Abstract
Esophageal cancer is one of the leading malignancies globally and long non-coding RNAs (lncRNAs) have been proved to have an important role in different malignancies including esophageal cancer. However their role in disease progression is still not clear. The objective of the study was to investigate the expression and role of LINC01234 in progression of esophageal cancer cells. LncRNA LINC01234 was found to be upregulated in esophageal cancer cells by chip sequencing. The expression level of LINC01234 was detected from different esophageal cancer cell lines by qRT-PCR. After this, the LINC01234 knockdown effects on cell proliferation, migration, invasion, and apoptosis were evaluated by cell proliferation assay, wound healing assay, invasion assay, and flow cytometric analysis in vitro. Expression of lncRNA LINC01234 was found to be markedly upregulated in the CEC2 cell line. Furthermore, cell proliferation, migration and invasion were significantly (P < 0.05) suppressed as compared to negative control while apoptotic rate was also found increased as a result of the knockdown of LINC01234. Significantly upregulated expression of LINC01234 in CEC2 cells and downregulated expression after knockdown is observed. The impact of LINC01234 knockdown on cell migration, invasion, proliferation and apoptosis indicated that LINC01234 may represent a new marker and a potential therapeutic target for esophageal cancer.
Keywords: LINC01234, esophageal cancer, cell proliferation, invasion, long noncoding RNA, flow cytometry
Introduction
Esophageal cancer is one of the most aggressive cancers worldwide with a high death rate and is ranked as one of the top five deadliest malignancies in China 1-3. Its two subtypes; esophageal squamous cell carcinoma (ESCC) and esophageal adreno-carcinoma (EAC) have distinct pathological and aetiological characters. Of all esophageal cancer cases, more than 90% of patients in China suffer from ESCC 4. Despite the improvement in diagnostic techniques as well as advancement in therapeutics, it is usually diagnosed at a late stage. Late diagnosis and aggressiveness of the disease have made it lethal and the overall five-year survival rate is around 15%-25% 5,6. Early diagnosis and effective therapeutic modalities are the critical issues, and thus can help to reduce the mortality rate. Identification of novel biological markers can play an important role in achieving this goal. It is therefore necessary to understand the disease pathogenesis and all molecular changes which occur. Lack of specificity and sensitivity are the problems associated with already identified biological markers. Therefore, the identification of new biological markers which can be used for early diagnosis and prognosis of esophageal cancer is urgently required.
Non-coding RNAs were identified for the first time in 2002. A large spectrum of RNAs is produced as a result of transcription including protein-coding RNAs and non-protein coding RNAs. Research evidence has showed that non-coding RNAs are important biological regulators which affect the phenotype of the cell from normal to cancerous 7-9. A new class of non-coding RNAs has been identified as long non-coding RNAs (lncRNAs). These RNA molecules are longer than 200 nucleotides with limited or no protein-coding potential but play important roles in different biological pathways and processes like growth, differentiation, proliferation etc. 10-12. Research evidence indicates that variation in expression level of lncRNAs takes place due to stimuli while the role of lncRNAs as a scaffold at DNA level is also documented showing that they cause modification in chromatin and regulates transcription and translation processes. They regulate the expression of many genes and their epigenetic role is documented in different studies 13-15. Dysregulated expression of lncRNAs is linked with many human diseases and cancers, including esophageal cancer. They play an important role in modulation of tumor-suppression as well as oncogenic pathways. Most of the lncRNAs have shown tissue-specific and cancer-specific expression 13,16,17. This advancement has opened a new research direction for researchers to investigate their role in different diseases, especially in cancer. H19 was the first lncRNA identified and described as a tumor suppressor gene and its role has been investigated in different cancers such as breast cancer, lung cancer, cervical cancer, esophageal cancer and bladder cancer 18,19. CCAT2, POU3F3, SPRY4-IT1, and HOTAIR are few important potential biomarkers which have been studied and have shown prognostic value for ESCC 20-23. Previous research regarding the functions of lncRNAs in different cancers have proved that lncRNAs are important regulators of cancer initiation and progression by affecting cell proliferation, cell cycle regulation, migration, and invasion 24-26.
One of the previous studies investigated the expression profile of LINC01234 along with two other lncRNAs and identified the three-lncRNA signature as a prognostic marker for ESCC patients. This study found a significant difference in expression, in esophageal squamous cell carcinoma (ESCC) tissues and normal esophageal epithelial tissues 27. Differential expression of LINC01234 has also been reported in lung cancer tissues and normal lung tissues as well as in head and neck cancerous and normal tissues 28. There is no research about the role of LINC01234 in cancer cell progression therefore, there is need to explore the role of this lncRNA in different biological processes.
In the present study, we investigated the expression of LINC01234 in esophageal cancer cell lines as well as in an immortalized HaCat cell line for comparison of its expression and to see the expression difference between normal and cancer cells. Human embryonic esophageal epithelial cells were transformed into malignant cells by HPV18- E6E7 in our laboratory 29. After this, upregulated expression of LINC01234 was found in Gene Chip showing that it may have some role in causing esophageal cancer 30. Therefore, we investigated the expression of LINC01234 in different cell lines and also studied its role in cell proliferation, migration, invasion, and apoptosis in esophageal cancer cell line. We know about lncRNAs as biological regulators but their functions are still not clearly understood so future efforts can help to use lncRNAs as therapeutic targets for cancer especially esophageal cancer.
Materials and Methods
Cell Lines and Culture Conditions
The human esophageal cancer cell lines KYSE-450, EC-109, CEC2, EC-9706, and KYSE-180 were obtained from Chinese Center for Disease Control and Prevention along with immortalized cell line HaCaT. These cell lines were maintained in DMEM (Hyclone, Thermo Scientific) supplemented with 10% FBS (Gibco, Life Technologies), 100 units/ml penicillin and 100 mg/ml streptomycin. All the cell lines were maintained in humidified and sterile atmosphere of an incubator at 37 °C with CO2 (5%).
Transformation of Normal cells to Malignant cells
Embryonic esophageal epithelial cells of human were transformed into malignant cells in our laboratory with the help of HPV18-E6/E7- Adeno-associated virus (AAV). In the first step, epithelial cells were infected with HPV18 E6E7 then tumor-promoting factor TPA was added as the second step 29. After 76 passages, the transformation of normal esophageal cells into cancer cells was observed. Elevated expression of LINC01234 was detected in Affymetrix cDNA Chip result of these epithelial cells provided evidence that it may play some important role in causing esophageal cancer 30.
RNA Extraction and qRT-PCR
Total RNA was extracted from Cell lines and DNA was removed according to manufacturer's instructions, by using Trizol (Invitrogen, Life Technologies) and DNase kit (Promega) respectively. cDNA from RNA was synthesized by using Reverse Transcription kit (Promega according to manufacturer's protocol. Applied Biosystems ViiA™ 7 (Life Technologies) was used for amplification of cDNA by quantitative real-time PCR (qRT-PCR) with SYBR Select Master Mix (Life Technologies). 2ul cDNA was used with a total reaction volume of 8ul (per well) for qRT-PCR in 384 well block plate. For qRT-PCR, two sets of primers were designed by Beacon Designer 7 Software and which are given in supplementary material as Table S1. For the relative expression levels of LINC01234, GAPDH was used as an endogenous control while HaCaT was used as reference sample. Each sample was used in triplicates for qRT-PCR. The following reactions conditions were used for amplification: denaturation at 95 °C (10 min), followed by 40 PCR cycles at 95 °C (15s) and 60 °C (1 min).
Transfection
To silence the lncRNA LINC01234, three small interfering RNAs (siRNAs) and one negative control (si-NC) were constructed (RiboBio, China). Cells were collected after trypsinization and 25×104 cells per well were seeded on six-well plates. After one day, cells were transfected with three siRNAs and the negative control. For this purpose, FuGENE HD Transfection Reagent (Promega) was used. After 48 hours of transfection, qRT-PCR was used to determine the interfering efficiency of siRNA, and the siRNAs showing maximum silencing efficacy were used for all experiments. Sequences of siRNAs are provided in supplementary material as Table S2.
Wound Healing Assay
To determine cell migration, esophageal cancer cells transfected with si-LINC01234 or si-NC were seeded into 6-well plates and allowed to grow to 90-95% confluence. Wounds were introduced into monolayer cells using a 200ul sterile pipette tip. Wounded monolayer cells were washed three times with PBS to remove cell debris and then cultured. The speed of wound closure was monitored and photographed at different time intervals.
Transwell Invasion Assay
Costar chambers with transwell insert (Corning Incorporated, USA) having 8 μm pore size were used for cell invasion assay. 100uL of diluted Matrigel (Corning) was used to coat the permeable support. After two hours, 5 × 105 / mL cells transfected with si-LINC01234 or si-NC were suspended in serum-free medium and 200 μl were seeded in the upper chamber coated with matrigel, while 750uL DMEM with FBS (20%) was added in the lower chamber. Then cell invasion chambers were placed in the incubator for 24 hours. After incubation, methanol was used to fix the invaded cells and crystal violet (0.1%) was used to stain the cells. Non-invading cells in the upper chamber were carefully removed with the help of wet cotton swabs. Then invaded cells were imaged and counted at three different fields under an inverted microscope.
Cell Proliferation Assay
Cells were transfected with si-LINC01234 or si-NC in 6-well plate. After 48 hours, cells were trypsinized and seeded on 96-well plates (2000/well). Cell counting kit8 (Dojindo) was used according to the manufacturer's protocol for the cell proliferation assay. The proliferated cells were quantified in triplicate at 450 nm after every 12 hours intervals.
For colony forming experiments, CEC2 cells at a density of 1000 cells/well were seeded onthe six-well plate after transfection with si-LINC01234 or si-NC, maintained in DMEM medium containing FBS (15%). The medium was changed after 3 days interval. After 12 days, methanol and crystal violet (0.1%) was used to fix and stain the colonies respectively. Visible colonies were counted manually after taking photographs.
Flow-Cytometric Analysis
After 48 h transfection, cells were trypsinized and harvested from six-well plates then collected after centrifugation. Cell pellets were washed with PBS (cold) and then cells were stained with FITC-Annexin V and propidium iodide (4A Biotech, China). Viable and dead cells, as well as early & late apoptotic cells were quantified and analyzed by FACS Calibur and Cell Quest Pro software (BD Bioscience, USA). The negative control group was used to compare relative ratios of the early and late apoptotic cells with knockdown LINC01234 CEC2 cells.
Statistical Analysis
All biological experiments were performed in triplicate and data analysis was performed by using GraphPad Prism 7. Data are shown as mean ± SD. Statistical significance was assessed using the Student T-test. *p < 0.05 was defined to be the significant difference.
Results
Elevated expression of LINC01234 in malignant cells
Gene Chip (Affymetrix) reported up-regulation of LINC01234 when normal esophageal cells were transformed into cancerous cells after 76 passages through HPV18 E6E7-AAV (Gene chip result is provided in supplementary material as Figure S1). This predicts that HPV may have some relationship with esophageal cancer and some role in up-regulation of LINC01234. This also provided the evidence that LINC01234 may have some regulatory role for causing esophageal cancer.
Expression of LINC01234 in different Cell Lines
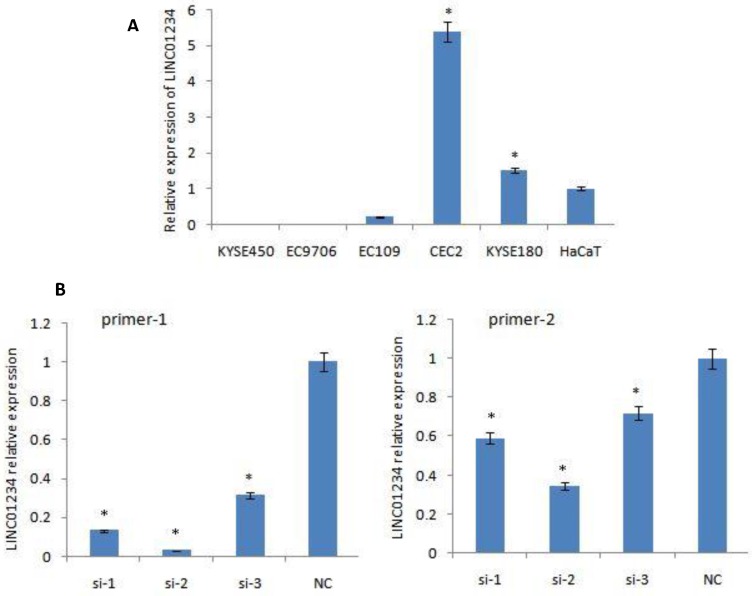
Quantitative RT-PCR was used to measure the expression of LINC01234. CEC2 cell line showed the highest level of LINC01234 expression so CEC2 (esophageal cancer cell line) can be used as a model cell line for the study of LINC01234. KYSE-180 have also shown high expression of LINC01234 as compared to HaCaT cell line (Figure 1A, P < 0.05) showing that LINC01234 may be related to esophageal cancer pathogenesis.
Figure 1.
Expression of LINC01234 in different Cell Lines. (A) Expression of LINC01234 in different Esophageal cancer cell lines compared with immortalized cell line (HaCat) (B) Expression level of LINC01234 in CEC2 cells following treatment with three si-LINC01234 and si-NC. (*P<0.05).
Expression of LINC01234 was also checked separately in cytoplasm and nucleus and has shown expression both in cytoplasm as well as in nucleus 30.
Expression of LINC01234 after LINC01234 Knockdown
We down-regulated the expression of LINC01234 with the help of silencing its expression in CEC2 cell line, using three small interfering RNAs (siRNAs) and one scrambled siRNA (si-NC). At 48 h post-transfection, the efficiency of the siRNAs was checked by qRT-PCR using two primers for LINC01234. SiRNA-2 was observed to have the highest knockdown efficiency with both primers. Therefore, siRNA-2 was chosen for further experiments. LINC01234 expression was down-regulated by approximately 70% in CEC2 cells by si-2 when compared with the siNC (Figure 1B, P < 0.05).
Wound Healing Assay
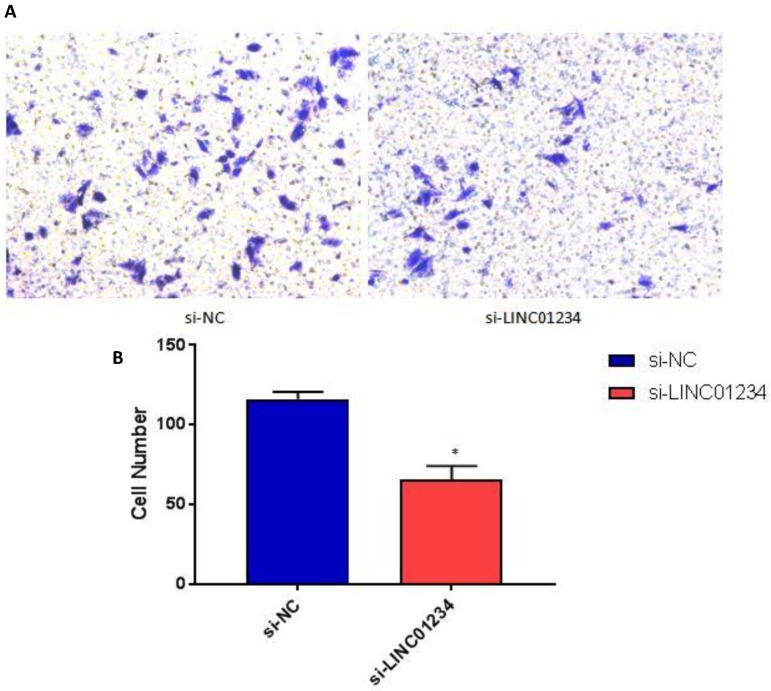
We performed cell wound healing assay to investigate the role of LINC01234 in the regulation of cell migration in human esophageal cancer cells. The migration ability of cells was assessed after specific time intervals with treatment (si-LINC01234) and without treatwment (si-NC). Wound healing assays showed that the migratory rate of esophageal cancer cells transfected with si2-LINC01234 was significantly less compared with si-NC (Figure 2).
Figure 2.
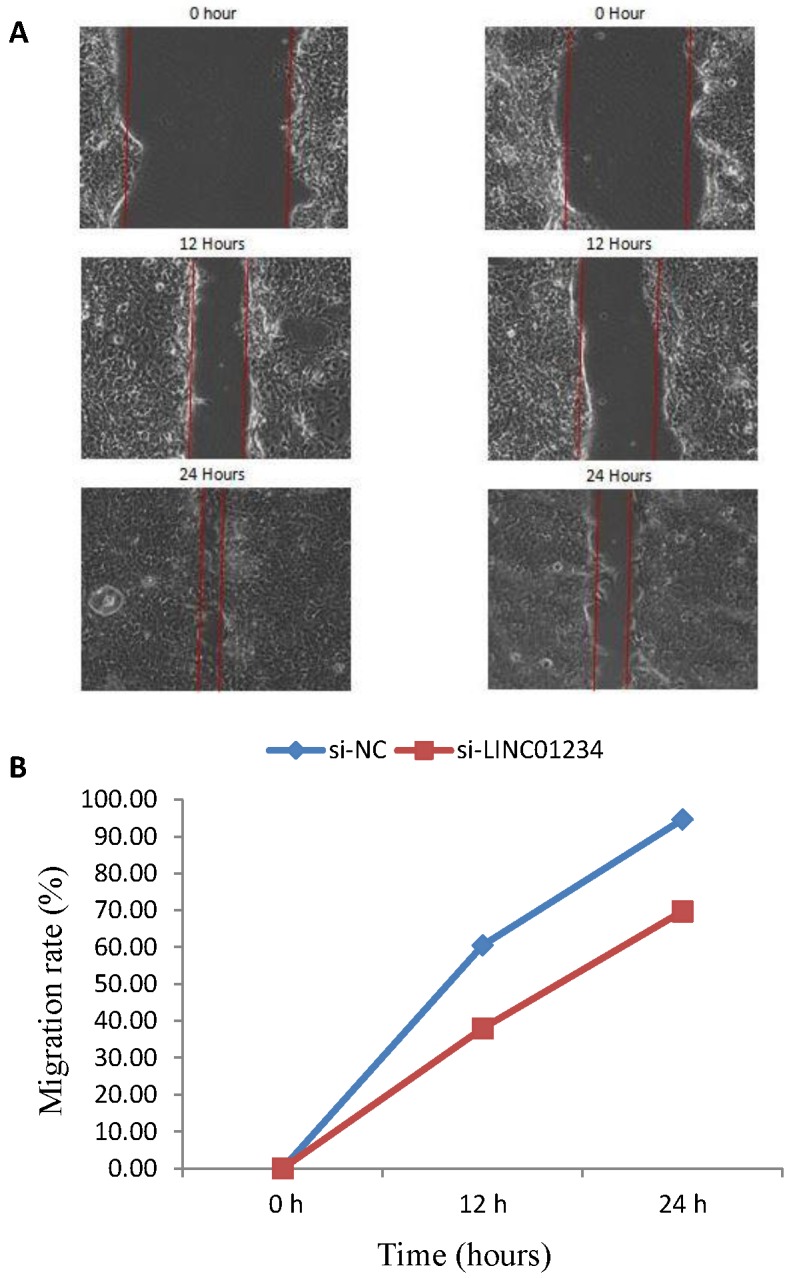
Effect of LINC01234 Knockdown on cell migration in CEC2 cells. (A) Representative images of cell migration. (B) Migration rate decreases after LINC01234 knockdown as compared to NC in CEC2 cells.
Transwell Invasion Assay
We performed transwell invasion assays to investigate the role of LINC01234 in the regulation of cell invasion in human esophageal cancer cells CEC2. Transwell invasion assay showed that the invasion of esophageal cancer cells transfected with si-LINC01234 was notably decreased compared with si-NC group (Figure 3, P < 0.05).
Figure 3.
Knockdown of LINC01234 decreases invasion capacity in CEC2 cells. (A) Representative images showing transwell invasion of CEC2 cells. (B) Bar chart represented the number of invasive cells. (*P<0.05).
Knockdown of LINC01234 Decreases Esophageal Cancer Cell Proliferation in Vitro
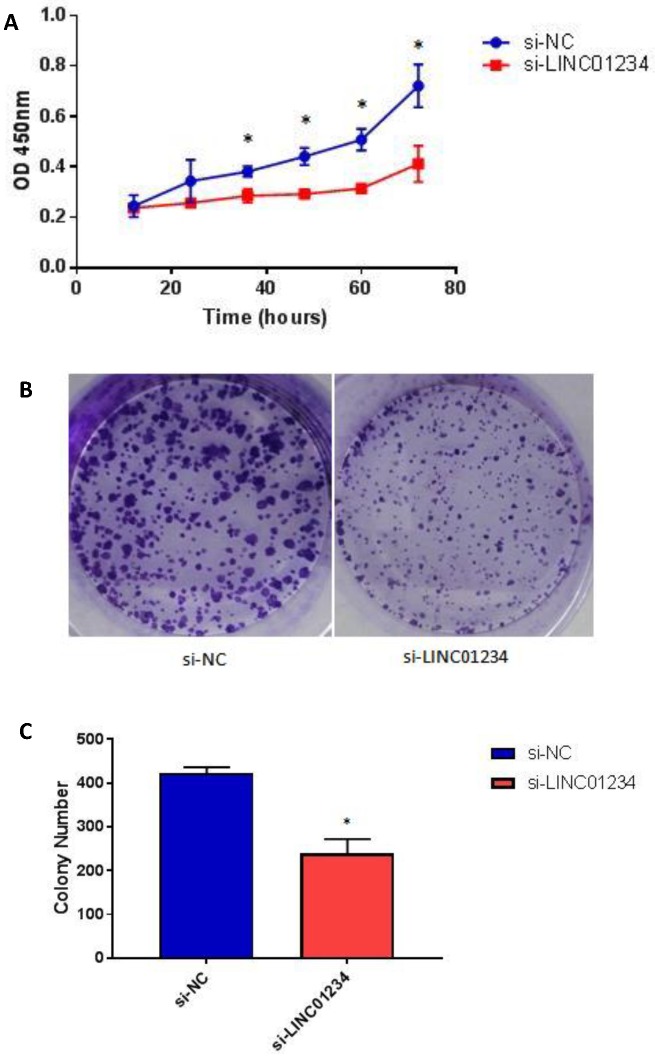
To assess the biological role of LINC01234 in esophageal cancer, we investigated the effect of LINC01234 on cell proliferation. CCK8 assay revealed that cell growth was significantly decreased in esophageal cancer cell lines after Knockdown compared with the NC group (Figure 4A, P < 0.05). The result indicated that downregulation of LINC01234 decreases esophageal cancer cell proliferation. The colony formation ability in CEC2 was also decreased by silencing the LINC01234 (Figure 4B, P < 0.05).
Figure 4.
Knockdown of LINC01234 suppresses the cell proliferation (A) Knockdown of LINC01234 suppresses the growth in vitro (in CEC2 cell line) as compared to NC when assessed by CCK8 (B) Cell proliferation was also assessed by colony forming assay (C) Colony number is significantly decreased in CEC2 cells after knockdown of LINC01234 as compared to NC (*P<0.05).
Effect of Knockdown of LINC01234 on Apoptosis
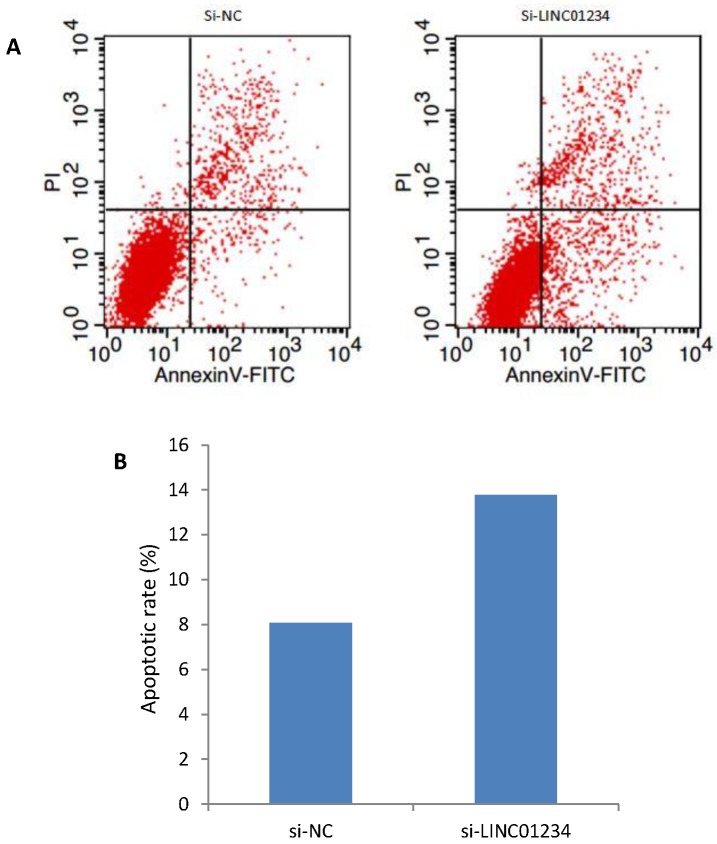
We investigated the role of lncRNA LINC01234 in apoptosis with the help of flow cytometric analysis. Apoptotic rate of CEC2 cells transfected with si-LINC01234 was elevated as compared to si-NC. We used to compare relative ratios of the early and late apoptotic cells. (Figure 5).
Figure 5.
Effects of LINC01234 Knockdown on Cell apoptosis determined by flow cytometric analysis. (A) Representative images of cell apoptosis by flow cytometry (B) Bar chart represents the percentage of apoptotic cells in si-NC and si-LINC01234.
Discussion
Esophageal cancer is one of the leading causes of death due to cancer, so it is important to find new molecular targets for its diagnosis, prognosis and treatment which can then play an important role for disease diagnosis, prognosis, and therapeutics. In the past decade, more and more data is providing strong evidence of the importance of non-protein coding regulatory RNAs along with protein-coding RNAs 15.
lncRNAs have gained attention in recent years due to evidence indicating their role in carcinogenesis in different studies. They are associated with disease progression and development by playing a role in migration, invasion, and proliferation. Abnormal expressions of lncRNAs have been studied in different cancers. Unfortunately, we have little knowledge about lncRNAs and their role in esophageal cancer because study regarding their functions in different cancers is still at its initial stage. Only a few lncRNAs have been studied in detail and most of them are still to be explored more in detail 10,12,31.
We found upregulated expression of LINC01234 in gene chip after malignant transformation of human embryonic esophageal epithelial cells as a result of HPV18-E6E7 infection 29,30; showing that lncRNA may play a key role in the pathogenesis of esophageal cancer. Before this, a lncRNA profile study associated three lncRNA signature with the survival of esophageal squamous cell carcinoma patients providing evidence for the role of LINC01234 in esophageal cancer for the first time 27. Gene chip analysis was the second line of evidence supporting the role of LINC01234 in esophageal cancer. Differential expression of lncRNA LINC01234 was revealed in lung cancer in another study through transcriptome sequencing. In the same study, its differential expression was also observed in head and neck squamous cell carcinoma 28. To the best of our knowledge, this is the first study exploring the role of LINC01234 in esophageal cancer. In this study, we focused on investigating the role of LINC01234 in cell proliferation, migration and invasion by using esophageal cancer cell lines. For this purpose, we did qRT-PCR by using five esophageal cancer cell lines (CEC2, EC-109, KYSE-180, KYSE450, EC-9706), and one immortalized cell line HaCaT. Our qRT-PCR result showed significantly high expression of LINC01234 in only two esophageal cancer cell lines (CEC2 and KYSE180) as compared to the immortalized HaCaT cell line.
Further results after siRNA mediated knockdown of lncRNA LINC01234 showed marked decrease in cell proliferation, migration, and invasion while the rate of apoptosis was increased. This shows that the silencing of lncRNA may inhibit esophageal cancer migration, invasion, and proliferation ability in vitro. Our findings indicated that lncRNA LINC01234 may have potential to function as an important contributor to cancer progression and its elevated expression may contribute to esophageal cancer development. One of the previous studies used plasma samples of esophageal cancer patients and detected different lncRNA but expression level of LINC01234 was found to be less than 60% in the plasma samples 32; while the first study reporting LINC01234 lncRNA used esophageal cancer patients' tissues for lncRNA profile study 27. We investigated the role of LINC01234 in esophageal cancer for the first time, using esophageal cancer cell lines.
This study identifies that lncRNA LINC01234 has the potential of carcinogenesis in esophageal cancer that may act by cell proliferation, migration, invasion, and apoptosis. To the best of our knowledge, there is no article reporting the role of LINC01234 in cell proliferation, migration, and invasion for esophageal carcinoma. However, the downstream regulator involved in LINC01234-mediated cell migration and invasion of esophageal cancer cells still needs to be explored. Thus, much more work is still required to determine the detailed mechanisms of its functions in ESCC and the potentiality of LINC01234 as a therapeutic target for esophageal carcinoma.
One limitation of this study is the sole use of cell lines for the investigation and evaluation of the potential lncRNA LINC01234.Therefore, further extensive investigation is needed to evaluate its potency for esophageal carcinoma by using esophageal cancer tissues and normal adjacent tissues along with esophageal cancer cell lines. Thus, the present study provides initial but important information about the role of lncRNAs LINC01234 in esophageal carcinoma.
In conclusion, our findings indicate that LINC01234 has oncogenic potential in esophageal carcinoma cells in vitro. We demonstrated for the first time that lncRNA LINC01234 was upregulated in esophageal cancer cell lines and significantly associated with cell proliferation, migration, invasion and apoptosis. Furthermore, the upregulation of LINC01234 may play a key role in esophageal cancer progression. These results suggest that lncRNA LINC01234 can be a promising biomarker and a therapeutic target for esophageal cancer in future. However, there is still need for much more research to explore the detailed mechanisms of its functions in esophageal carcinoma.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This study was supported by Beijing Natural Science Foundation (Grant No. 5162003), Beijing University of Technology Foundation (Grant No. 015000514314004), Development Program of China (Grant No. 2011SLKID103) and National Key Technology Support Program (Grant No. 2006BAI19B03).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Hu L, Wu Y, Tan D. et al. Up-regulation of long noncoding RNA MALAT1 contributes to proliferation and metastasis in esophageal squamous cell carcinoma. J Exp Clin Cancer Res. 2015;34:7. doi: 10.1186/s13046-015-0123-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen W, Zheng R, Zhang S. et al. Esophageal cancer incidence and mortality in China, 2010. Thorac Cancer. 2014;5(4):343–8. doi: 10.1111/1759-7714.12100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liang H, Fan JH, Qiao YL. Epidemiology, etiology, and prevention of esophageal squamous cell carcinoma in China. Cancer Biol Med. 2017;14(1):33–41. doi: 10.20892/j.issn.2095-3941.2016.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arnal MJD, Arenas AF, Arbeloa AL. Esophageal cancer: Risk factors, screening and endoscopic treatment in Western and Eastern countries. World J Gastroenterol. 2015;21(26):7933–43. doi: 10.3748/wjg.v21.i26.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toh Y, Egashira A, Yamamoto M. Epigenetic alterations and their clinical implications in esophageal squamous cell carcinoma. Gen Thorac Cardiovasc Surg. 2013;61(5):262–9. doi: 10.1007/s11748-013-0235-3. [DOI] [PubMed] [Google Scholar]
- 7.Okazaki Y, Furuno M, Kasukawa T. et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420(6915):563–73. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 8.Mattick JS. RNA regulation: a new genetics. Nat Rev Genet. 2004;5(4):316–23. doi: 10.1038/nrg1321. [DOI] [PubMed] [Google Scholar]
- 9.Guttman M, Donaghey J, Carey BW. et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477(7364):295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spizzo R, Almeida MI, Colombatti A. et al. Long non-coding RNAs and cancer: a new frontier of translational research. Oncogene. 2012;31(43):4577–87. doi: 10.1038/onc.2011.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauli A, Rinn JL, Schier AF. Non-coding RNAs as regulators of embryogenesis. Nat Rev Genet. 2011;12(2):136–49. doi: 10.1038/nrg2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1(5):391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, Wu Z, Fu X. et al. Long noncoding RNAs: insights from biological features and functions to diseases. Med Res Rev. 2013;33(3):517–53. doi: 10.1002/med.21254. [DOI] [PubMed] [Google Scholar]
- 14.Mercer TR, Dinger ME, Sunkin SM. et al. Specific expression of long noncoding RNAs in the mouse brain. Proc Natl Acad Sci USA. 2008;105(2):716–21. doi: 10.1073/pnas.0706729105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev. 2009;10(3):155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 16.Kotake Y, Nakagawa T, Kitagawa K. et al. Long non-coding RNA ANRIL is required for the PRC2 recruitment to and silencing of p15(INK4B) tumor suppressor gene. Oncogene. 2011;30(16):1956–62. doi: 10.1038/onc.2010.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Werven FJV, Neuert G, Hendrick N. et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150(6):1170–81. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barsyte-Lovejoy D, Lau SK, Boutros PC. et al. The c-Myc oncogene directly induces the H19 noncoding RNA by allele-specific binding to potentiate tumorigenesis. Cancer Res. 2006;66(10):5330–7. doi: 10.1158/0008-5472.CAN-06-0037. [DOI] [PubMed] [Google Scholar]
- 19.Tanos V, Ariel I, Prus D. et al. H19 and IGF2 gene expression in human normal, hyperplastic, and malignant endometrium. Int J Gynecol Cancer. 2004;14(3):521–5. doi: 10.1111/j.1048-891x.2004.014314.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhang X, Xu Y, He C. et al. Elevated expression of CCAT2 is associated with poor prognosis in esophageal squamous cell carcinoma. J Surg Oncol. 2015;111(7):834–9. doi: 10.1002/jso.23888. [DOI] [PubMed] [Google Scholar]
- 21.Li W, Zheng J, Deng J. et al. Increased levels of the long intergenic non-protein coding RNA POU3F3 promote DNA methylation in esophageal squamous cell carcinoma cells. Gastroenterology. 2014;146(7):1714–26. doi: 10.1053/j.gastro.2014.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Xie HW, Wu QQ, Zhu B. et al. Long noncoding RNA SPRY4-IT1 is upregulated in esophageal squamous cell carcinoma and associated with poor prognosis. Tumour Biol. 2014;35(8):7743–54. doi: 10.1007/s13277-014-2013-y. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Wu Z, Mei Q. et al. Long non-coding RNA HOTAIR, a driver of malignancy, predicts negative prognosis and exhibits oncogenic activity in oesophageal squamous cell carcinoma. Br J Cancer. 2013;109(8):2266–78. doi: 10.1038/bjc.2013.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Liu H, Shi X. et al. The long non-coding RNA HNF1A-AS1 regulates proliferation and metastasis in lung adenocarcinoma. Oncotarget. 2015;6(11):9160–72. doi: 10.18632/oncotarget.3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z, Yu X, Shen J. ANRIL: a pivotal tumor suppressor long non-coding RNA in human cancers. Tumour Biol. 2016;37(5):5657–61. doi: 10.1007/s13277-016-4808-5. [DOI] [PubMed] [Google Scholar]
- 26.Yang X, Song JH, Cheng Y. et al. Long non-coding RNA HNF1A-AS1 regulates proliferation and migration in oesophageal adenocarcinoma cells. Gut. 2014;63(6):881–90. doi: 10.1136/gutjnl-2013-305266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li J, Chen Z, Tian L. et al. LncRNA profile study reveals a three-lncRNA signature associated with the survival of patients with oesophageal squamous cell carcinoma. Gut. 2014;63(11):1700–10. doi: 10.1136/gutjnl-2013-305806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White NM, Cabanski CR, Silva-Fisher JM. et al. Transcriptome sequencing reveals altered long intergenic non-coding RNAs in lung cancer. Genome Biol. 2014;15(8):429. doi: 10.1186/s13059-014-0429-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shen Z, Cen S, Shen J. et al. Study of immortalization and malignant transformation of human embryonic esophageal epithelial cells induced by HPV18 E6E7. J Cancer Res Clin Oncol. 2000;126(10):589–94. doi: 10.1007/pl00008469. [DOI] [PubMed] [Google Scholar]
- 30.Ghaffar M, Khodahemmati S, Li J, et al. Expression of CASC9 and LINC01234 in Different Cell Lines. MSHH 2017. ISBN: 978-1-60595-472-1. http://dpi-proceedings.com/index.php/dtbh/article/view/
- 31.Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38. doi: 10.1186/1476-4598-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tong YS, Wang XW, Zhou XL. et al. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.