Abstract
Glial fibrillary acidic protein (GFAP) is an intermediate filament that provides mechanical support to astrocytes. Rs2070935 is a single nucleotide polymorphism (SNP) located in the promoter region of the GFAP gene. The aim of this pilot study is to investigate GFAP expression at mRNA, protein levels and rs2070935 polymorphism in 50 different grade human astrocytoma samples. GFAP expression at mRNA level was measured using quantitative reverse transcription polymerase chain reaction (qRT-PCR) with SYBR Green dye, whereas the translational activity of the following gene was detected using western blot assay. Furthermore, genotypes of rs2070935 were identified using qPCR with TaqMan probes. As a result, GFAP mRNA and protein expression was found to be declining with increasing astrocytoma grade (p < 0.05). A tendency was observed between increased GFAP mRNA expression and shorter grade IV astrocytoma patient survival (p = 0.2117). The rs2070935 CC genotype was found to be associated with increased GFAP translational activity in grade II astrocytoma (p = 0.0238). Possible links between rs2070935 genotypes and alternative splicing of GFAP were also observed. The rs2070935 AA genotype was found to be associated with poor clinical outcome for grade IV astrocytoma patients (p = 0.0007), although the following data should be checked in a larger sample size of astrocytoma patients.
Keywords: GFAP, rs2070935, glioblastoma
Introduction
Astrocytomas are tumors originating from astrocyte cells of the central nervous system (CNS) 1. The World Health Organization (WHO) classifies astrocytomas into 4 grades based on histological properties 2. A higher astrocytoma grade is associated with increased tumor invasiveness and shorter patient survival 3. The highest, grade IV, astrocytomas are defined as glioblastoma 4. Astrocytomas are attributed to gliomas, a larger group of CNS tumors, which originate from neuroglial cells. Gliomas are often considered to be the most difficult tumors to treat due to molecular and cellular heterogeneity between as well as within CNS tumors 5.
Glial fibrillary acidic protein (GFAP) is an intermediate filament, which provides mechanical support to cells. GFAP is mostly detected in astrocyte cells of the CNS 6. GFAP gene is expressed in various isoforms, which feature structural and functional differences. The activity of GFAP expression has a significant effect on various astrocyte properties, such as morphology, growth and cell division 7. Furthermore, the GFAP promoter region contains single nucleotide polymorphisms (SNP), such as rs2070935, which is part of the binding site for the transcription factor AP-1. Alleles of rs2070935 were observed to have an impact on the transcription factors ability to bind, thus affecting GFAP expression in commercial glioma cell lines 8. No scientific research was found regarding the interaction between rs2070935 polymorphism and GFAP expression in human astrocytoma tissue.
GFAP translational activity was reported to experience changes in human astrocytoma 9. Since GFAP expression is a biomarker for astrocyte maturity, a decrease in the total amount of the following filament might be a result of cellular dedifferentiation in brain tumor tissue 10. Although GFAP expression is considered to decrease with higher tumor grade, specific GFAP isoforms have been detected in increased amounts, as the abundance of GFAP-δ isoform has been identified in glioblastoma cells using immunohistochemistry 11. However, while protein quantities of GFAP have been thoroughly researched, studies regarding GFAP transcriptional activity in different grades of human astrocytoma are scarce.
The objective of this pilot study is to investigate GFAP expression at mRNA and protein levels in astrocytomas of varying degrees. Accordingly, another task is to identify the genotypes of rs2070935 in an attempt to detect novel associations between the following SNP and GFAP expression. The last objective is the analysis of the clinical significance of GFAP expression and rs2070935 genotypes for grade IV astrocytoma patients. On the whole, the main purpose of this small-scale study is to determine, whether GFAP expression and rs2070935 polymorphism is of interest for an extensive, large sample size clinical investigation of astrocytoma patients.
Materials and Methods
Sample collection
In total 50 glioma specimens of astrocytic origin were collected from the Department of Neurosurgery, Hospital of Lithuanian University of Health Sciences, Kaunas Clinics, between 2015 and 2017. Tumor sample collection and written informed consent procedures are in accordance with the Lithuanian regulations and the Helsinki Declaration. Database closure was in November 2017. Diagnoses were established by pathologists at the Hospital of Lithuanian University of Health Sciences, Kaunas Clinics according to the World Health Organization (WHO) classification. Tumor samples were frozen and stored in liquid nitrogen until experimentation. 12 specimens were identified as diffuse astrocytomas (grade II), while 3 were attributed to anaplastic astrocytomas (grade III) and 35 to glioblastoma (grade IV) respectively. All patients provided written informed consent before the commencement of the surgery. The overall survival of the patient was calculated from the date of the operation to the date of death or the last recorded contact with the live patient (censored). None of the patients had received chemotherapy or radiation before surgery.
Detection of GFAP transcriptional activity
Tumor mRNA was extracted using TRIzol® (Invitrogen). Afterwards, cDNA samples were synthesized from total mRNA using RevertAid H Minus First Strand cDNA Synthesis Kit (Catalog#: K1631, Thermo Fisher Scientific). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was performed using an Applied Biosystems 7,500 real-time PCR system. The 12 μl PCR reaction consisted of 3 μl of cDNA (15 ng), 6 μl of SYBR Green mix, 2.6 μl of H2O and 0.2 μl of forward (F) and reverse (R) primers for GFAP (F: 5`-ACCTGCAGATTCGAGAAACC-3`, R: 5`‑CTCCTTAATGACCTCTCCATCC-3`). Beta‑actin cDNA (F: 5`-AGAGCTACGAGCTGCCTGAC-3`, R: 5`‑AGCACTGTGTTGGCGTACAG-3`) was used as an endogenous control. Samples were incubated in a thermocycler for 10 min at 95°C, 40 cycles (30 sec each at 95°C, 60°C and 72°C), then held at 4°C. Each sample was examined in triplicates to calculate mean gene expression. A negative water control was used in every assay. The acquired data for GFAP transcriptional activity is in relation to the reference human brain control (Catalog#: AM6050, Ambion) and processed using the log-transformed expression (Log2(2-ΔΔCt)) method.
Detection of GFAP translational activity
Translational activity of the GFAP gene was identified using western blot analysis. Protein lysates were prepared from frozen tumor tissue samples, as brief sonication and denaturation in the heat of 95°C for 5 min was included to ensure that only primary protein structures would remain in the samples. The protein specimens were loaded on to 10 % polyacrylamide gels containing 10 % SDS, 30 % acrylamide/Bis-acrylamide solution (Carl Roth™, GmbH), 1.5 M TRIS-HCl, and 10 % APS, TEMED reagents (Carl Roth™, GmbH). SDS‑PAGE running buffer was used for the polyacrylamide electrophoresis running at 120 V, 300 mA, 3W. Afterwards, samples were transferred onto nitrocellulose membranes using Trans-Blot™ SD Semi‑Dry Transfer Cell (Bio-Rad) for 35 min at 17 V. Membranes were blocked using 10 % milk PBS solution for 2 h at room temperature and then incubated with primary GFAP antibodies (Catalog#: ab4648, Abcam) in 5% milk with PBS at 4°C overnight. After washing with PBST (1x PBS, 0.05 % Tween), the membranes were incubated for one hour at room temperature with secondary anti-mouse antibodies (Catalog#: 31430, Thermo Fisher Scientific). Protein Bands were visualized using tetramethylbenzidine substrate and scanned with a flatbed scanner to produce digital images. Afterwards, polyclonal anti-Beta-actin (Catalog#: PA1-46296, Thermo Fisher Scientific) and anti‑rabbit (Catalog#: 31460, Thermo Fisher Scientific) antibodies were utilized to detect the reference Beta-actin protein. The scanned figures were processed using Adobe Photoshop and analyzed with ImageJ software. GFAP translational activity was normalized to the quantitative values of Beta-actin protein.
Identification rs2070935 genotypes
DNA from tumor samples was extracted using ZR Genomic DNA™ Tissue MiniPrep (Zymo Research). Afterwards, qPCR was performed with SNP genotyping assay (Assay ID: C__15868049_10, Thermo Fisher Scientific). A 9 μl reaction volume consisted of 1 μl of genomic tumor DNA (20 ng/μl), 5 μl of TaqMan® Universal PCR Master Mix (Thermo Fisher Scientific), 0.2 μl of TaqMan® SNP Genotyping Assay, and 3.8 μl of H2O. Negative water control was used in every assay. Samples were incubated in a qPCR thermocycler for 10 min at 95°C, 40 cycles (15 sec at 92°C, 1 min at 60°C), 1 min at 60°C, then held at 4°C.
Statistical analysis and data visualization
Statistical analysis was performed using IBM SPSS Statistics 22 and GraphPad Prism 7, which was additionally utilized to draw the graphs presented in the article. In gene expression graphs the upper and bottom whiskers mark the 75th and 25th percentiles accordingly, while the line in between the whiskers marks the mean value of the respective group. Quantitative data is presented with standard deviation (SD). Mann‑Whitney U test was carried out to compare means of sample groups. χ² test was used to check for Hardy‑Weinberg equilibrium. Spearman's Rho test was performed to measure correlation, which is considered to be strong, if the correlation coefficient r is above 0.75 or below -0.75. Survival time was calculated from the time of the initial surgery to the death of the patient. The Kaplan-Meier method was implemented to estimate patient survival rates and the Wilcoxon rank sum test was utilized to measure statistical significance. Measurements were considered to be statistically significant, when the probability value p was below 0.05.
Results
GFAP expression in human astrocytoma
GFAP transcriptional activity was detected in 45 astrocytoma samples via qRT-PCR assay. Mean transcriptional activity for GFAP (fig 1A) was significantly higher in grade II tumor samples (mean ± SD: 2.068 ± 1.513), compared to that of grade IV (mean ± SD: -0.06347 ± 1.329, p < 0.0001). The calculated mean and medial GFAP transcriptional activity in grade II astrocytoma (median: 2.028) was approximately 2 Log2(2‑ΔΔCt) higher than the reference human brain control sample. In contrast, the mean and medial GFAP mRNA expression in grade IV tumors (median: -0.06421) was nearly the same as that of the reference human brain control sample. Since only 3 grade III astrocytoma samples (mean ± SD: 1.279 ± 1.224) were analyzed, no statistically significant findings were detected regarding the following group of tumors. Spearman correlation test revealed a negative correlation between GFAP mRNA levels and astrocytoma grade (r = ‑0.6469, p < 0.0001).
Figure 1.
GFAP expression varies in different grades of human astrocytoma. A) Transcriptional activity of GFAP, as detected by qRT-PCR assay and grouped by tumor grade. B) Kaplan‑Meier curve for grade IV astrocytoma patient overall survival in accordance to GFAP mRNA expression. Increased GFAP expression - patient group, exhibiting higher GFAP mRNA expression than the reference human brain control (Log2(2‑ΔΔCt) > 0). Decreased GFAP expression - patient group, featuring lower GFAP mRNA expression than the reference human brain control (Log2(2‑ΔΔCt) < 0) . C) Translational activity of GFAP, as observed by western blot assay and grouped by tumor grade. D) GFAP and Beta-actin protein bands, as captured by western blot assay. The molecular weight of GFAP isoforms ranges from 50 to 38 kDa, whereas Beta-actin protein is at approximately 42 kDa. In all graphs GFAP expression values at mRNA and protein levels were normalized to that of the reference gene (Beta-actin). GFAP transcriptional activity for tumor samples is in relation to the reference human brain control.
Clinical characteristics of astrocytoma patients are provided in Table 1. Kaplan-Meier test was carried out to compare overall patient survival between increased and decreased GFAP transcriptional activities with grade IV astrocytoma samples (fig 1B). Patients, who exhibited increased GFAP mRNA levels, had a median survival of 37 weeks, whereas lower gene expression was attributed to a median survival of 51 weeks (p = 0.2117).
Table 1.
Astrocytoma patient gender and age characteristics
| Astrocytomas | Grade II (n = 12) |
Grade III (n = 3) |
Grade IV (n = 35) |
Total (n = 50) |
|
|---|---|---|---|---|---|
| Gender | Female | 2 (17 %) | 0 (0 %) | 19 (54 %) | 21 (42 %) |
| Male | 10 (83 %) | 3 (100 %) | 16 (46 %) | 29 (58 %) | |
| Age (years) | Median | 35 | 32 | 61 | 52.5 |
| Mean | 36.75 | 37.33 | 59.59 | 52.63 | |
| Minimum | 25 | 32 | 38 | 25 | |
| Maximum | 63 | 48 | 82 | 82 | |
GFAP translational activity was detected in 44 astrocytoma samples using western blot assay (fig 1C). Multiple GFAP isoforms were detected (fig 1D). Higher quantities of mean GFAP protein were observed in grade II tumor specimens (mean ± SD: 16.69 ± 8.347), compared to that of grade IV (mean ± SD: 6.671 ± 4.605, p = 0.0001). Alternatively, the detected mean GFAP protein expression for grade III astrocytoma (mean ± SD: 20.98 ± 9.972) was higher than that of grade IV (p = 0.0077) and II (p = 0.3706) tumors respectively. Spearman correlation test was performed to identify a negative correlation between GFAP protein levels and astrocytoma grade (r = -0.5934, p < 0.0001).
GFAP transcriptional and translational activities were compared using Spearman correlation test. 42 astrocytoma samples (10 II, 3 III, 29 IV), which featured detectable signals of GFAP expression in both western blot and qRT-PCR assays, were used. Spearman correlation test revealed a positive correlation between GFAP mRNA and protein levels (r = 0.6546, p < 0.0001).
Interaction between rs2070935 and GFAP expression
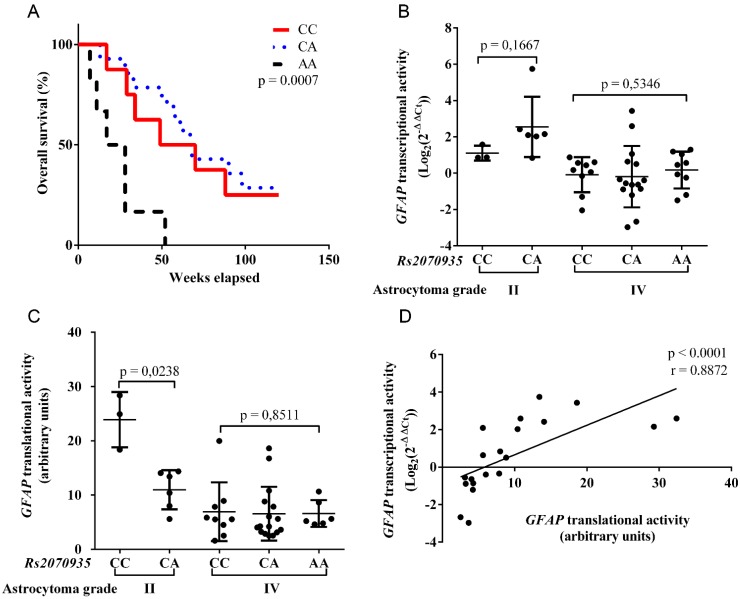
Rs2070935 genotypes were identified in 47 astrocytoma samples using qPCR assay. Frequencies of Rs2070935 are presented in Table 2. Statistical comparison of observed and estimated rs2070935 genotypes resulted in p > 0.05, which indicates that the examined groups did not deviate from Hardy‑Weinberg equilibrium. Kaplan-Meier test, which was performed with grade IV astrocytoma samples (fig 2A), revealed an association between rs2070935 AA genotype and shorter median patient survival (p = 0.0007). Median patient survival for CC, CA and AA genotypes of rs2070935 was 57, 48 and 24 weeks respectively.
Table 2.
Rs2070935 characteristics
| Astrocytomas | Grade II (n = 10) |
Grade III (n = 3) |
Grade IV (n = 34) |
Total (n = 47) |
|
|---|---|---|---|---|---|
| Observed rs2070935 genotypes | CC | 3 (30 %) | 0 (0 %) | 10 (29 %) | 13 (28 %) |
| CA | 7 (70 %) | 1 (33 %) | 15 (44 %) | 23 (49 %) | |
| AA | 0 (0 %) | 2 (67 %) | 9 (27 %) | 11 (23 %) | |
| Expected rs2070935 genotypes | CC | 4.23 | 0.08 | 9.01 | 12.77 |
| CA | 4.55 | 0.83 | 16.99 | 23.46 | |
| AA | 1.22 | 2.08 | 8.01 | 10.77 | |
| Rs2070935 alleles | C | 65 % | 17 % | 51 % | 52 % |
| A | 35 % | 83 % | 49 % | 47 % | |
| χ² test for observed and expected genotypes | p | 0.559 | 0.833 | 0.889 | 0.988 |
Figure 2.
Rs2070935 polymorphism is linked with varying GFAP expression and different grade IV astrocytoma patient survival. A) Kaplan‑Meier curve for grade IV astrocytoma patient overall survival in accordance to rs2070935 genotypes. B) GFAP transcriptional activity, as detected by qRT-PCR assay and subdivided by genotypes of rs2070935 as well as tumor grade. C) GFAP translational activity, as observed by western blot assay and grouped by rs2070935 polymorphism as well as astrocytoma grade. D) Correlation between GFAP transcriptional and translational activities in the group of rs2070935 CA heterozygotes. In all graphs GFAP expression values at mRNA and protein levels were normalized to that of the reference gene (Beta-actin). GFAP transcriptional activity for tumor samples is in relation to the reference human brain control.
GFAP transcriptional and translational activities were grouped in accordance to rs2070935 genotypes and tumor grade. In grade II astrocytoma, mean GFAP mRNA expression (fig 2B) for CC homozygotes (mean ± SD: 1.107 ± 0.41) tended to be lower, compared to CA heterozygotes (mean ± SD: 2.549 ± 1.662, p = 0.1667). Opposite results were observed for GFAP translational activity (fig 2C), as CC genotypes (mean ± SD: 23.9 ± 5.091) featured higher protein quantities than that of CA (mean ± SD: 10.98 ± 3.612, p = 0.0238) in grade II astrocytoma. No vast differences regarding GFAP expression and rs2070935 polymorphism were detected in grade IV tumors.
GFAP transcriptional and translational activities were compared in the group of 20 rs2070935 CA heterozygotes, regardless of tumor grade. Spearman correlation test (fig 2D) was performed, which resulted in a strong correlation between different expression levels of GFAP (r = 0.8872, p < 0.0001).
Using data acquired from western blot experiments, GFAP protein samples were grouped by astrocytoma grade and rs2070935 polymorphism (fig 3). Samples of CC genotype featured a considerably greater portion of 38 kDa GFAP protein isoforms (mean ± SD: 23.9 ± 5.091 %), compared to specimens, exhibiting either the CA or AA genotype of rs2070935 (mean ± SD: 10.98 ± 3.612 %, p = 0.0238).
Figure 3.
Rs2070935 polymorphism is associated with different GFAP isoform quantities in human astrocytoma. 38 kDa proportion (%) - a percentile value, which represents the signal intensity of the 38 kDa GFAP isoform, compared to the signal intensity of the whole sample.
Discussion
In our study novel observations were made regarding GFAP expression at mRNA and protein levels in multiple grades of human astrocytoma. Using qRT-PCR and western blot assays, we found GFAP expression to correlate negatively with tumor grade, as mRNA and protein levels of the following gene are significantly lower in grade IV, compared to grade II or III astrocytoma. In previous studies similar results were obtained using immunohistochemistry 10 9. However, our findings further expand on these studies, as our data indicates that GFAP mRNA expression in grade II and III astrocytoma is considerably higher than in the reference human brain control, which represents a healthy CNS. Nevertheless, our research further expands on these findings, as we have implemented a reference human brain control in our qRT-PCR assay to detect that in some grade IV astrocytoma patients GFAP transcriptional activity was lower than in healthy brain tissue.
We also observed a linkage between GFAP transcriptional and translational activities across different grades of human astrocytoma. Nevertheless, it is noteworthy that the REpository for Molecular BRAin Neoplasia DaTa (REMBRANDT) data set does not depict any links between brain tumor grade and GFAP transcriptional activity, as measured by microarrays 12. The following discrepancies could have been influenced by different samples sizes as well as including non-astrocytic tumors in the analysis or reliance on the microarray method, which might be less accurate for measuring quantitative gene expression than qRT-PCR assay.
The precise causes for different GFAP expression in multiples grades of human astrocytoma are still unknown. One possible explanation was proposed by Huang et al. (2014) that the least differentiated astrocytoma cells might feature the lowest GFAP expression levels 10. Likewise, malignant glial cells of grade IV glioma are prone to undergo dedifferentiation 13. A decline of GFAP transcriptional and translational activities in grade IV astrocytoma might be due to the loss of differentiation properties in astrocytes. Accordingly, increased GFAP mRNA and protein expression levels in grade II and III brain tumors could be a result of greater amounts of differentiated astrocytes in tumor tissue.
The observed tendency between shorter grade IV astrocytoma patient survival and increased GFAP transcriptional activity could be linked with pathological mechanisms. Hagemann et al. identified an association between elevated GFAP expression and the formation of Rosenthal fibers 14. The following fibers are attributed to Alexander's disease, which involves the destruction of myelin 15. Moreover, Sugita et al. detected Rosenthal fibers in human brain tumor tissue. Increased GFAP transcriptional activity might facilitate the development of Rosenthal fibers that could further complicate the medical condition of grade IV astrocytoma patients 16. Likewise, usig qRT-PCR assay we observed a tendency between higher GFAP mRNA levels and shorter grade IV astrocytoma patient survival. We checked out our data with the REMBRANDT data set and found also features a link between longer median glioma patient survival and decreased GFAP mRNA expression, as detected by microarrays 12. We believe these findings to be of clinical significance, as our data indicates that GFAP might be used a prognostic marker for astrocytoma outcome.
Apart from GFAP expression, we observed rs2070935 polymorphism to be associated with different grade IV astrocytoma patient survival. In regards to experiments with rs2070935, Bachetti et al. used plasmids with different alleles of rs2070935 and a luciferase assay to draw a conclusion that GFAP transcriptional activity changes based on the polymorphism of rs2070935 8. However, our findings validate these results by utilizing western blot assay to correlate the different genotypes of rs2070935 with varying amounts of GFAP. Instead of a cell culture model used by Bachetti et al. 8, we ran experiments on human astrocytoma tissue, which also resulted in a statistically significant association between different genotypes of rs2070935 and varying survival of grade IV astrocytoma patients. At the moment, no other clinical cancer studies exist for rs2070935, making our findings relevant for future scientific investigations. For the first time the AA genotype was found to be associated with a significantly shorter astrocytoma patient median survival, compared to other variants of the following SNP. It is difficult to determine the clinical significance of rs2070935 polymorphism in human brain tumors, as previous research was conducted in commercial cell lines 8. Nevertheless, our data indicates that rs2070935 has the potential to be used as a prognostic factor for grade IV astrocytoma patient survival, if further research with a considerably larger sample size confirms the associations, detected in this study.
The processes of GFAP alternative splicing are not fully understood 7. Analysis of western blot data suggests that GFAP variants are not in equal proportion throughout different grades of astrocytoma. The most noticeable instance of isoform disequilibrium is the considerably greater amount of 38 kDA GFAP protein isoforms in samples, which feature the CC genotype of rs2070935, compared to other polymorphisms of the same SNP. The 38 kDA protein fragment of the GFAP gene is attributed to GFAPΔEx6 isoform 17. Accordingly, since a stronger correlation of GFAP mRNA and protein levels was observed in the group of CA genotypes, compared to that of all astrocytoma samples, rs2070935 might be linked with the process of GFAP alternative splicing, although further isoform-specific research is required for confirmation.
A statistically significant link between rs2070935 genotypes and GFAP translational activity was observed. Bachetti et al. reported that the A allele creates a novel binding site for the transcription factor AP-1, resulting in a lower expression of GFAP in commercial glioma cell lines 8. Our study demonstrates for the first time that the CC genotype of rs2070935 is associated with increased GFAP translational activity in grade II human astrocytoma. As for transcriptional activity, it is important to note that qRT-PCR measures GFAP mRNA, which is localized within the neural cells, whereas western blot analysis identifies GFAP proteins in the cells as well as in the extracellular matrix. The following features of the analytical methods could have caused the observed disparity between GFAP transcriptional and translational activities according to rs2070935 genotypes in grade II astrocytoma. Nonetheless, polymorphisms of rs2070935 were not found to be associated with different GFAP expression in grade IV astrocytoma.
In summary, a negative correlation between GFAP expression and human astrocytoma grade was observed. Rs2070935 AA genotype was found to be a sign of poor clinical outcome for grade IV astrocytoma patients, making the following SNP a potential prognostic factor. An association was identified in grade II astrocytoma between increased GFAP protein levels and the CC genotype of rs2070935. Indications were observed that suggest a possible interaction between rs2070935 polymorphism and alternative splicing of GFAP in multiple grades of human astrocytoma. The following findings indicate that GFAP expression and rs2070935 polymorphism are relevant biomarkers for large‑scale scientific research of astrocytoma patients.
Acknowledgments
This research was funded by grant no. MIP-052/2015 from the Research Council of Lithuania and Faculty of Medicine of Lithuanian University of Health Sciences.
Authors' contributions
Experimentation, data analysis and writing of the manuscript - Mantas Sereika. Collection of astrocytoma samples and patient data - Arimantas Tamasauskas. Preparation of astrocytoma protein samples and technical support - Ruta Urbanaviciute. Conception, design of study and critical revision of the manuscript - Paulina Vaitkiene and Daina Skiriute. All authors have read and approved this article.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Consent for publication
No patient personal data are provided in the manuscript.
Compliance with ethical standards
All patients provided written informed consent before the commencement of the tumor surgery. Permission (no. P2-9/2003) to undertake the study was obtained from the Kaunas Regional Biomedical Research Ethics Committee.
References
- 1.Appin CL, Brat DJ. Molecular genetics of gliomas. Cancer J. 2014;20:66–72. doi: 10.1097/PPO.0000000000000020. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 3.De Groot JF. High-grade gliomas. Continuum. Lifelong Learn. Neurol. 2015;21:332–344. doi: 10.1212/01.CON.0000464173.58262.d9. [DOI] [PubMed] [Google Scholar]
- 4.Ostrom QT, Bauchet L, Davis FG, Deltour I, Fisher JL, Langer CE, Pekmezci M, Schwartzbaum JA, Turner MC, Walsh KM, Wrensch MR, Barnholtz-Sloan JS. The epidemiology of glioma in adults: A state of the science review. Neuro. Oncol. 2014;16:896–913. doi: 10.1093/neuonc/nou087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aum DJ, Kim DH, Beaumont TL, Leuthardt EC, Dunn GP, Kim AH. Molecular and cellular heterogeneity: the hallmark of glioblastoma. Neurosurg. Focus. 2014;37:E11. doi: 10.3171/2014.9.FOCUS14521. [DOI] [PubMed] [Google Scholar]
- 6.Yang Z, Wang KKW. Glial fibrillary acidic protein: From intermediate filament assembly and gliosis to neurobiomarker. Trends Neurosci. 2015;38:364–374. doi: 10.1016/j.tins.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hol EM, Pekny M. Glial fibrillary acidic protein (GFAP) and the astrocyte intermediate filament system in diseases of the central nervous system. Curr. Opin. Cell Biol. 2015;32:121–130. doi: 10.1016/j.ceb.2015.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Bachetti T, Di Zanni E, Lantieri F, Caroli F, Regis S, Filocamo M, Rainero I, Gallone S, Cilia R, Romano S, Savoiardo M, Pareyson D, Biancheri R, Ravazzolo R, Ceccherini I. A Novel Polymorphic AP-1 Binding Element of the GFAP Promoter is Associated with Different Allelic Transcriptional Activities. Ann. Hum. Genet. 2010;74:506–515. doi: 10.1111/j.1469-1809.2010.00614.x. [DOI] [PubMed] [Google Scholar]
- 9.Qin J, Liu Z, Zhang H, Shen C, Wang X, Tan Y, Wang S, Wu X, Tian J. Grading of Gliomas by Using Radiomic Features on Multiple Magnetic Resonance Imaging (MRI) Sequences. Med. Sci. Monit. 2017;23:2168–2178. doi: 10.12659/MSM.901270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang S, Chen G, Dang Y, Chen LH. Overexpression of DcR3 and its significance on tumor cell differentiation and proliferation in glioma. Sci. World J; 2014. p. 605236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brehar FM, Arsene D, Brinduse LA, Gorgan MR. Immunohistochemical analysis of GFAP-δ and nestin in cerebral astrocytomas. Brain Tumor Pathol. 2015;32:90–98. doi: 10.1007/s10014-014-0199-8. [DOI] [PubMed] [Google Scholar]
- 12.Scarpace L, Flanders AE, Jain R, Mikkelsen T. Andrews DW 2015. Data From REMBRANDT. The Cancer Imaging Archive. http://doi.org/10.7937/K9/TCIA. 2015. 588OZUZB.
- 13.Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K. Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis. 2015;2:152–163. doi: 10.1016/j.gendis.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagemann TL, Gaeta SA, Smith MA, Johnson DA, Johnson JA, Messing A. Gene expression analysis in mice with elevated glial fibrillary acidic protein and Rosenthal fibers reveals a stress response followed by glial activation and neuronal dysfunction. Hum. Mol. Genet. 2005;14:2443–2458. doi: 10.1093/hmg/ddi248. [DOI] [PubMed] [Google Scholar]
- 15.Messing A, Brenner M, Feany MB, Nedergaard M, Goldman JE. Alexander disease. J Neurosci. 2012;32:5017–5023. doi: 10.1523/JNEUROSCI.5384-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sugita Y, Nakashima S, Ohshima K, Terasaki M, Morioka M. Anaplastic astrocytomas with abundant Rosenthal fibers in elderly patients: A diagnostic pitfall of high-grade gliomas. Neuropathology. 2013;33:533–540. doi: 10.1111/neup.12025. [DOI] [PubMed] [Google Scholar]
- 17.Kamphuis W, Mamber C, Moeton M, Kooijman L, Sluijs JA, Jansen AHP, Verveer M, de Groot LR, Smith VD, Rangarajan S, Rodríguez JJ, Orre M, Hol EM. GFAP isoforms in adult mouse brain with a focus on neurogenic astrocytes and reactive astrogliosis in mouse models of Alzheimer disease. PLoS One; 2012. p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.