The glycoside hydrolase family 3 β-glucosidase Cel3A from Neurospora crassa (NcCel3A) is highly specific for cellobiose. The crystal structure of NcCel3A has been solved to 2.25 Å resolution. The structure is a dimer and exhibits a high degree of N-glycosylation.
Keywords: glycoside hydrolase family 3, β-glucosidase, biodegradation, crystal structure, Neurospora crassa, NcCel3A
Abstract
The glycoside hydrolase family 3 (GH3) β-glucosidases are a structurally diverse family of enzymes. Cel3A from Neurospora crassa (NcCel3A) belongs to a subfamily of key enzymes that are crucial for industrial biomass degradation. β-Glucosidases hydrolyse the β-1,4 bond at the nonreducing end of cellodextrins. The hydrolysis of cellobiose is of special importance as its accumulation inhibits other cellulases acting on crystalline cellulose. Here, the crystal structure of the biologically relevant dimeric form of NcCel3A is reported. The structure has been refined to 2.25 Å resolution, with an R cryst and R free of 0.18 and 0.22, respectively. NcCel3A is an extensively N-glycosylated glycoprotein that shares 46% sequence identity with Hypocrea jecorina Cel3A, the structure of which has recently been published, and 61% sequence identity with the thermophilic β-glucosidase from Rasamsonia emersonii. NcCel3A is a three-domain protein with a number of extended loops that deepen the active-site cleft of the enzyme. These structures characterize this subfamily of GH3 β-glucosidases and account for the high cellobiose specificity of this subfamily.
1. Introduction
Fungal degradation of cellulose is considered to be accomplished by four primary enzymatic activities, which act synergistically to overcome the recalcitrance of the cellulose polymer (Payne et al., 2015 ▸). Endoglucanases [EGs; endo-(1,4)-β-d-glucanhydrolases; EC 3.2.1.4] cleave exposed cellulose chains randomly, which introduces new chain ends, and release shorter cello-oligosaccharides of varying lengths. Cellobiohydrolases [CBHs; exo-(1,4)-β-d-glucan cellobiohydrolases; EC 3.2.1.91 and EC 3.2.1.176] processively traverse a cellulose chain while successively releasing cellobiose units. β-Glucosidases (BGLs; EC 3.2.1.21) hydrolyse the soluble oligosaccharides and cellobiose to primarily produce glucose. Lytic polysaccharide monooxygenases (LPMOs) are a more recently discovered nonhydrolytic class of polysaccharide-degrading enzymes (Harris et al., 2010 ▸). LPMOs break polysaccharide chains in an oxygen- and electron-dependent process; they break glycosidic bonds by directly oxidizing the C1 or C4 carbon of a glycopyranose ring, apparently without the need for depolymerization (Meier et al., 2018 ▸). In the classification system of carbohydrate-active enzymes, the Carbohydrate-Active enZYmes Database (CAZy; Lombard et al., 2014 ▸), β-glucosidases can be found in glycoside hydrolase (GH; Henrissat & Davies, 1997 ▸) families GH1, GH3, GH5, GH9, GH30 and GH116. The enzymes in all of these GH families except for GH9 perform hydrolysis by a double-displacement reaction mechanism with net retention of the configuration at the anomeric carbon (Gebler et al., 1992 ▸). All such β-glucosidases have a common (α/β)8 TIM-barrel fold. Glycoside hydrolase family 3 (GH3) is one of the larger families in the CAZy classification and currently contains over 13 600 annotated protein sequences. The family groups together several exo-acting activities and includes enzyme members with broad substrate specificity with respect to the type of monosaccharide, linkages and chain length of the substrate.
The filamentous fungus Neurospora crassa is an ascomycete that decomposes and consumes dead plant material in nature. It has been widely used as a model organism in the field of eukaryotic biology (Davis & Perkins, 2002 ▸) and produces and secretes a full suite of carbohydrate-degrading enzymes (Romero et al., 1999 ▸; Eberhart et al., 1964 ▸, 1977 ▸; Yazdi et al., 1990 ▸). These enzymes are able to completely decrystallize and depolymerize cellulose as well as other plant cell-wall polysaccharides in an orchestrated fashion (Tian et al., 2009 ▸). There are at least seven genes encoding GH3 enzymes in the genome of N. crassa. Three of these genes have a signal peptide and are expected to produce secreted GH3 enzymes: GH3-1 (Bgl7, NCU03641), Cel3A (GH3-3, Bgl6, NCU08755) and GH3-4 (Bgl2, NCU04952). All three gene products are upregulated when wild-type N. crassa is grown using cellulose as the main carbon source (Wu et al., 2013 ▸), but only Cel3A and GH3-4 have been experimentally characterized as true β-glucosidases (Tian et al., 2009 ▸; Bohlin et al., 2010 ▸), and it has recently been shown that Cel3A exhibits a high affinity for cellobiose compared with longer β-1,4-gluco-oligosaccharides (Colabardini et al., 2016 ▸). Cel3A was identified in the conidia cell walls of N. crassa (Maddi et al., 2009 ▸) and GH3-4 was identified in the supernatant of N. crassa grown on Avicel and Miscanthus by mass spectrometry (Tian et al., 2009 ▸). Cel3A is homologous to several enzymes with recently published crystal structures: Rasamsonia emersonii Cel3A (ReCel3A; Gudmundsson et al., 2016 ▸), Aspergillus aculeatus BGLI (AaBGLI; Suzuki et al., 2013 ▸), Aspergillus oryzae Cel3A (AoCel3A; Agirre et al., 2016 ▸) and Aspergillus fumigatus Cel3A (AfCel3A; Agirre et al., 2016 ▸). Both ReCel3A and AaBGLI have also been shown to exhibit the properties of dedicated cellobiases.
There are currently nine structural models of fungal GH3 enzymes available in the Protein Data Bank. Six are characterized as β-1,4-glucosidases and the remaining three are a β-1,3-1,4-glucanase (Varghese et al., 1999 ▸), a β-N-acetylglucosaminidase (Qin et al., 2015 ▸) and a β-1,4-xylosidase (PDB entry 5a7m; Mikkelsen et al., unpublished work). These structures highlight the modularity of the GH3 enzymes, which has become apparent since the first structure of a GH3 enzyme was solved, that of the exo-β-1,3-1,4-glucanase Hordeum vulgare ExoI (HvExoI). HvExoI is composed of two domains: an N-terminal (α/β)8 TIM-barrel domain containing the catalytic nucleophile aspartate and an (α/β)6 sandwich domain containing the catalytic acid/base glutamate. This two-domain structure and the position of the catalytic residues are a core feature of GH3 enzymes and are retained in the multidomain GH3 enzymes that are now known. In 2010 the first three-domain GH3 structure was published, that of the β-glucosidase Thermotoga neapolitana Bgl3B (TnBgl3B; Pozzo et al., 2010 ▸). The C-terminal FnIII-like third domain straddles the barrel and sandwich domains on the opposite side to the active site of the enzyme. The function of the third domain is unknown, although it has been suggested that it stabilizes the TIM-barrel domain, which has an incomplete/collapsed fold in all three-domain GH3 enzymes with known structure (Gudmundsson et al., 2016 ▸). A fourth domain is present in Kluyveromyces marxianus BglI (KmBglI; Yoshida et al., 2010 ▸) and Pseudoalteromona sp. ExoP (PsExoP; Nakatani et al., 2012 ▸). KmBglI has a PA14 domain that extends the active site, while PsExoP has a highly mobile CBM-like domain of unknown function.
In this study, we present the crystallization and structure determination of a GH3 β-glucosidase from N. crassa (NcCel3A) solved to 2.25 Å resolution. These results are discussed in the light of differences from and similarities to other GH3 enzymes with known structure. NcCel3A is most similar to the recently solved crystal structures of the GH3 β-glucosidases AaBGL1 and ReCel3A (PDB entries 4iib and 5ju6; Suzuki et al., 2013 ▸; Gudmundsson et al., 2016 ▸). These three GH3 structures all have certain major structural features in common. A high number of N-glycosylations can be observed, with over 40 modelled glycans per protein molecule, which are peculiarly localized only on one face of the proteins. These enzymes also all have an extended C-terminal loop protruding from the C-terminal domain covering large parts of the first domain and several of its N-glycosylations. The linkers connecting the three domains of these enzymes extend much further towards the active-site cleft of the enzyme compared with HjCel3A and TnBgl3B, and unlike HjCel3A and TnBgl3B this class of GH3 β-glucosidases have all been shown to exist as dimers in solution (Murray et al., 2004 ▸; Gudmundsson et al., 2016 ▸; Suzuki et al., 2013 ▸; Agirre et al., 2016 ▸).
2. Materials and methods
2.1. Macromolecule production
The gh3-3 gene encoding NcCel3A (GenBank EAA26868.1) was overexpressed in an H. jecorina strain with eight genes coding for cellulases (cbh1, cbh2, egl1, egl2, egl3, egl4, egl5 and egl6) deleted and one gene coding for a mannanase (man1) deleted. The target gene was cloned into the pTrex3G vector (amdSR, ampR, Pcbh1; Foreman et al., 2005 ▸) and used to transform H. jecorina. Transformants of H. jecorina were picked from Vogel’s minimal medium plates (Vogel, 1956 ▸) containing acetamide after seven days of incubation at 37°C and were grown in Vogel’s minimal medium with a mixture of glucose and sophorose as carbon sources. The overexpressed protein appeared as a dominant protein in the culture supernatants.
Culture filtrate from the production of NcCel3A in H. jecorina (obtained using Sarstedt Filtropur 0.2 µm filters) was diluted tenfold with 25 mM sodium acetate pH 4.0 and incubated at 37°C for 30 min. The sample was desalted using a Sephadex G-25M column (GE Healthcare, Piscataway, New Jersey, USA) equilibrated with acetate buffer and concentrated using a centrifugal concentrator with a 10 kDa cutoff (Vivascience, Littleton, Massachusetts, USA). The protein solution was loaded onto a gel-filtration column (Superdex 200 HiLoad 16/60) with 20 mM sodium acetate pH 7.5, 150 mM NaCl as the running buffer and eluted at a flow rate of 0.5 ml min−1 on an ÄKTApurifier (GE Healthcare Biosciences, Sweden) at room temperature. The fractions containing NcCel3A were pooled, washed and concentrated to 15 mg ml−1 in 20 mM sodium acetate buffer pH 5.0, 20 mM NaCl using Vivaspin 20 centrifuge concentration tubes with a 30 kDa molecular-mass cutoff (Sartorius Stedim Biotech, France). The purity of the NcGH3 protein was greater than 95% as judged by SDS–PAGE. The protein concentration was determined by measuring the absorbance of the protein solution at 280 nm using a calculated extinction coefficient for NcCel3A of 160 130 M −1 cm−1.
2.2. Crystallization
Crystals of NcCel3A were grown using the hanging-drop vapour-diffusion method at 20°C. Initial crystallization trials were carried out using a Mosquito crystallization robot (TTP Labtech, Cambridge, England). To identify the best crystallization condition, several commercially available crystallization screens such as PEG/Ion (Hampton Research), Crystal Screen and Crystal Screen 2 (Molecular Dimensions, UK) and the JCSG+ Suite (Qiagen, Germany) were utilized. Using a 96-well plate, drops (0.3 µl) were prepared by mixing protein solution at ∼15 mg ml−1 with an equal amount of well solution. The best crystallization condition was obtained from the PEG/Ion screen and consisted of 0.2 M ammonium citrate dibasic pH 5.1, 20%(w/v) polyethylene glycol 3350. Crystals that were large enough for X-ray data collection grew within a week. Prior to X-ray data collection, crystals of NcCel3A were transferred into a cryoprotectant solution containing 40% PEG 3350 and cooled in liquid nitrogen.
2.3. Data collection and structure refinement
Data were collected at a wavelength of 1.0 Å at 100 K on beamline I911-3 at MAX-lab, Lund, Sweden. The data were processed using XDS (version of 3 February 2010; Kabsch, 2010 ▸) and scaled by the scaling program SCALA v.3.3.16 (Evans, 2006 ▸) via the CCP4 v.6.5.0 program suite (Winn et al., 2011 ▸). A set of 5% of the reflections was put aside and used to calculate the quality factor R free (Brünger, 1992 ▸). Details of data collection and processing are presented in Table 1 ▸. The crystal structure of NcCel3A was determined by molecular replacement using Phaser v.2.1.4 (McCoy et al., 2007 ▸). The molecular-replacement search molecule consisted of one molecule of β-glucosidase 1 from A. aculeatus (AaBGL1; PDB entry 4iib; Suzuki et al., 2013 ▸). Rigid-body refinement of individual molecules using data between 20 and 3 Å resolution was performed and the resulting 2F o − F c and F o − F c electron-density maps showed continuous density for two NcCel3A molecules in the asymmetric unit. Throughout the refinement, 2mF o − DF c and mF o − DF c σA-weighted maps (Pannu & Read, 1996 ▸) were inspected and the models were manually adjusted during repetitive cycles of iterative model building using Coot v.0.8.7 (Emsley et al., 2010 ▸; Emsley & Cowtan, 2004 ▸; Krissinel & Henrick, 2007 ▸) and maximum-likelihood refinement and TLS refinement using REFMAC5 v.5.8.0135 (Murshudov et al., 1997 ▸, 2011 ▸) until no further improvement of structural parameters could be observed. Water molecules were added using ARP/wARP v.7.1 (Perrakis et al., 1997 ▸) and manually using Coot. Figures were prepared using PyMOL v.1.5.0.4 (DeLano, 2002 ▸). Root-mean-square deviation values (r.m.s.d.s) were calculated using the SSM function (Krissinel & Henrick, 2004 ▸) in Coot. Carbohydrates were modelled via cyclical building in Coot and refinement with REFMAC5 with torsion-angle restraints enabled. Validation of correct stereochemistry and low-energy conformation of carbohydrates between refinement cycles was performed with Privateer v.MKIII (Agirre et al., 2015 ▸). Coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with PDB code 5nbs.
Table 1. Data collection and processing.
Values in parentheses are for the outer shell.
| Data collection | |
| Diffraction source | I911-3, MAX-lab |
| Wavelength (Å) | 1.0 |
| Temperature (K) | 100 |
| Detector | MAR Mosaic 225 |
| Crystal-to-detector distance (mm) | 198.59 |
| Rotation range per image (°) | 0.25 |
| Total rotation range (°) | 97.5 |
| Space group | P21212 |
| a, b, c (Å) | 142.9, 286.8, 58.0 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution range (Å) | 47.00–2.25 |
| Total No. of reflections | 217078 |
| No. of unique reflections | 113592 |
| Completeness (%) | 99.2 |
| Multiplicity | 1.9 (1.9) |
| 〈I/σ(I)〉 | 22.9 (4.3) |
| R merge † | 0.028 (0.38) |
| R p.i.m. ‡ | 0.028 (0.38) |
| R r.i.m. § | 0.040 (0.53) |
| CC1/2 ¶ | 0.998 (0.825) |
| Overall B factor from Wilson plot (Å2) | 29.7 |
| Structure refinement | |
| Resolution range (Å) | 286.8–2.25 |
| Completeness (%) | 99.2 |
| No. of reflections, working set | 107929 (7878) |
| No. of reflections, test set | 5627 (417) |
| Final R cryst †† | 0.18 (0.30) |
| Final R free †† | 0.22 (0.35) |
| No. of non-H atoms | |
| Total | 15120 |
| Protein | 13147 |
| Carbohydrate | 1021 |
| Water | 954 |
| Model quality | |
| R.m.s. deviations | |
| Bonds (Å) | 0.012 |
| Angles (°) | 1.625 |
| Ramachandran plot‡‡ | |
| Most favoured (%) | 97 |
| Allowed (%) | 2.9 |
| Pyranose conformations (total/percentage)§§ | |
| Lowest energy conformation | 83/100 |
| Higher energy conformations | 0.0/0 |
R
merge = 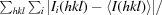
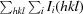 , where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl and 〈I(hkl)〉 is the mean intensity of I
i(hkl) for all i measurements.
, where I
i(hkl) is the intensity of the ith measurement of an equivalent reflection with indices hkl and 〈I(hkl)〉 is the mean intensity of I
i(hkl) for all i measurements.
R p.i.m. is the precision-indicating (multiplicity-weighted) R r.i.m. (Diederichs & Karplus, 1997 ▸; Weiss, 2001 ▸).
CC1/2 is the correlation coefficient of the mean intensities between two random half-sets of data (Karplus & Diederichs, 2012 ▸; Evans, 2012 ▸).
R
cryst = 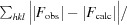
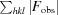 ; R
free is calculated in an identical manner using a randomly selected 5% of the reflections which were not included in the refinement.
; R
free is calculated in an identical manner using a randomly selected 5% of the reflections which were not included in the refinement.
Calculated using a strict-boundary Ramachandran definition given by Kleywegt & Jones (1996 ▸).
3. Results and discussion
3.1. Expression, purification and crystallization of NcCel3A
The purified and concentrated NcCel3A crystallized in the orthorhombic space group P21212, with unit-cell parameters a = 142.9, b = 286.8, c = 58.1 Å. The molecular-replacement solution gave a best solution with two protein molecules (molecular weight 93.6 kDa) in the asymmetric unit, with a calculated V M of 3.17 Å3 Da−1 (Matthews, 1968 ▸) and a solvent content of 61%. The NcCel3A structure was refined at 2.25 Å resolution to final R cryst and R free values of 17.9 and 21.6%, respectively. The final NcCel3A structure is composed of two noncrystallographic symmetry (NCS)-related molecules with 842 and 843 amino-acid residues, respectively, 875 water molecules and 85 carbohydrate residues. No gaps were found in the protein chains. There are eight cis-peptides and eight cysteines, of which six form disulfide bonds, in each protein molecule in the structure. Chains A and B have 45 and 38 modelled N-glycans (N-acetyl-β-d-glucosamine, α-d-mannopyranose and β-d-mannopyranose), respectively. Additional X-ray data-collection and refinement statistics are presented in Table 1 ▸.
3.2. The fold and structure of NcCel3A
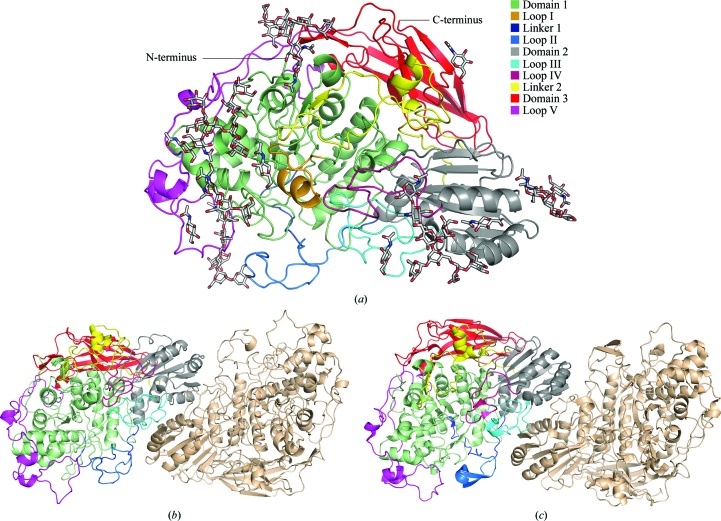
The NcCel3A crystal structure model is composed of two NCS-related protein molecules in the asymmetric unit. NcCel3A chain A contains 843 amino-acid residues and the first modelled residue is Ser34 of the deposited NcCel3A DNA sequence (GenBank EAA26868.1), while chain B contains 842 residues and the first modelled residue is Leu35. The last modelled residue in both protein chains is Pro875. The signal-peptide cleavage site of the EAA26868.1 sequence predicted by SignalP 4.1 (Petersen et al., 2011 ▸) is at position 18. The N-terminal residues 1–16 are most likely not visible owing to high flexibility. The overall structure of NcCel3A is composed of three separate domains connected by two linkers and has a high degree of N-glycosylation. The two NcCel3A protein chains form a dimer similar to those observed in ReCel3A (Gudmundsson et al., 2016 ▸) and AaBGL1 (Suzuki et al., 2013 ▸).
There are 15 NXT/S N-glycosylation sites in NcCel3A and all are found on or near the surface of the protein molecule. 12 sites in chain A and ten sites in chain B possessed sufficient electron density to allow the modelling of N-glycan moieties. The glycans ranged in length from single N-acetylglucosamine (GlcNAc) residues to longer Man7GlcNAc2 chains. The majority of the long N-glycan chains are positioned on domain 1 and domain 2, and are also asymmetrically distributed onto the same face of the protein. This pattern of N-glycans seems to be conserved among this subclass of GH3 BGLs. We have previously theorized (Gudmundsson et al., 2016 ▸) that these extensive and partially buried N-glycans serve a function in stabilizing the collapsed TIM-barrel fold of domain 1 by binding to hydrophobic patches in conjunction with the C-terminal loop V. Also of interest is that protein glycans have a potential binding affinity for polysaccharides such as cellulose, as has been proposed by Payne et al. (2013 ▸). This could suggest a functionality of the N-glycans in conferring binding to cellulose, which would be in the process of being degraded by other cellulases and thus be where β-glucosidic enzyme activity would be most needed by the organism. For additional analysis of GH3-BGL N-glycosylation, see Gudmundsson et al. (2016 ▸) and Agirre et al. (2016 ▸).
The structure of NcCel3A is highly homologous to several GH-family 3 β-glucosidase structures, ReCel3A, AaBGL1, AfCel3A and AoCel3A, with 61% sequence identity to the first three proteins and 59% to AoCel3A. Structural alignments highlight the same high similarities, with r.m.s.d. values of 0.74 Å with regard to ReCel3A and AfCel3A, 0.76 Å for AoCel3A and 0.82 Å for AaBGL1. All three-domain GH3 enzymes fall into subcluster C2 as specified by Cournoyer & Faure (2003 ▸).
3.3. Domain 1
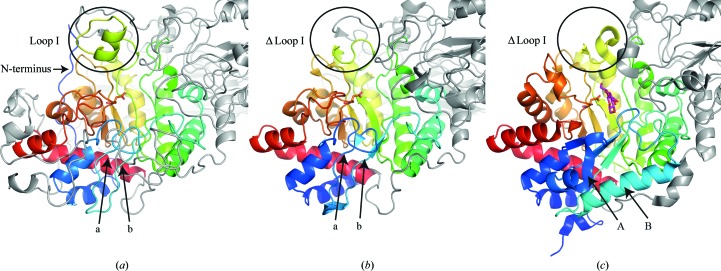
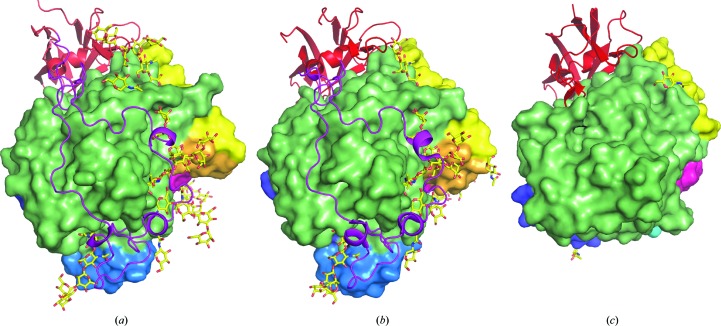
The first domain of NcCel3A (residues 34–340; coloured light green in Fig. 1 ▸) is composed of a collapsed TIM-barrel fold [or ββ(β/α)6 fold], which was described for the first time in the structure of the GH family 3 β-glucosidase TnBgl3B (Pozzo et al., 2010 ▸) and more recently by other groups presenting new structures of GH3 β-glucosidases (Yoshida et al., 2010 ▸; Suzuki et al., 2013 ▸; Karkehabadi et al., 2014 ▸; Gudmundsson et al., 2016 ▸; Agirre et al., 2016 ▸). Domain 1 of NcCel3A contains the catalytic nucleophile Asp276, as well as the majority of residues comprising substrate-binding subsite −1, with the catalytic centre being located between subsites −1 and +1. The subsite nomenclature of glycoside hydrolases is described by Biely et al. (1981 ▸) and Davies et al. (1997 ▸). All three-domain GH3 structures, where the C-terminal domain is an FNIII-like domain, lack two α-helices compared with the canonical TIM barrel of the barley GH3 structure HvExoI (Varghese et al., 1999 ▸; Fig. 2 ▸). The loss of overall protein stability owing to the lack of central secondary elements may be mitigated by the introduction of a disulfide bridge between the two strands (Cys69 and Cys85 in NcCel3A). The missing loops allow a wider binding cleft in HjCel3A, whereas in NcCel3A this space is occupied, and extended, primarily by loops I and II (Fig. 1 ▸ a). The structure of NcCel3A, as well as those of ReCel3A and AaBGL1, has a protruding loop (loop I in Fig. 1 ▸) which is not present in the three-domain GH3 structures HjCel3A (Fig. 1 ▸ b), KmBGLI and TnBgl3B. This loop extends one side of the active-site cleft, whereas loop II in linker 1 (Fig. 2 ▸) extends the opposite side of the cleft. The α-helical part of loop I introduces three residues towards the active-site cleft: Glu200, Asp203 and Tyr204. In the ReCel3A structure there is a two-amino-acid deletion compared with NcCel3A and AaBgl1. The lack of these two amino-acid residues results in a loss of α-helical structure that leaves a slightly wider active-site cleft.
Figure 1.
(a) Cartoon representation of the NcCel3A structure displayed in ribbon representation. The three domains of the protein are coloured green (domain 1), grey (domain 2) and red (domain 3). Loops and linkers are highlighted in colours according to the legend in the top-right corner. N-Glycosylations are shown as grey sticks. (b, c) The quaternary structures of NcCel3A (b) and ReCel3A (Gudmundsson et al., 2016 ▸) (c) showing the dimer formation found in these two structures.
Figure 2.
Cartoon representation of domain 1 of (a) N. crassa Cel3A (NcCel3A), (b) H. jecorina Cel3A (HjCel3A; PDB entry 3zyz; Karkehabadi et al., 2014 ▸) and (c) H. vulgare ExoI (PDB entry 1iex; Hrmova et al., 2001 ▸) shown in ribbon representation. Loops a and b in NcCel3A and HjCel3A highlight the deleted helices which are present and marked A and B in HvExoI. The position of loop I is highlighted by a circle.
3.4. Linker 1 and loop II
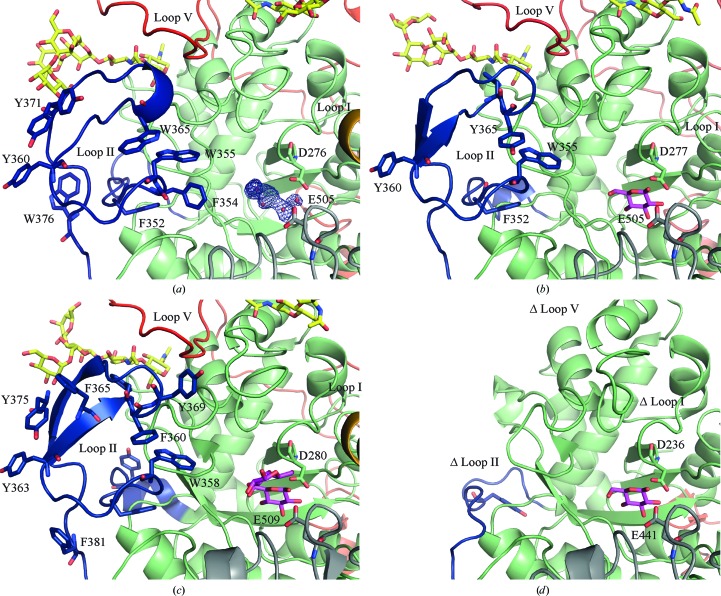
In NcCel3A, domain 1 is connected to domain 2 by a 42-residue linker (residues 341–383). In HjCel3A and HvExoI this linker is only 18 and 16 residues in length, respectively. The insertion of 25 residues (residues 351–376) observed in NcCel3A, as well as in ReCel3A and AaBgl1, and denoted as loop II in Fig. 3 ▸, has previously been described as a hydrophobic linker that activates T. aurantiacus BGLII in organic solvents (Hong et al., 2006 ▸) and is present with a similar fold in the ReCel3A and AaBgl1 structures (Yoshida et al., 2010 ▸; Suzuki et al., 2013 ▸; Karkehabadi et al., 2014 ▸; Gudmundsson et al., 2016 ▸). Loop II is not present in the other fungal GH3 structures HjCel3A (Fig. 3 ▸ d) and KmBGLI, but in the NcCel3A structure it extends the opposite side of the active-site cleft compared with loop I. In NcCel3A, loop II has two tryptophan residues and one phenylalanine (Trp355, Trp365 and Phe354), which constitute one side of the putative substrate-binding subsites +1 and +2 (discussed in more detail in Sections 3.7 and 3.8). Loop II also contains two tyrosine residues and a tryptophan (Tyr360, Tyr371 and Trp376), which are positioned outside the active-site cleft. Interestingly, this part of loop II resembles the flat binding surface of a carbohydrate-binding molecule type 1 (CBM1), which also consists of two tyrosines and a tryptophan (Mattinen et al., 1998 ▸). There seems to be high variability in loop II among GH3 β-glucosidases. The two aromatic residues Phe352 and Trp355 are the most conserved residues in this loop, and these two residues are also present in the R. emersonii and A. aculeatus structures (Figs. 3 ▸ a, 3 ▸ b and 3 ▸ c). All of the GH3 structures that include loop II have certain common features. For instance, Tyr360 and an aromatic residue at the position of Trp366 are present in these structures. A second common feature is the presence of an N-glycan chain emanating from Asn57 in NcCel3A, which is wedged in between loop II and the C-terminal loop V. This apparent feature of using N-glycans as a structural element could be unique to this class of enzymes.
Figure 3.
Cartoon ribbon representation of loop II (blue). (a) shows NcCel3A, (b) ReCel3A (PDB entry 5ju6; Gudmundsson et al., 2016 ▸), (c) AaBgl1 (PDB entry 4iih; Suzuki et al., 2013 ▸) and (d) HjCel3A (PDB entry 3zyz; Karkehabadi et al., 2014 ▸). Domain 1 is coloured green, domain 2 grey, loop V red and loop I orange; active-site ligands are shown as magenta sticks, active-site residues in stick representation, electron density as a blue mesh and N-glycosylations as yellow sticks.
3.5. Loops in domain 2 and the second linker
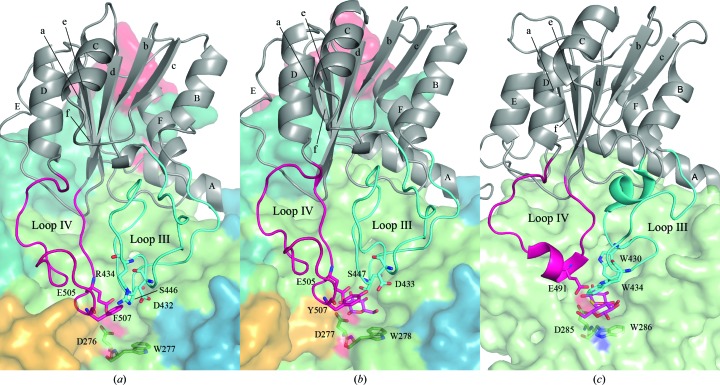
The second domain of the NcCel3A structure (residues 383–584) has an (α/β)6 sandwich fold, which is structurally well preserved among all GH3 enzymes with known structure that contain at least two domains, except in the bacterial TnBgl3B, in which one edge β-strand (strand c in Fig. 4 ▸) is substituted by an additional α-helix and a flexible loop (Pozzo et al., 2010 ▸). Domain 2 of NcCel3A has two loops, III and IV, that encompasses residues 421–455 and 501–524, respectively. These two loops constitute one side of the active-site cleft (Fig. 4 ▸), in between loops I and II. Loop III of NcCel3A is a 34-residue loop that extends between strand b and helix B. The loop folds back over itself and is stabilized by a disulfide bridge formed by Cys430 and Cys435. Ser446 and Asp432 are two conserved residues in loop III that are directed towards the active site. Ser446 is especially important as it is positioned pointing directly towards the hexose ring of a substrate bound in the −1 subsite. In HvExoI this position is occupied by Trp343 (Fig. 4 ▸ c), which is a very common motif in carbohydrate-binding domains. Trp343 constitutes half of the ‘coin-slot’ binding pocket of HvExoI (Varghese et al., 1999 ▸), which is not present in any three-domain GH3 structures. Loop IV is a 23-residue loop in which the catalytic acid/base Glu505 is located. NcCel3A has a phenylalanine at position 507, whereas the other related GH3 structures have a tyrosine located at this position.
Figure 4.
Overview of domain 2 (grey) in ribbon representation. (a) shows NcCel3A, (b) ReCel3A (PDB entry 5ju6; Gudmundsson et al., 2016 ▸) and (c) HvExoI (PDB entry 1iex; Hrmova et al., 2001 ▸). Loops III and IV are highlighted in cyan and magenta, respectively. Other domains are represented by surfaces coloured according to the scheme presented in Fig. 1 ▸.
Domain 2 of the NcCel3A structure is followed by the second linker region (residues 585–649). This linker is extended compared with that described as a C-terminal extension in the HvExoI structure. This extension covers and stabilizes loops I and IV. These loops appear to be a large part of the interface between domains 2 and 3, but are not present in the barley enzyme HvExoI.
3.6. Domain 3 and loop V
Domain 3 is an FnIII-like or immunoglobulin s-type domain (residues 650–857) and was first observed in GH3 in the TnBgl3B structure (Pozzo et al., 2010 ▸). The FnIII domain is a β-sandwich composed of two layers of β-sheets of three and four β-strands, respectively (Fig. 5 ▸). The extended loop V, which was first observed in the structures of AaBGLI and ReCel3A, encompasses domain 1 and interacts with loop II on the opposite side of the molecule from domain 3. Loop V folds over three large N-glycan chains (bound to Asn61, Asn311 and Asn318). Several conserved aromatic residues π-stack with GlcNAc residues in loop V (Tyr706, Tyr708, Tyr723 and Phe730). Lima and coworkers proposed that the homologous loop V from Aspergillus niger Bgl1 (AnBgl1) is flexible and allows the FNIII domain to extend and bind to lignin, thus explaining the tadpole-like structure that was observed in SAXS experiments carried out with AnBgl1 (Lima et al., 2013 ▸). We argue, however, that the domain reorganization speculated upon by Lima and coworkers is unlikely. Firstly, many of the conserved residues form seemingly crucial stacking interactions with N-glycans, a fact that is not accounted for in their model. Secondly, the presence of flexibility within a protein crystal usually results in poor or even no electron density. This is observed for instance in the PsExoP structure, the only published GH3 structure in which electron density is completely missing for one highly flexible domain even though it was expressed as part of the protein. We thus believe it is unlikely that loop V has this degree of flexibility in NcCel3A and other GH3 proteins that contain this loop.
Figure 5.
Domain 3 and loop V displayed in ribbon representation (red and magenta, respectively) for (a) NcCel3A, (b) ReCel3A (PDB entry 5ju6; Gudmundsson et al., 2016 ▸) and (c) HjCel3A (PDB entry 3zyz; Karkehabadi et al., 2014 ▸). The N-glycans attached to the three structures are displayed as yellow sticks and other domains are represented as surfaces.
3.7. Subsite −1 and catalytic residues
The location of the catalytic centre subsite −1 of NcCel3A is positioned at the carboxyl side of domain 1. The two catalytic residues of NcCel3A were identified based on homology to other GH3 structures. The nucleophile is Asp276 and the acid/base is Glu505 (Fig. 3 ▸ a). No distinct density corresponding to a bound glucose was observed in subsite −1 of NcCel3A, in contrast to several other GH family 3 structures. Extra electron density is present in the −1 subsite that is insufficient for interpretation. This density may be owing to a partially bound buffer molecule, a polyethylene glycol (PEG) molecule and/or partial density of a glucose molecule.
3.8. Putative +1 and +2 subsites
The putative +1 subsite of the NcCel3A structure is very similar to those of ReCel3A and AaBGL1, but differs from the suggested +1 subsite in HvExoI, which is lined by two tryptophan residues (Fig. 4 ▸ c). Hrmova and coworkers proposed this to be the basis of the broad substrate specificity of HvExoI (Hrmova et al., 2002 ▸). Trp277 in NcCel3A corresponds to one of these tryptophan residues, but the side chain has shifted to become an essential part of the −1 subsite rather than the +1 subsite as in HvExoI. This shift causes a rearrangement of the core residues and contributes to the collapse of the TIM-barrel fold described above. In the collapsed TIM-barrel fold, a feature that seems to be shared by many fungal and bacterial β-glucosidases, the second barrel β-strand is shorter and antiparallel, which makes the −1 subsite wider compared with the active site in GH3 enzymes with a complete TIM-barrel fold. Similar to the structures of ReCel3A and AaBGL1, one side of the +1 subsite is formed by Phe301, which stacks with the side chain of Trp64, which is only slightly further away from the active site. Phe507 is situated on the opposite side of the +1 subsite and the active-site entrance. This phenylalanine seems to correspond to the ‘coin-slot’ Trp434 side chain in HvExo1. The corresponding residues in ReCel3A and AaBGL1 have almost the same side-chain conformations but are tyrosines (Tyr507 and Tyr511, respectively) in both these enzymes. In both the ReCel3A and the AaBGL1 structures the hydroxyl group of the tyrosine makes hydrogen-bond interactions with Asp433 and Asp437 in a potential +2 subsite (Fig. 4 ▸). In the NcCel3A structure the corresponding residue (Asp432) interacts with the side chain of Arg434, which should stabilize the aspartate residue and compensate for the slight increase of hydrophobicity in the +1 binding site. This arginine is not present in the ReCel3A and AaBgl1 enzymes. It thus seems as if not only the presence of the aspartate residue but also its flexibility/stability may be important for substrate and/or product interaction in this class of β-glucosidases. Arg196 and Gln197 are two conserved residues in ReCel3A and AaBGL1 that make potentially important interactions with the substrate, forming part of the +1 subsite.
Previously, we have shown that ReCel3A prefers cellobiose to cellotriose, while HjCel3A prefers the hydrolysis of slightly longer cello-oligosaccharides such as cellotriose and cellotetraose (Karkehabadi et al., 2014 ▸; Gudmundsson et al., 2016 ▸). In analogy with the ReCel3A structure, the plane of the Trp64 side chain in the NcCel3A structure has rotated almost 90° in the structure when compared with the corresponding tryptophan residue in the HjCel3A structure, and stacks with the side chain of Phe301. This puts the phenylalanine residue in subsite +1 rather than in a tentative +2 subsite, as in the structure of HjCel3A. Thus, similar to ReCel3A the existence of a +2 subsite is less pronounced in NcCel3A than in HjCel3A and the enzyme may also have a substrate specificity similar to that of ReCel3A.
4. Conclusions
We have determined the structure of a glycoside hydrolase family 3 β-glucosidase, Cel3A from N. crassa, at 2.2 Å resolution and show that this β-glucosidase is structurally similar to two other fungal β-glucosidases: A. aculeatus BglI and R. emersonii Cel3A. These structures share several features that may be unique to this class of GH3 β-glucosidases. Most pronounced among these features are the likely dimeric form of the active enzyme and the large and seemingly conserved glycosylations. The structural analysis further showed that NcCel3A should have a similar substrate specificity to the previously structurally and biochemically characterized ReCel3A.
Supplementary Material
PDB reference: Neurospora crassa Cel3A, 5nbs
References
- Agirre, J., Ariza, A., Offen, W. A., Turkenburg, J. P., Roberts, S. M., McNicholas, S., Harris, P. V., McBrayer, B., Dohnalek, J., Cowtan, K. D., Davies, G. J. & Wilson, K. S. (2016). Acta Cryst. D72, 254–265. [DOI] [PMC free article] [PubMed]
- Agirre, J., Davies, G., Wilson, K. & Cowtan, K. (2015). Nature Chem. Biol. 11, 303. [DOI] [PubMed]
- Biely, P., Krátký, Z. & Vršanská, M. (1981). Eur. J. Biochem. 119, 559–564. [DOI] [PubMed]
- Bohlin, C., Olsen, S. N., Morant, M. D., Patkar, S., Borch, K. & Westh, P. (2010). Biotechnol. Bioeng. 107, 943–952. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Colabardini, A. C., Valkonen, M., Huuskonen, A., Siika-Aho, M., Koivula, A., Goldman, G. H. & Saloheimo, M. (2016). Mol. Biotechnol. 58, 821–831. [DOI] [PubMed]
- Cournoyer, B. & Faure, D. (2003). J. Mol. Microbiol. Biotechnol. 5, 190–198. [DOI] [PubMed]
- Davies, G. J., Wilson, K. S. & Henrissat, B. (1997). Biochem. J. 321, 557–559. [DOI] [PMC free article] [PubMed]
- Davis, R. H. & Perkins, D. D. (2002). Nature Rev. Genet. 3, 397–403. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL. http://www.pymol.org.
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol. 4, 269–275. [DOI] [PubMed]
- Eberhart, B. M., Beck, R. S. & Goolsby, K. M. (1977). J. Bacteriol. 130, 181–186. [DOI] [PMC free article] [PubMed]
- Eberhart, B., Cross, D. F. & Chase, L. R. (1964). J. Bacteriol. 87, 761–770. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, P. (2012). Science, 336, 986–987. [DOI] [PubMed]
- Foreman, P., Goedegebuur, F., Van Solingen, P. & Ward, M. (2005). Patent WO/2005/001036.
- Gebler, J., Gilkes, N. R., Claeyssens, M., Wilson, D. B., Béguin, P., Wakarchuk, W. W., Kilburn, D. G., Miller, R. C. Jr, Warren, R. A. & Withers, S. G. (1992). J. Biol. Chem. 267, 12559–12561. [PubMed]
- Gudmundsson, M., Hansson, H., Karkehabadi, S., Larsson, A., Stals, I., Kim, S., Sunux, S., Fujdala, M., Larenas, E., Kaper, T. & Sandgren, M. (2016). Acta Cryst. D72, 860–870. [DOI] [PMC free article] [PubMed]
- Harris, P. V., Welner, D., McFarland, K. C., Re, E., Navarro Poulsen, J. C., Brown, K., Salbo, R., Ding, H., Vlasenko, E., Merino, S., Xu, F., Cherry, J., Larsen, S. & Lo Leggio, L. (2010). Biochemistry, 49, 3305–3316. [DOI] [PubMed]
- Henrissat, B. & Davies, G. (1997). Curr. Opin. Struct. Biol. 7, 637–644. [DOI] [PubMed]
- Hong, J., Tamaki, H. & Kumagai, H. (2006). Appl. Microbiol. Biotechnol. 73, 80–88. [DOI] [PubMed]
- Hrmova, M., De Gori, R., Smith, B. J., Fairweather, J. K., Driguez, H., Varghese, J. N. & Fincher, G. B. (2002). Plant Cell, 14, 1033–1052. [DOI] [PMC free article] [PubMed]
- Hrmova, M., Varghese, J. N., De Gori, R., Smith, B. J., Driguez, H. & Fincher, G. B. (2001). Structure, 9, 1005–1016. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Karkehabadi, S., Helmich, K. E., Kaper, T., Hansson, H., Mikkelsen, N. E., Gudmundsson, M., Piens, K., Fujdala, M., Banerjee, G., Scott-Craig, J. S., Walton, J. D., Phillips, G. N. Jr & Sandgren, M. (2014). J. Biol. Chem. 289, 31624–31637. [DOI] [PMC free article] [PubMed]
- Karplus, P. A. & Diederichs, K. (2012). Science, 336, 1030–1033. [DOI] [PMC free article] [PubMed]
- Kleywegt, G. J. & Jones, T. A. (1996). Structure, 4, 1395–1400. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lima, M. A., Oliveira-Neto, M., Kadowaki, M. A., Rosseto, F. R., Prates, E. T., Squina, F. M., Leme, A. F., Skaf, M. S. & Polikarpov, I. (2013). J. Biol. Chem. 288, 32991–33005. [DOI] [PMC free article] [PubMed]
- Lombard, V., Golaconda Ramulu, H., Drula, E., Coutinho, P. M. & Henrissat, B. (2014). Nucleic Acids Res. 42, D490–D495. [DOI] [PMC free article] [PubMed]
- Maddi, A., Bowman, S. M. & Free, S. J. (2009). Fungal Genet. Biol. 46, 768–781. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Mattinen, M. L., Linder, M., Drakenberg, T. & Annila, A. (1998). Eur. J. Biochem. 256, 279–286. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Meier, K. K., Jones, S. M., Kaper, T., Hansson, H., Koetsier, M. J., Karkehabadi, S., Solomon, E. I., Sandgren, M. & Kelemen, B. (2018). Chem. Rev. 118, 2593–2635. [DOI] [PMC free article] [PubMed]
- Murray, P., Aro, N., Collins, C., Grassick, A., Penttilä, M., Saloheimo, M. & Tuohy, M. (2004). Protein Expr. Purif. 38, 248–257. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nakatani, Y., Cutfield, S. M., Cowieson, N. P. & Cutfield, J. F. (2012). FEBS J. 279, 464–478. [DOI] [PubMed]
- Pannu, N. S. & Read, R. J. (1996). Acta Cryst. A52, 659–668.
- Payne, C. M., Knott, B. C., Mayes, H. B., Hansson, H., Himmel, M. E., Sandgren, M., Ståhlberg, J. & Beckham, G. T. (2015). Chem. Rev. 115, 1308–1448. [DOI] [PubMed]
- Payne, C. M., Resch, M. G., Chen, L., Crowley, M. F., Himmel, M. E., Taylor, L. E. II, Sandgren, M., Ståhlberg, J., Stals, I., Tan, Z. & Beckham, G. T. (2013). Proc. Natl Acad. Sci. USA, 110, 14646–14651. [DOI] [PMC free article] [PubMed]
- Perrakis, A., Sixma, T. K., Wilson, K. S. & Lamzin, V. S. (1997). Acta Cryst. D53, 448–455. [DOI] [PubMed]
- Petersen, T. N., Brunak, S., von Heijne, G. & Nielsen, H. (2011). Nature Methods, 8, 785–786. [DOI] [PubMed]
- Potterton, L., Agirre, J., Ballard, C., Cowtan, K., Dodson, E., Evans, P. R., Jenkins, H. T., Keegan, R., Krissinel, E., Stevenson, K., Lebedev, A., McNicholas, S. J., Nicholls, R. A., Noble, M., Pannu, N. S., Roth, C., Sheldrick, G., Skubak, P., Turkenburg, J., Uski, V., von Delft, F., Waterman, D., Wilson, K., Winn, M. & Wojdyr, M. (2018). Acta Cryst. D74, 68–84. [DOI] [PMC free article] [PubMed]
- Pozzo, T., Pasten, J. L., Karlsson, E. N. & Logan, D. T. (2010). J. Mol. Biol. 397, 724–739. [DOI] [PubMed]
- Qin, Z., Xiao, Y., Yang, X., Mesters, J. R., Yang, S. & Jiang, Z. (2015). Sci. Rep. 5, 18292. [DOI] [PMC free article] [PubMed]
- Romero, M. D., Aguado, J., González, L. & Ladero, M. (1999). Enzyme Microb. Technol. 25, 244–250.
- Suzuki, K., Sumitani, J.-I., Nam, Y.-W., Nishimaki, T., Tani, S., Wakagi, T., Kawaguchi, T. & Fushinobu, S. (2013). Biochem. J. 452, 211–221. [DOI] [PubMed]
- Tian, C., Beeson, W. T., Iavarone, A. T., Sun, J., Marletta, M. A., Cate, J. H. & Glass, N. L. (2009). Proc. Natl Acad. Sci. USA, 106, 22157–22162. [DOI] [PMC free article] [PubMed]
- Varghese, J. N., Hrmova, M. & Fincher, G. B. (1999). Structure, 7, 179–190. [DOI] [PubMed]
- Vogel, H. J. (1956). Microb. Genet. Bull. 13, 42–43.
- Weiss, M. S. (2001). J. Appl. Cryst. 34, 130–135.
- Winn, M. D., Ballard, C. C., Cowtan, K. D., Dodson, E. J., Emsley, P., Evans, P. R., Keegan, R. M., Krissinel, E. B., Leslie, A. G. W., McCoy, A., McNicholas, S. J., Murshudov, G. N., Pannu, N. S., Potterton, E. A., Powell, H. R., Read, R. J., Vagin, A. & Wilson, K. S. (2011). Acta Cryst. D67, 235–242. [DOI] [PMC free article] [PubMed]
- Wu, W., Kasuga, T., Xiong, X., Ma, D. & Fan, Z. (2013). Arch. Microbiol. 195, 823–829. [DOI] [PubMed]
- Yazdi, M. T., Woodward, J. R. & Radford, A. (1990). Enzyme Microb. Technol. 12, 116–119. [DOI] [PubMed]
- Yoshida, E., Hidaka, M., Fushinobu, S., Koyanagi, T., Minami, H., Tamaki, H., Kitaoka, M., Katayama, T. & Kumagai, H. (2010). Biochem. J. 431, 39–49. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Neurospora crassa Cel3A, 5nbs