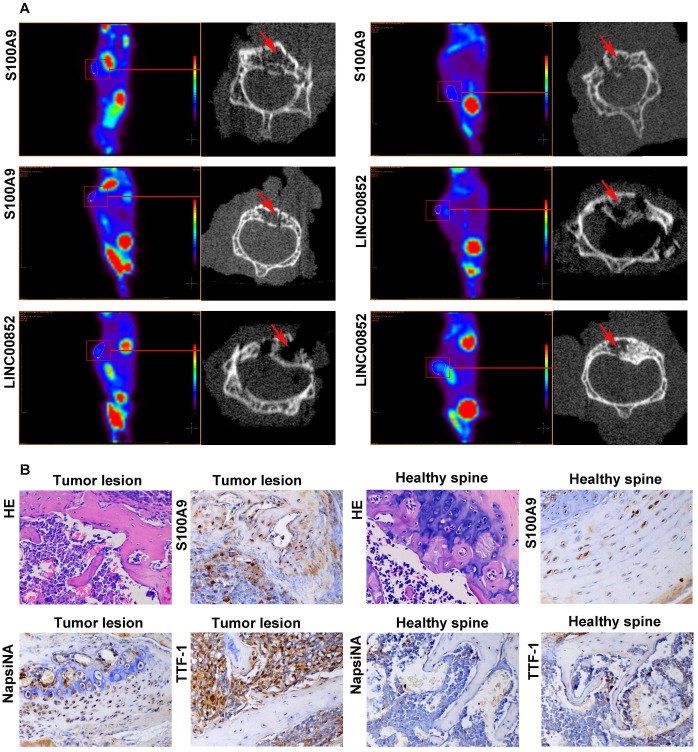
PET-CT scans showed metastases in 4 out of the 10 (40%) nude mice in the LINC00852 group: one case involving the liver, one involving both colon and spine, and two involving the spine only. One of the spinal lesions was limited to two vertebral segments, while the remaining two lesions each involved more than two segments. In the si-LINC00852 group, 1 of the 10 (10%) nude mice exhibited metastasis involving the pancreas. In the S100A9 group, 5 of the 10 (50%) nude mice developed metastasis: one case involving the femur and spine, one involving the neck, one an adrenal lesion, and two involving the spine only. Moreover, two of the spinal lesions involved two vertebral segments, while the others involved more than three segments. In the si-S100A9 group, 1 of the 10 (10%) nude mice showed metastasis involving the spleen. (Figure 5A and Figure S3). The differences in SM occurrence between the LINC00852 and si-LINC00852 group, and between the S100A9 and si-S100A9 group, were not statistically significant (P > 0.05). Finally, all the suspected metastases were verified by pathological examination, including hematoxylin-eosin staining and immunohistochemistry (Figure 5B).
Figure 5.
After 6-8 weeks, the model mice received a PET scan. If a suspected SM was found, the lesion underwent a further micro-CT scan and pathological examination. (A) Micro-CT scan showing local destruction (arrow) in the affected vertebrae of the tumorigenic mice. (B) Hematoxylin-eosin staining and immunohistochemical staining of S100A9, NapsiNA, and TTF-1 are shown in tumor tissues (400×).
Discussion
lncRNA dysregulation is involved in cancer progression and metastasis18,19. lncRNAs regulate the expression of nearby coding genes indirectly by modulating transcription20, or directly by playing an enhancer-like role21,22. Their functional roles in these processes are only beginning to emerge, owing to their relatively complex structures and extensive regulation mechanisms. However, the identification of cancer-associated lncRNAs may enable the development of novel therapeutic methods or potential prognostic biomarkers for cancers.
Here, we report, for the first time, an association between LINC00852 and lung adenocarcinoma SM and propose a preliminary description of the molecular mechanism by which this lncRNA promotes SM. LINC00852 expression levels were significantly increased in lung adenocarcinoma SM, and the extent of the increase was associated with the involvement of vertebral segments. An RNA pull-down assay was performed to determine the effector molecule targeted by this lncRNA. After the complex was eluted, western blotting and mass spectrometry were used to detect the interactions of specific RNA-binding proteins with LINC00852. Among these, S100A9 has been reported to be associated with lung cancer progression23,24, which is consistent with the results of the present study of clinical specimens. Therefore, the role of S100A9 in lung adenocarcinoma SM was further analyzed.
Recent studies have identified S100A9 as a ligand of the receptor for advanced glycation end products (RAGE), which plays a regulatory role in pro-inflammatory responses 25,26. Moreover, S100A9 has been shown to be closely related to the MAPK signaling pathway, which promotes cancer development and tumor spread via inflammatory pathways8,27; however, the mechanism underlying the regulatory role of S100A9 in SM progression was unclear. In this study, we found that RAGE exhibited differential expression in the three different lung adenocarcinoma cell types (Figure 2A). At the functional level, the S100A9-knockdown A549 cells exhibited a strong decline in cell migration, as described previously in other malignant cells28,29. Previous studies have demonstrated that the MAPK pathway plays a critical role in metastasis in lung cancer30,31. In the present work, signal pathway prediction demonstrated that the MAPK pathway was enriched in SM tissues. Therefore, we examined the change in kinase phosphorylation after S100A9 was upregulated with or without inhibitor in A549 and SPC-A1 cells; the results confirmed that S100A9 activated the MAPK pathway mainly by phosphorylating P38 and ERK1/2 in lung cancer cells. In the xenograft study, no statistical difference in terms of SM was observed between the LINC00852 and si-LINC00852 groups. This may have been attributable to the limited sample size, or because the inoculated cells did not undergo bone tropism induction as in a previous study17; however, the rate of SM and the involvement of pathological vertebral segments to be higher in the LINC00852 group than in the si-LINC00852 group.
The present findings strongly suggest that LINC00852 targets S100A9 to activate the MAPK signaling pathway, thereby contributing to the formation of a metastatic microenvironment, enhancing lung adenocarcinoma cell migration and invasion, and eventually facilitating SM. This lncRNA should be considered a potential target for the early prevention of SM in lung adenocarcinoma.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81572629 and 81772855) and China Postdoctoral Science Foundation (2017M621362). Peng Liu and Houlei Wang contributed equally to the present study and are co-first authors of this manuscript.
References
- 1.Zagryazhskaya A, Gyuraszova K, Zhivotovsky B. Cell death in cancer therapy of lung adenocarcinoma. Int J Dev Biol. 2015;59:119–129. doi: 10.1387/ijdb.150044bz. [DOI] [PubMed] [Google Scholar]
- 2.Schulman KL, Kohles J. Economic burden of metastatic bone disease in the U.S. Cancer. 2007;109:2334–2342. doi: 10.1002/cncr.22678. [DOI] [PubMed] [Google Scholar]
- 3.Sarabia-Estrada R, Zadnik PL, Molina CA. et al. A rat model of metastatic spinal cord compression using human prostate adenocarcinoma: histopathological and functional analysis. Spine J. 2013;13:1597–1606. doi: 10.1016/j.spinee.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 4.Silva GT, Bergmann A, Thuler LC. Incidence, associated factors, and survival in metastatic spinal cord compression secondary to lung cancer. Spine J. 2015;15:1263–1269. doi: 10.1016/j.spinee.2015.02.015. [DOI] [PubMed] [Google Scholar]
- 5.Vicent S, Perurena N, Govindan R, Lecanda F. Bone metastases in lung cancer. Potential novel approaches to therapy. Am J Respir Crit Care Med. 2015;192:799–809. doi: 10.1164/rccm.201503-0440SO. [DOI] [PubMed] [Google Scholar]
- 6.Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64. doi: 10.1210/er.2014-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang R, Xia LQ, Lu WW, Zhang J, Zhu JS. LncRNAs and cancer. Oncol Lett. 2016;12:1233–1239. doi: 10.3892/ol.2016.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ehrchen JM, Sunderkotter C, Foell D, Vogl T, Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J Leukoc Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 9.Liu S, Yan G, Zhang J, Yu L. Knockdown of long noncoding RNA (lncRNA) metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) inhibits proliferation, migration, and invasion and promoted apoptosis by targeting miR-124 in retinoblastoma. Oncol Res. 2017 doi: 10.3727/096504017X14953948675403. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Poggi A, Musso A, Dapino I, Zocchi MR. Mechanisms of tumor escape from immune system: role of mesenchymal stromal cells. Immunol Lett. 2014;159:55–72. doi: 10.1016/j.imlet.2014.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 12.Jolly LA, Massoll N, Franco AT. Immune suppression mediated by myeloid and lymphoid derived immune cells in the tumor microenvironment facilitates progression of thyroid cancers driven by HrasG12V and Pten Loss. J Clin Cell Immunol. 2016;7:451–471. doi: 10.4172/2155-9899.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Veirman K, De Beule N, Maes K. et al. Extracellular S100A9 protein in bone marrow supports multiple myeloma survival by stimulating angiogenesis and cytokine secretion. Cancer Immunol Res. 2017;5:839–846. doi: 10.1158/2326-6066.CIR-17-0192. [DOI] [PubMed] [Google Scholar]
- 14.Zhao F, Hoechst B, Duffy A. et al. S100A9 a new marker for monocytic human myeloid-derived suppressor cells. Immunology. 2012;136:176–183. doi: 10.1111/j.1365-2567.2012.03566.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Basso D, Fogar P, Plebani M. The S100A8/A9 complex reduces CTLA4 expression by immature myeloid cells: Implications for pancreatic cancer-driven immunosuppression. Oncoimmunology. 2013;2(e):24441. doi: 10.4161/onci.24441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han L, Zhang K, Shi Z. et al. LncRNA profile of glioblastoma reveals the potential role of lncRNAs in contributing to glioblastoma pathogenesis. International Journal of Oncology. 2012;40:2004–2012. doi: 10.3892/ijo.2012.1413. [DOI] [PubMed] [Google Scholar]
- 17.Zadnik P, Sarabia-Estrada R, Groves ML. et al. A novel animal model of human breast cancer metastasis to the spine: a pilot study using intracardiac injection and luciferase-expressing cells. J Neurosurg Spine. 2013;18:217–225. doi: 10.3171/2012.11.SPINE12325. [DOI] [PubMed] [Google Scholar]
- 18.Wu Y, Liu H, Shi X, Yao Y, Yang W, Song Y. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6:9160–9172. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutschner T, Hammerle M, Eissmann M. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013;73:1180–1189. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mattick JS, Gagen MJ. The evolution of controlled multitasked gene networks: the role of introns and other noncoding RNAs in the development of complex organisms. Mol Biol Evol. 2001;18:1611–1630. doi: 10.1093/oxfordjournals.molbev.a003951. [DOI] [PubMed] [Google Scholar]
- 21.Mattick JS. Linc-ing Long noncoding RNAs and enhancer function. Dev Cell. 2010;19:485–486. doi: 10.1016/j.devcel.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 22.Orom UA, Derrien T, Beringer M. et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai H, Minamiya Y, Takahashi N. Prognostic impact of S100A9 overexpression in non-small cell lung cancer. Tumour Biol. 2011;32:641–646. doi: 10.1007/s13277-011-0163-8. [DOI] [PubMed] [Google Scholar]
- 24.Feng PH, Lee KY, Chang YL. et al. CD14(+)S100A9(+) monocytic myeloid-derived suppressor cells and their clinical relevance in non-small cell lung cancer. Am J Respir Crit Care Med. 2012;186:1025–1036. doi: 10.1164/rccm.201204-0636OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao H, Hou J, Meng H, Zhang X, Zheng Y, Peng L. Proinflammatory effects and mechanisms of calprotectin on human gingival fibroblasts. J Periodontal Res. 2017;56:975–983. doi: 10.1111/jre.12465. [DOI] [PubMed] [Google Scholar]
- 26.Gao H, Zhang X, Zheng Y, Peng L, Hou J, Meng H. S100A9-induced release of interleukin (IL)-6 and IL-8 through toll-like receptor 4 (TLR4) in human periodontal ligament cells. Mol Immunol. 2015;67:223–232. doi: 10.1016/j.molimm.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 27.Srikrishna G. S100A8 and S100A9: new insights into their roles in malignancy. J Innate Immun. 2012;4:31–40. doi: 10.1159/000330095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng S, Zhang X, Huang N, Qiu Q, Jin Y, Jiang D. Down-regulation of S100A9 inhibits osteosarcoma cell growth through inactivating MAPK and NF-kappaB signaling pathways. BMC Cancer. 2016;16:253–265. doi: 10.1186/s12885-016-2294-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Duan L, Wu R, Ye L. et al. S100A8 and S100A9 are associated with colorectal carcinoma progression and contribute to colorectal carcinoma cell survival and migration via Wnt/beta-catenin pathway. PLoS One. 2013;8(e):62092. doi: 10.1371/journal.pone.0062092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo Y, Jiang M, Zhao X. et al. Cyclophilin A promotes non-small cell lung cancer metastasis via p38 MAPK. Thorac Cancer. 2018;9:120–128. doi: 10.1111/1759-7714.12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada M, Canals D, Adada M. et al. P38 delta MAPK promotes breast cancer progression and lung metastasis by enhancing cell proliferation and cell detachment. Oncogene. 2017;36:6649–6657. doi: 10.1038/onc.2017.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.