Summary
Evaluating progression risk and determining optimal therapy for ulcerative colitis (UC) is challenging as many patients exhibit incomplete responses to treatment. As part of the PROTECT (Predicting Response to Standardized Colitis Therapy) Study, we evaluated the role of the gut microbiome in disease course for 405 pediatric, new-onset, treatment-naive UC patients. Patients were monitored for one year upon treatment initiation, and microbial taxonomic composition was analyzed from fecal samples and rectal biopsies. Depletion of core gut microbes and expansion of bacteria typical of the oral cavity were associated with baseline disease severity. Remission and refractory disease were linked to species-specific temporal changes that may be implicative of therapy efficacy, and a pronounced increase in microbiome variability was observed prior to colectomy. Finally, microbial associations with disease-associated serological markers suggest host-microbial interactions in UC. These insights will help improve existing treatments and develop therapeutic approaches guiding optimal medical care.
Graphical Abstract:

eTOC blurb:
Many patients exhibit incomplete responses to ulcerative colitis (UC) therapy. Schirmer et al. investigate the gut microbiome's role in pediatric UC treated with two conventional therapies. Baseline and longitudinal microbial trends are implicated in disease severity and progression, including remission and colectomy requirement. These insights may motivate new therapeutic approaches.
Introduction
Recent insights into the molecular pathogenesis of inflammatory bowel disease (IBD) suggest a complex interplay of susceptibility genes, aberrant mucosal immune responses and environmental factors and implicate gut bacteria and their products (Xavier and Podolsky, 2007). Aberrant immune responses to commensal microbes likely result in lesions of the intestinal mucosal layer involving extensive epithelial damage, immune infiltration, crypt abscesses, and chronic inflammation – hallmarks of ulcerative colitis (UC). Pediatric UC incidence rates are rising, and over 50% of patients present with extensive colitis (Peloquin et al., 2016). Converging evidence for genetic- and cell type-specific defects suggest that the UC spectrum encompasses multiple disease and immune phenotypes.
Microbial organisms and products impact immune homeostasis by affecting immune education, development and response (Atarashi et al., 2013; Kugathasan et al., 2017; Sartor and Wu, 2017; Schirmer et al., 2016; Wlodarska et al., 2015). The IBD gut is characterized by reduced microbiome diversity and a depletion of protective bacteria such as short-chain fatty acid (SCFA)-producing Ruminococcaceae and Lachnospiracaea (Duvallet et al., 2017; Gevers et al., 2014) that coincides with an expansion of pro-inflammatory microbes such as Enterobacteriaceae, including Escherichia coli, and Fusobacteriaceae (Gevers et al., 2014; Morgan et al., 2012; Shaw et al., 2016). Blooms of Ruminococcus gnavus strains and an increase in facultative anaerobes co-occur with more severe IBD activity (Hall et al., 2017). Faecalibacterium prausnitzii, a major producer of the SCFA butyrate, displays anti-inflammatory effects and is reduced in UC patients. In genetically susceptible gnotobiotic mice, oral pathobionts such as Klebsiella strains induce T helper 1 cells and elicit severe gut inflammation (Atarashi et al., 2017). Microbes also show protective effects in mouse models: a mix of 17 Clostridium strains inhibits colitis through induction of regulatory T cells (Atarashi et al., 2013), and Bacteroides fragilis mono-colonization protects against induced colitis (Chiu et al., 2014). While these studies implicate specific bacteria and bacterial products in intestinal health, the IBD microbiome fluctuates more compared to healthy controls, highlighting the need for longitudinal sampling of new-onset patients. Microbial organisms also impact drug availability and treatment efficacy (Ananthakrishnan et al., 2017; Haiser et al., 2013; Sivan et al., 2015; Vetizou et al., 2015). Further, UC is pathophysiologically distinct from other types of IBD and may arise from different mechanisms. To date, few studies have focused specifically on the UC microbiome.
Current UC therapies inhibit immune system overactivation (Wahl et al., 1998), but little is known about their ability to restore gut microbial balance or the impact of the microbiome on treatment efficacy. We investigated the microbiome’s role in disease progression during the first year of UC therapy as part of the PROTECT (Predicting Response to Standardized Colitis Therapy) Study (Hyams et al., 2017). We monitored 405 pediatric, new-onset, treatment-naive UC patients (mean age 12.8±3.3 years) upon receiving either 5-ASA mesalamine or corticosteroids (CS) followed by mesalamine. Patients refractory to these conventional approaches received immunomodulators or anti-TNF biologic therapy as a part of a pre-specified treatment algorithm (Fig. S1A). We observed taxonomic shifts prior to treatment initiation that were associated with disease severity and progression, including remission and colectomy. Analysis of follow-up samples further identified altered microbial abundances associated with treatment, temporal changes linked to disease severity and increased variability of microbial composition in colectomy patients.
Results
Gut microbiota changes are associated with disease progression
Fecal samples were collected at baseline (week 0, prior to treatment) and 3 follow-up time points (4, 12 and 52 weeks after treatment initiation), and paired rectal biopsies were collected at baseline and week 52, resulting in a total of 1,212 samples. Extensive clinical metadata, including treatment, disease activity and serology, were collected for each patient. We analyzed microbial taxonomic composition based on 16S rRNA gene amplicon sequencing to evaluate the role of the gut microbiome in disease progression and treatment response.
Treatment initiation improved disease activity and markers of inflammation, such as fecal calprotectin levels, for many patients (Fig. 1). Remission was not a consistent outcome based on initial disease severity or treatment strategy (Fig. 1A, Fig. S1B). A clear shift in taxonomic composition accompanied changes in disease activity and fecal calprotectin levels following therapy (Fig. 1B+C). Fecal calprotectin levels increased (Wilcoxon, mild-inactive p=2*10−16, moderate-mild p=3*10−5, severe-moderate p=3*10−3) with more severe disease, and microbial community diversity decreased (Wilcoxon, all p<3*10−3) with moderate or severe compared to mild or inactive disease (Fig. 1D, Fig. S1C+D; fecal calprotectin (mcg/g): meaninactive=833, meanmild=2003, meanmoderate=2590, meansevere=3298). Sample type accounted for some observed variation in community composition and was considered as a covariate for all subsequent analyses (Fig. S1E).
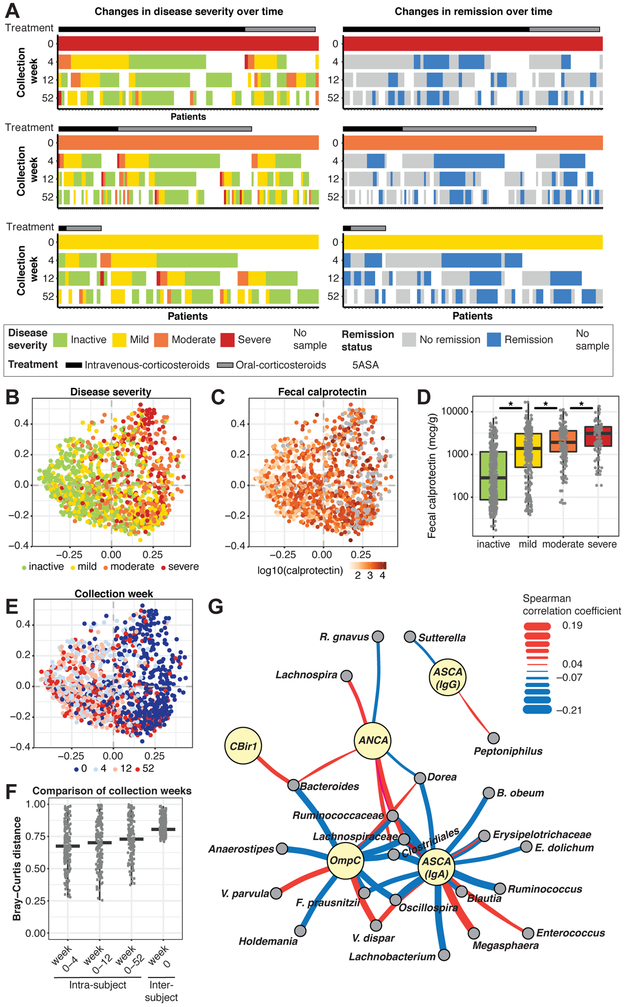
Figure 1: Microbial and serological changes in relation to disease severity and treatment efficacy.
(A) Patients with a baseline (week 0) and at least one follow-up sample were included (week 0: nsevere=90, nmoderate=139, nmild=75). Ordination analysis (PCoA on Bray-Curtis distance) over all samples revealed marked stratification by disease severity (B) and fecal calprotectin level (C), which were significantly associated (D; sequential Wilcoxon tests, all p<0.01). Stratification closely followed the longitudinal sequence of samples within patients, consistent with improvement over time (E). (F) Microbial profiles significantly changed (Bray-Curtis) over time within individuals (Wilcoxon; all p<0.05 except for changes from week 0-4 to week 0-12). (G) Network of significant associations between microbial clades in baseline samples (gray nodes) and serological markers (yellow nodes) determined by linear model coefficients (MaAsLin). Edges summarize the association strength by Spearman correlation (red, positive; blue, negative; heavier edge weight implies greater strength). See also Fig. S1 and Tables S1+S6.
Taxonomic composition within patients changed substantially over time, highlighting the importance of examining treatment-naive patients to identify relevant early microbial shifts (Fig. 1E+F). After 4 weeks, the mean Bray-Curtis distance between samples from the same patients was 0.68 (by week 12: 0.69). This further increased to 0.73 by week 52 and was comparable to unrelated samples (mean across all time points: 0.81).
Serological markers are implicated in disease progression and associated with microbial factors
Serological measurements are commonly used to support IBD diagnosis. Despite high specificity, serological biomarkers lack high sensitivity (Peyrin-Biroulet et al., 2007; Ruemmele et al., 1998). We measured anti-Saccharomyces cerevisiae antibody (ASCA) immunoglobulin A (IgA), ASCA immunoglobulin G (IgG), antineutrophil cytoplasmic antibodies (ANCA), anti-flagellin antibodies (CBir1) and anti-outer membrane porin C (OmpC) at baseline. ASCA IgA, OmpC and ANCA levels were decreased in patients with mild disease (Fig. S1F; Wilcoxon; ASCA IgA: all p<2*10−4, OmpC: all p<0.03, ANCA p<10−2), whereas CBir1 was increased (p<0.04). ASCA IgA levels were lower in patients who achieved remission without a colectomy, whereas OmpC levels increased in colectomy patients. In contrast, baseline ANCA antibodies were increased in patients that achieved CS-free remission by week 52 (Fig. S1G-J).
We next tested for microbial associations with these biomarkers (FDR<0.2), and those associated with disease progression yielded the most associations: ASCA IgA (28 associations), OmpC (12) and ANCA (8) (Fig. 1G, Table S1). Most ASCA IgA associations involved Lachnospiraceae (12 associations) and Ruminococcaceae (5 associations) species, including negative correlations with F. prausnitzii and positive correlations with 2 Veillonellaceae, Megasphaera and Veillonella dispar. The most significant negative correlation with OmpC (lowest FDR, see Methods) involved F. prausnitzii and positive correlations included 2 Veillonella species (V. dispar and V. parvula). The most significant ANCA correlation was a negative association with R. gnavus. Overall, 36 negative and 16 positive microbial associations with disease-specific changes in antibody levels were detected. All but 2 of these operational taxonomic units [OTUs] (Lachnospiraceae and V. dispar) were uniquely associated with a specific antibody, suggesting potential interactions between the gut microbiome and the immune system at the antibody level in disease progression.
Stool microbial composition overlaps with rectal biopsy profile
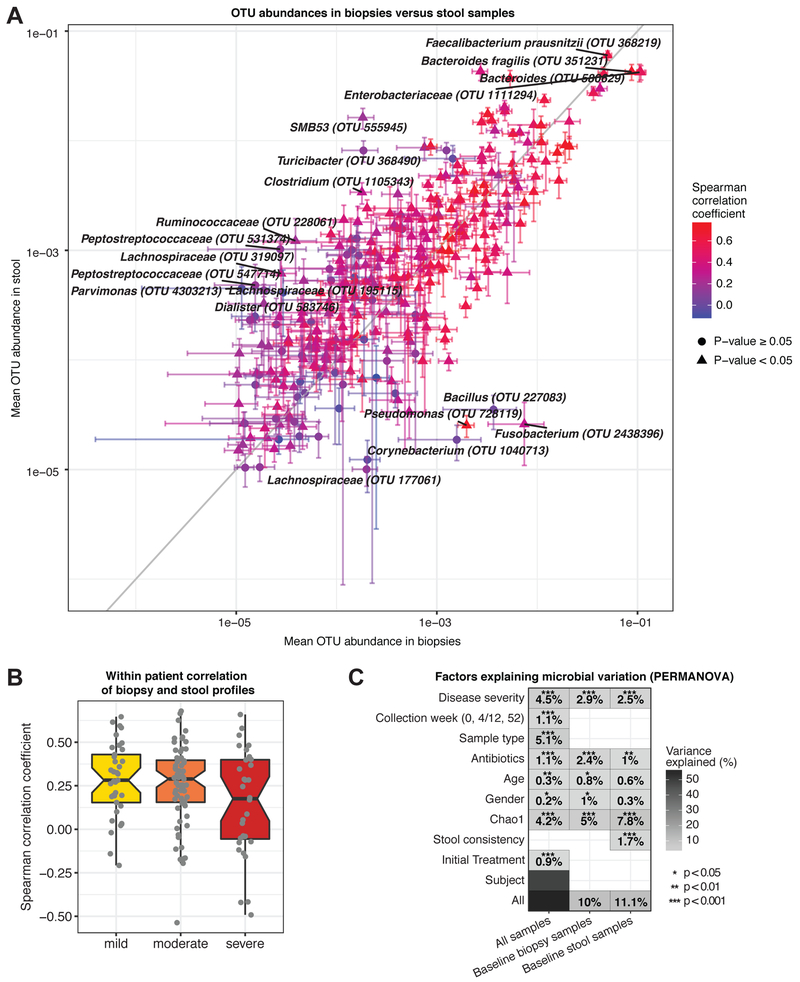
We compared paired fecal and rectal biopsy profiles within each patient at baseline to determine the extent of community structure overlap (Fig. 2). Although profiles could substantially vary for individual patients (Fig. S2A+B), the abundance of each OTU on average was comparable in fecal samples and biopsies across patients, with more abundant OTUs showing higher correlations (Fig. 2A). Fusobacterium, a known mucin degrader (Tailford et al., 2015), showed the largest difference at twice the abundance in biopsies.
Figure 2: Factors associated with variation in microbial community composition.
(A) Comparison of paired microbial profiles in biopsies and stool samples from treatment-naive patients (n=132, week 0). OTUs were required to occur in ≥20 biopsy or stool samples with mean abundance >10−5. Error bars specify SEM, color indicates Spearman correlation coefficient for each OTU across all samples. The 15 OTUs with the largest difference between sample types (outliers from the x=y line) and the 5 most abundant OTUs (top right) are labeled. (B) Intra-patient correlation of biopsy versus stool samples at week 0, stratified by disease severity (min. abundance: 10−4). Correlation coefficients decreased with increasing disease severity (median Spearman rmild=0.28, rmoderate=0.28, rsevere=0.17), however, this trend was not statistically significant. (C) Microbial taxonomic variation explained by various factors (y-axis) for all or only baseline samples (permANOVA, nperm=1,999). Percentages indicate variance explained by each variable in single-variable models. Total variation explained by all variables is indicated in the final row. See also Fig. S2.
IBD microbiomes display a higher degree of intra-individual variability than healthy microbiomes (Halfvarson et al., 2017). Our data suggests that dissimilarity increases with disease severity (Fig. 2B), however, organisms present in both sample types showed high correlation (Fig. S2C). Stool samples generally showed higher diversity than biopsies, even though sequencing depth was comparable (Fig. S2D+E). No effect of alpha diversity on the stool-biopsy correlation was noticed (Fig. S2F). Overall, sample type accounted for 5.1% of the observed variation (Fig. 2C, Fig. S2G). As expected, inter-individual variation accounted for most taxonomic variation. Collection week and patient age and gender were also significant contributors explaining ~1% of microbiota differences. Disease severity had a more substantial effect, accounting for up to 4.5% of variation, and differences in community diversity (Chao1) explained as much as 7.8% in baseline stool samples. The effect of initial treatment (0.9%) was comparable to the effect of antibiotic usage (1.1%), further highlighting the importance of examining treatment-naive samples.
Extensive microbial depletion and expansion of bacteria typical of the oral cavity are linked to severe disease in treatment-naive patients
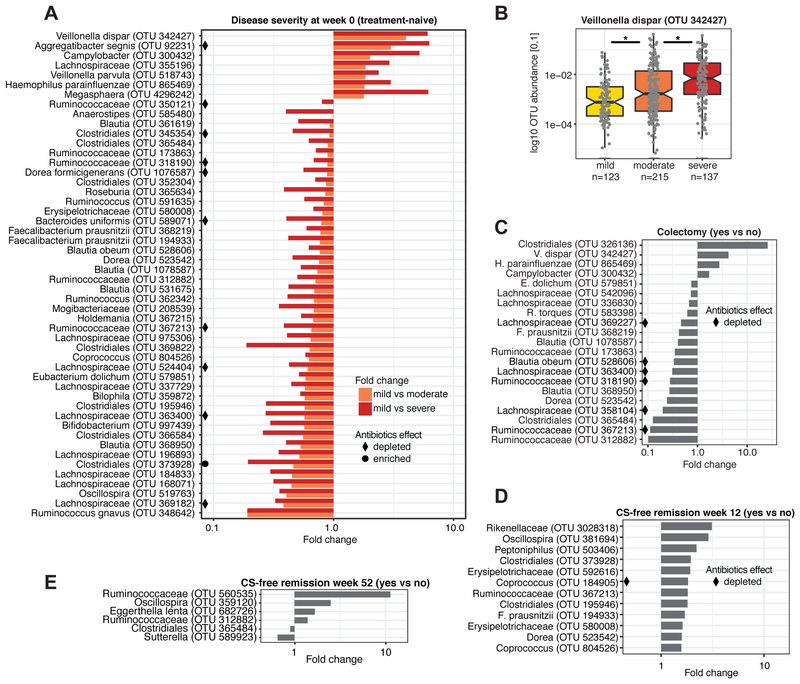
In addition to community-wide shifts, specific OTUs in baseline samples were linked to changes in disease severity and progression during the first year after diagnosis (Fig. 3). In baseline samples, 50 OTUs were associated with initial disease severity and showed a continuous increase or decrease with worsening disease (FDR<0.05). The majority of these OTUs (43/50) were depleted in patients with more severe disease and represented Ruminococcaceae and Lachnospiraceae (Fig. 3A). Lachnospiraceae were previously implicated in IBD, including Clostridium clusters IV and XIVa, which are involved in maintenance of gut health and can induce T regulatory cells (Atarashi et al., 2013). Considering only rectal biopsies at baseline, 9 significant associations were identified, where Streptococcus anginosus was uniquely differentially abundant in biopsies and substantially increased in patients with severe compared to mild disease (14-fold).
Figure 3: Microbial associations with disease severity, colectomy, and remission.
(A) Significant (FDR<0.05) associations between microbial abundance and disease severity in treatment-naive (week 0) stool samples and biopsies (nmild=123, nmoderate=215, nsevere=137). Any OTU significantly altered in moderate or severe disease compared to mild was included (see also Table S5). Associations are summarized by fold change in mean OTU abundance for mild vs. moderate (orange) and mild vs. severe (red) disease. Black diamonds/circles indicate OTUs associated with antibiotics. (B) V. dispar was the most depleted species in mild disease (Wilcoxon, mild-moderate: p=0.003, moderate-severe: p=0.0005). (C) Significant (FDR<0.2) associations between microbial abundance in baseline samples and refractory disease requiring colectomy. Associations are summarized by fold change in mean OTU abundances in the colectomy group (n=19) vs. patients not requiring colectomy (n=304). Significant (FDR<0.2) associations between microbial abundance and CS-free remission at week (D) 12 and (E) 52. See also Fig. S3 and Tables S2 and S5.
Strikingly, all OTUs increased in severe disease (V. dispar, Aggregatibacter segnis, Campylobacter, Lachnospiraceae, V. parvula, Haemophilus parainfluenzae, Megasphaera) represent bacteria typical of the oral cavity (de Vries et al., 2008; Nallabelli et al., 2016; Segata et al., 2012). V. dispar, the OTU with the largest decrease between mild and severe disease, is implicated in Crohn's disease (CD) (Fig. 3B) (Jacobs et al., 2016). Haemophilus and Veillonella spp. can induce dendritic cell (DC) maturation, which may enable bacteria-exposed DCs to prime T cell responses (Larsen et al., 2012). H. parainfluenzae was on average (mean relative abundance across all samples) the most abundant of the negatively implicated oral OTUs (Fig. S3A).
Twenty OTUs were depleted in patients with extensive disease involvement or pancolitis compared to patients with proctosigmoiditis or left-sided colitis (Fig. S3B+C). The most substantial increased OTUs in extensive or pancolitis, V. parvula and V. dispar, were also increased in severe disease at baseline. While disease severity does not necessarily imply the extent of disease involvement, our analysis suggests that the microbial signature associated with extensive or pancolitis is similar to the signature associated with more severe disease (Fig. S3D).
We also predicted metagenomic functional potential based on 16S rRNA profiles. Comparing severe and mild disease samples, 126 KEGG pathways were significantly increased (n=64) or decreased (n=62) (Table S2). Overall fold-changes (severe/mild) were less substantial compared to shifts in taxonomic composition.
Treatment-naive microbiome is linked to refractory and responsive disease
A subset of patients with baseline samples (19/343) progressed to refractory disease and required colectomy in the first year after diagnosis (compared to 304 patients with no colectomy, 20 unknowns; Fig. S1B). At baseline, 21 OTUs were associated with the later requirement of colectomy (Fig. 3C). Among the increased OTUs were 3 oral taxa also associated with more severe disease, and 12 of the 17 OTUs decreased in colectomy patients were depleted in more severe disease. This suggests that a subset of microbes associated with disease severity may be indicative of patients at risk for medically refractory disease.
Depletion of certain microbes at baseline was associated with sustained disease and indicative of CS-free remission at weeks 12 and 52 (Fig. 3D+E). Twelve OTUs were positively associated with week 12 CS-free remission, including 9 Clostridia and 2 Erysipelotrichaceae. Commensal Clostridia populate the mucosa in close proximity to epithelial cells and play a vital role in maintaining gut barrier function (Pryde et al., 2002). Both Erysipelotrichaceae and Clostriales are decreased in treatment-naive CD patients (Gevers et al., 2014). Further, 3 Clostridia taxa were positively associated with week 52 CS-free remission, and 1 was slightly decreased in patients that achieved remission. A decrease in Sutterella and an increase in Eggerthella lenta were also positively associated with week 52 CS-free remission.
Age of disease onset did not play a major role in disease course. We investigated age-related differences (patients were 4-17 years old) with regards to remission at weeks 4, 12 and 52, colectomy, initial treatment distribution and disease severity. While patients that achieved remission at week 12 were significantly younger than patients who did not (Wilcoxon, p=0.04), no other comparisons were associated with age of disease onset (all p>0.05).
Microbes associated with UC severity are antibiotic-responsive
Antibiotics are used to treat septic complications and mild flares in IBD but can substantially alter gut microbial community composition and reduce microbial diversity. Many microbes associated with antibiotic usage (up to 27 days prior to baseline sample collection; FDR<0.2) were implicated in disease severity and progression (Fig. 3A, C-E, Fig. S3E). Most OTUs depleted with antibiotic usage were Lachnospiraceae, Ruminococcaceae and Clostridiales, including several F. prausnitzii (Fig. S3E). These taxa include many SCFA-producing bacteria with essential roles in maintaining intestinal epithelial barrier function, antimicrobial defense and mucus production (Kelly et al., 2015), suggesting that antibiotics may adversely impact the IBD microbiome.
Corticosteroids significantly impact gut microbial composition
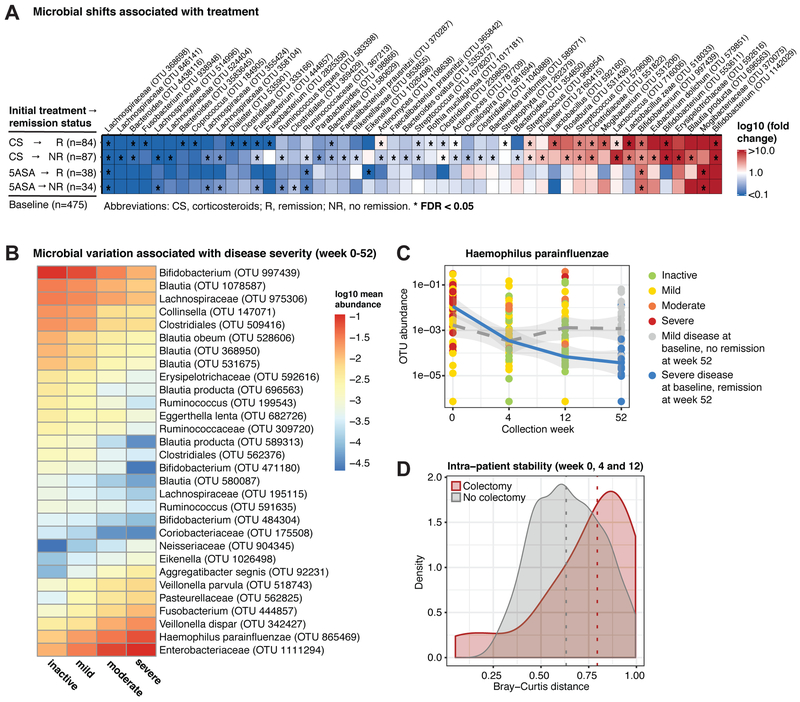
Longitudinal analysis showed that initial 5ASA or CS (oral or intravenous [IV]) treatment was significantly associated with altered gut microbial abundances (Fig. 4A). We compared treatment-naive baseline to week 4 samples grouped by initial treatment and remission status. Analysis was restricted to the first follow-up sample as treatment succession was highly patient-specific. Initial treatment is likely to exhibit the strongest effect on week 4 samples as most patients remained on 5ASA or CS during this timeframe. For patients with severe disease, IV CS were often followed by oral CS unless escalation to anti-TNF was required. In total, 47 OTUs were associated with CS and mostly showed a similar increase or decrease in the week 4 CS remission and no remission groups. Opposite trends were observed for 2 OTUs (fold changes <1 vs >1). Actinomyces abundance increased in CS-treated patients that achieved week 4 remission but decreased in those with sustained disease. The reverse was observed for a Clostridium OTU. Further, 7 species showed significant differences in their mean abundance between the remission and no-remission group (Wilcoxon, p<0.05), including 2 Fusobacteria (Table S3). These results notably differ from previous cross-sectional analyses involving CS (Morgan et al., 2012), suggesting that treatment response may be highly personalized and baseline-sensitive.
Figure 4: Temporal variation in microbial profiles linked to treatment efficacy and disease progression.
(A) Microbial changes between week 0 and 4 were associated with initial treatment efficacy within each treatment group (CS and 5ASA). Asterisks indicate patient groups for which the association was significant (FDR<0.05). (B) Microbial associations with disease severity in biopsies and stool samples from all time points (ninactive=447, nmild=337, nmoderate=275, nsevere=152). Mean abundances of each disease severity group for the 30 most significant OTUs (FDR<10−10). (C) H. parainfluenzae (OTU 865469) levels showed opposing temporal trends in connection with CS-free remission at week 52. Patients with initially mild disease that failed to achieve remission (dotted grey line) displayed consistent levels, while patients with initially severe disease that achieved remission (blue line) showed a substantial reduction in H. parainfluenzae. (D) Intra-patient stability was significantly lower (Wilcoxon, p=0.02) in the colectomy group. We compared intra-patient similarity of microbial community composition (Bray-Curtis; fecal samples only) from the first 3 time points. Dotted lines indicate group medians. See also Fig. S4 and Tables S3+S4.
With initial 5ASA treatment, 11 OTUs were associated with remission status, all showing the same trends in the week 4 remission and no-remission group. Differences (Wilcoxon, p<0.05) in abundance with regards to remission status were observed for 5 OTUs, including a Fusobacteria implicated in CS treatment efficacy (Table S3).
A decline in gut bacteria typical of the oral cavity over time is linked to improved disease severity and potentially favors remission
Trends in associations with disease severity observed in baseline samples persisted in all samples longitudinally, however the microbial signature differed. We identified 91 OTUs with a continuous increase or decrease across all disease severity categories (inactive, mild, moderate and severe; FDR<0.05; Table S4; top 30 associations in Fig. 4B). The most significant changes were observed for H. parainfluenzae (FDR=2*10−28) and Bifidobacterium (FDR=2*10−27) (Fig. S4A+B). Overall, 32% (29/91) of these OTUs were associated with disease severity at baseline (Fig. 3A) and 79% (72/91) were decreased in patients with more severe disease.
In total, 75% (54/72) of OTUs decreased with increasing disease severity were from the order Clostridiales, with 24 Lachnospiraceae and 18 Ruminococcaceae, suggesting a substantial depletion of SCFA-producing bacteria affecting epithelial barrier function. SCFA such as butyrate can inhibit stem cell proliferation (Kaiko et al., 2016), induce tolerogenic DCs, increase intestinal regulatory T cell percentages, and elevate intestinal IgA production (Goverse et al., 2017). Eight OTUs belonged to the Blautia genus, including many Clostridium cluster XIVa species capable of producing butyrate and secondary bile acids (Ridlon et al., 2014). Among OTUs with increased abundances in severe disease were several oral taxa, including 2 Aggregatibacter, Fusobacterium, 4 Veillonellaceae, 2 Enterobacteriaceae, 2 Neisseriaceae and H. parainfluenzae. Two Ruminococcus species, R. gnavus and R. torques, are human secretory mucin degraders previously observed to be increased in IBD (Png et al., 2010).
Temporal changes in H. parainfluenzae further highlighted interesting patterns associated with remission (Fig. 4C). Patients with initially severe disease displayed higher levels of H. parainfluenzae at baseline that continuously decreased through week 52. This contrasts consistent H. parainfluenzae levels in patients with initially mild disease that failed to achieve week 52 remission. Decreases in the abundance of certain bacteria, such as H. parainfluenzae, may be linked with improved disease outcome.
Comparing the distribution of intra-patient dissimilarity of longitudinal samples revealed a greater degree of heterogeneity in colectomy patients (Fig. 4D, Fig. S4C; Wilcoxon; p=0.02). All stool samples included in the analysis were collected prior to colectomy. Thus, general variability associated with the IBD microbiome is exacerbated in refractory disease and may be predictive of patients who will require more extreme interventions.
Discussion
We assessed microbial changes associated with disease progression in a large inception cohort of treatment-naive children with new-onset UC. Patients with inactive disease were used as a proxy for a control group, and treatment-naive baseline samples served as intra-patient controls to monitor patient-specific changes over time. A striking increase in bacteria typical of the oral cavity in patients with more severe disease was observed at baseline and follow-up time points. Healthy guts may be innately resistant to colonization by bacteria from the oral cavity; however, inflammation or strain-specific adaptation, including antibiotic resistance and virulence genes such as adhesion genes, may allow these microbes to colonize the gut mucosa in IBD and exacerbate inflammation. Increased oxygen levels in the IBD gut may favor expansion of aerotolerant bacteria from the oral cavity and facilitate strain-specific niche adaptation (Atarashi et al., 2017; Hall et al., 2017; Lloyd-Price et al., 2017). Altered, strain-specific immune responses were previously observed for Campylobacter concisus, where IBD strains showed upregulated surface expression of TLR4 and MD-2 in HT-29 cells (Ismail et al., 2013).
Temporal changes in microbial abundances were correlated with treatment efficacy. In particular, H. parainfluenzae was associated with disease severity and remission. Interestingly, H. parainfluenzae was detected in biopsies suggesting that it is embedded in the mucosa as an active member of the UC gut. Changes in mucosal microbial communities or a defective mucosal barrier may initiate or exacerbate intestinal inflammation. In mouse models, a spatial redistribution of bacterial groups in the large intestine was associated with inflammation (Swidsinski et al., 2005), and diet-mediated alterations of the gut microbiota caused functional mucosal defects that were preventable with the application of B. longum or inulin treatment (Schroeder et al., 2018). UC patients with acute inflammation display a thin, highly penetrable mucus layer (Johansson et al., 2014; Wlodarska et al., 2015), and mucin2 polymorphisms are associated with IBD risk. These findings underscore the importance of host-microbial interactions to maintain gut health.
Recent studies identified cross-reactivity between antibodies used as IBD biomarkers and bacterial and fungal antigens (Cohavy et al., 2000; Landers et al., 2002). Our analysis, which considered serological markers as a continuous variable, revealed several microbial associations with specific serological markers, such as OmpC and ASCA IgA. This suggests that similar mechanisms may disrupt host-microbial interactions in IBD. OmpC is required for adherent-invasive E. coli to thrive in the gut, and anti-OmpC antibodies are associated with ileal CD. Positive associations of OmpC with Veillonella and Dorea spp., but not E. coli, highlight potential microbial interactions within the changing UC microbiome. ASCA IgA targets mannan, a fungal cell wall component that mediates host interactions. Moreover, yeast α-mannans serve as nutrients for several Bacteroides species (Cuskin et al., 2015), and bacterial-fungal interactions potentially impact the immune response in IBD and impair protection against fungal pathogens. Potential cross-reactivity between yeast and bacteria remains a subject of future studies.
With current treatment efficacy rates at only 50%, choosing optimal treatment strategies for UC remains a challenge. Here, we observed baseline and longitudinal microbial trends implicated in progression of UC severity and remission. Although we do not know whether these microbial factors are a cause or consequence of UC, they are evidence that the microbiome impacts treatment efficacy and may serve as a potential therapeutic resource. Microbial biomarkers predictive of disease progression and treatment efficacy could transform clinical practices and inform optimal treatment strategies, such as a more aggressive treatment regimen for patients at higher risk of progressive disease.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for reagents may be directed to and will be fulfilled by Lead Contact Ramnik J. Xavier (xavier@molbio.mgh.harvard.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cohort and sample collection
The PROTECT study consists of 428 new-onset, pediatric UC patients recruited between July 2012 and April 2015 from 29 centers in the USA and Canada and monitored over the course of one year (Hyams et al., 2017). Patients were treatment-naive at baseline (week 0) and assigned one of two conventional treatment strategies: (1) 5-aminosalicylic acid (mesalamine) or (2) oral / intravenous (IV) corticosteroids (CS) followed by mesalamine. Treatment strategies were based on disease severity and progression (detailed outline in Fig. S1A).
For a subset of 405 patients, stool and rectal samples were collected for microbiome profiling. Stool samples were collected at baseline (week 0) and 3 follow-up time points (approximately 4, 12 and 52 weeks after initial treatment initiation) in addition to rectal biopsies collected at baseline and week 52, resulting in 1,212 samples in total. Metadata and clinical data were collected throughout the year, including age, gender, ethnicity, treatment, PUCAI, stool consistency, disease progression (colectomy and remission), extent of disease involvement, and fecal calprotectin. Subject were between 4 and 17 years of age (mean age 12.8+/−3.3 years) and 48% (195) were female. Overall, 138 / 136 / 131 patients received IV CS / oral CS / 5-aminosalicylic acid (5ASA) as initial treatment at week 0.
Inclusion and exclusion criteria
Eligibility criteria for patients were disease extent beyond the rectum (i.e., proctitis excluded), a baseline Pediatric Ulcerative Colitis Activity Index (PUCAI) (Turner et al., 2007) score of at least 10, no previous therapy for colitis, and stool culture negative for enteric bacterial pathogens (Salmonella, Shigella, Campylobacter, Escherichia coli 0157:H7) and Clostridium difficile toxin. A clinical, endoscopic, and histological diagnosis of ulcerative colitis was made in accordance with previously established criteria (Bousvaros et al., 2007). Patients were followed for a minimum of one year and up to three years with no change in diagnosis. None of these patients had granulomatous inflammation or small bowel disease other than possible backwash ileitis in the presence of cecal disease. Samples were randomized throughout the study, but researcher were not blinded with regards to phenotype. No technical replicates were obtained as 16S sequencing results have been shown to be highly reproducible within protocol (Sinha et al.). All sequencing datasets were required to have at least 3,000 reads after OTU assignment. (See details below on data processing.)
Sample number overview
| Baseline (week 0) | Week 4 | Week 12 | Week 52 | |
|---|---|---|---|---|
| No. of patients | 343 | 243 | 247 | 217 |
| No. of biopsies | 211 | - | - | 52 |
| No. of stool samples | 264 | 243 | 247 | 195 |
Ethics statement
The PROTECT study was approved by the local investigational review boards (IRBs) at each site. Inclusion of volunteers and experiments were conducted according to the principles expressed in the Declaration of Helsinki. Written assent was obtained for all children before any material was taken.
METHOD DETAILS
Biopsy DNA extraction
Rectal biopsies were obtained at the diagnostic colonoscopy and stored in RNALater at −80°C until further processing. Total DNA and RNA were isolated using the Qiagen AllPrep RNA/DNA Mini Kit. PolyA-RNA selection, fragmentation, cDNA synthesis, adaptor ligation, and library preparation were performed [details on library preparation outlined below; also see (Gevers et al., 2014)].
Stool DNA extraction
Total Nucleic Acid (TNA) was extracted via the Chemagic MSM I with the Chemagic DNA Blood Kit-96 from Perkin Elmer, combining chemical and mechanical lysis with magnetic bead-based purification. Prior to extraction on the MSM-I, TE buffer, Lysozyme, Proteinase K, and RLT Buffer with beta-mercaptoethanol were added to each sample. The lysate solution was subsequently vortexed and samples were then placed on the MSM I unit. M-PVA Magnetic Beads were added to the lysate solution and vortexed. The bead-bound TNA was then removed from solution via a 96-rod magnetic head and washed in three ethanol-based wash buffers. The beads were washed in a final water wash buffer and dipped in elution buffer to re-suspend the DNA sample in solution. The beads were then removed from solution, leaving purified TNA eluate, which was then split into two equal volumes (one for DNA, one for RNA). SUPERase-IN solution was added to the DNA samples, the reaction was cleaned up using AMPure XP SPRI beads. DNase was added to the RNA samples, and the reaction was cleaned up using AMPure XP SPRI beads. DNA samples were subsequently quantified using a fluorescence-based PicoGreen assay.
16S rRNA gene sequencing
The V4 variable 16S rRNA region was targeted and libraries were sequenced on the Illumina Miseq in paired-end mode (2×175bp). Briefly, genomic DNA was amplified using primers designed to incorporate Illumina adapters and sample barcodes (Primers: 515F and 806R) (Caporaso et al., 2012). Sample concentration was normalized to 1.5ng/μl for stool and 6ng/μl for biopsies. PCR mixtures contained 10 μl of sample template, 10 μl of 5PRIME HotMasterMix (Quantabio), and 5 μl of primer mix (IDT) at 2 μM concentration of each primer. The cycling conditions consisted of an initial denaturation of 94°C for 3 min, followed by 25-30 cycles of denaturation at 94°C for 45s, annealing at 50°C for 60s and extension at 72°C for 5 min, and a final extension at 72°C for 10 min. Amplicons were quantified according to the Caliper LabChip GX: DNA 5K Assay (PerkinElmer, Hopkinton, MA), and pooled in equimolar concentrations according to the 400bp amplicon. The pooled samples were then size selected by Pippin Prep 2% agarose protocol (Sage Science, Beverly, MA), removing non-specific amplification products from host DNA (375-425 bp). Libraries were sequenced on the Illumina MiSeq v2 platform, according to the manufacturer’s specifications with addition of 5% PhiX.
Serology measurements
Serological measurements at baseline were done at Cedars-Sinai Hospital (Los Angeles, CA, USA) using previously published methods (Dubinsky et al., 2008). Briefly, sera were analyzed for expression of anti-Saccharomyces cerevisiae antibodies (ASCA) immunoglobulin A (IgA), ASCA immunoglobulin G (IgG), anti-neutrophil cytoplasmic antibodies (ANCA), anti-flagellin antibodies (CBir1) and anti-outer membrane porin C (OmpC) antibodies in a blinded fashion by enzyme-linked immunosorbent assay (ELISA). Antibody levels were determined, and results were expressed as ELISA units (EU/mL).
Fecal calprotectin
Fecal calprotectin was measured centrally with ELISA (Buhlmann Laboratories, Schonenbuch, Switzerland) from stool samples collected around colonoscopy, or at 4, 12, and 52 weeks post diagnosis. Stool samples around colonoscopies were collected before colonoscopy cleanout or at least 2 days after colonoscopy, but not more than 3 days after initial ulcerative colitis treatment as previously described (Hyams et al., 2017).
Disease severity categories
Patients were grouped into four disease severity categories based on their baseline PUCAI score, including inactive (PUCAI: <10), mild (PUCAI: 10-30), moderate (PUCAI: 35-60), and severe (PUCAI: ≥65) disease (scores are increments of 5).
Remission at week 4/12/52
Clinical remission at week 4 was defined by PUCAI score <10 with no prior rescue therapy or colectomy. For remission at week 12 and 52, patients were also required to not be on corticosteroid therapy for a minimum of 14 days (for week 12) and 28 days (for week 52) prior to the assessment time. Rescue therapy included immunomodulators, calcineurin inhibitors, and TNF inhibitors.
QUANTIFICATION AND STATISTICAL ANALYSIS
OTU construction and taxonomic assignment
OTU clustering and taxonomic profiling of 16 rRNA gene amplicon sequencing data was performed with the 16S bioBakery workflow built with AnADAMA2 (McIver et al., 2017), which incorporates ea-utils and the UPARSE pipeline (version 8.1). Briefly, paired-end reads from all datasets were first merged, filtered and de-replicated. For quality-filtering the UPARSE threshold of Emax=1 was used, at which the most probable number of base errors per read is zero for filtered reads, and a truncation quality threshold of 15. Further reads were trimmed to a fixed length of 200bp. OTUs were then sorted by size, singletons were discarded and OTUs were clustered at 97% similarity. Subsequently, the representative sequences for each cluster were mapped against the Greengenes 16S rDNA database (version 13.5) to filter chimeras and obtain taxonomic assignment.
Filtering
All datasets were required to have at least 3,000 reads after OTU assignment. Datasets were subsequently normalized for library size and converted to relative abundances. Furthermore, OTU were required to occur in at least 20 samples resulting in 1,015 OTUs that were used in the downstream analysis. All OTUs, regardless of taxonomic classification level, were included for all analyses. Overall, 10% of the OTUs were classified at species, 44% at genus, 34% at family, 11% at order and less than 1% at class level.
Antibiotics analysis
Antibiotic exposure is relatively common in IBD patients and can have long-term, non-linear effects on the gut microbiome. For baseline (week 0) samples the recorded antibiotic usage was within 27 days of sample collection for all but one patient (334 days; Fig. S2H). For the followup samples we used an omnibus test (permANOVA) to determine the treatment window that explains the maximum amount of variation in the microbiota in connection with antibiotic exposure (Fig. S2I). A treatment window of 40 days was used to encode antibiotics as a binary variable.
Statistical association analysis
Statistical significance was established by boosted additive general linear models of OTU abundance as a function of sample metadata using MaAsLin (Morgan et al., 2012). To account for confounding effects, we included several covariates in the model. Typical covariates were age, gender, ethnicity and antibiotic usage, which were modeled as fixed effects in addition to the respective outcome variable (e.g., disease severity). MaAsLin accounts for multiple testing using Benjamini-Hochberg correction and the respective FDR values are provided throughout the manuscript. Significant associations with antibiotic exposure that overlapped with the outcome variable (i.e., disease severity or remission) were indicated in Fig. 3A, 3C and 3D. Furthermore, for the longitudinal analysis subject ID was added as a random effect. When stool and biopsy samples were analyzed concurrently, sample type was included as well. When evaluating remission or remission in connection with initial treatment, disease severity and treatment were also taken in account as an additional fixed effect. Treatment was otherwise not included in the linear models. Note, that for the association analysis with disease severity only OTUs that showed a continuous increase or decrease with increasing disease severity were considered (Fig. 3A and 4B).
Gender was taken in account as a covariate for all association analyses. The only significant associations (FDR < 0.05) with gender in baseline samples was Parabacteroides distasonis, which was increased in female patients (MaAsLin covariates: age, gender, ethnicity, antibiotic usage, sample type and PUCAI).
To assess the significance of the difference between averages of groups of samples, a two-sided Wilcoxon signed-rank test was used (R stats library).
Metabolic pathway predictions
OTU abundances were first normalized by their known/predicted 16S copy number prior to predicting metagenomic functional potential using the software PICRUSt (Langille et al., 2013). KEGG genes were further summarized into metabolic pathways (n=328). All samples were required to have at least 3,000 mapped reads and were normalized by library size. Only pathways (n=267) that occurred in at least 20 samples were subsequently included in the MaAsLin analysis to investigate functional differences at baseline comparing mild (nsamples=123) and severe (nsamples=137) disease (covariates: sample type, age, gender, ethnicity, antibiotics; random effects: subject ID).
DATA AND SOFTWARE AVAILABILITY
Raw 16S rRNA sequencing data is available through NCBI SRA Bioproject: PRJNA436359. The processed OTU table and metadata are available in the supplementary material (Table S6).
Supplementary Material
Differential metabolic potential between mild and severe disease at baseline. Related to Figure 3.
Differentially abundant organisms in the remission versus no remission groups of patients stratified by initial treatment. Related to Figure 4. See also Fig. S4.
Mean abundances of severity-associated OTUs within samples from patients with inactive/mild/moderate/severe disease. Related to Figure 4. See also Fig. S4.
Microbial associations with disease severity in baseline samples. Related to Figure 3. See also Fig. S3
Sample-specific metadata and final microbial OTU profiles. Related to Figure 1. See also Methods and Fig. S1.
Highlights:
Intestinal microbiome was temporally tracked in new-onset pediatric UC patients for 1-year
Treatment-naive UC microbiome is associated with disease progression and severity
Clostridiales depletion and oral pathobiont expansions in the gut link to disease course
Longitudinal microbial variation link to colectomy need and may impact treatment efficacy
Acknowledgements
We thank the PROTECT cohort for their participation. This study was supported by grants from the National Institutes of Health (NIH) 5U01DK095745, P30DK043351, R01AT009708, Center for Microbiome Informatics and Therapeutics and Crohn's Colitis Foundation to RJX and NIH grant U54DK102557 to CH and RJX. We thank Tiffany Poon (Broad Institute) for help in sequence production and sample management, Jason Bishai (Broad Institute) for help with sample processing, Bahar Sayoldin (Broad Institute) for making the data available through the SRA and Theresa Reimels (MGH) for invaluable editorial help.
Footnotes
Declaration of Interest
Jeffrey S. Hyams: Advisory Board, Janssen, Consultant, Abbvie, Takeda, Lilly, Boerhinger-Ingelheim, Allergan, Pfizer, Receptos, Astra Zeneca; Sonia M. Thomas: independent data monitoring committee, Lycera Corporation; Lee A. Denson: Grant Support, AbbVie and Janssen; Neal LeLeiko: Consultant, Abbvie; Ashish Patel: Speakers Bureau Abbvie, Janssen; James Markowitz: Consultant for Janssen, Celgene, Lilly; Maria Oliva-Hemker: Abbott Immunology-research grant, Janssen-research grant, Hoffman LaRoche-consultant; Anne Griffiths: Research support Abbvie, Consultant Abbvie, Celgene, Janssen, Lilly, Pfizer, Takeda, Speaker Abbvie, Janssen, Shire; Joel Rosh: Consultant, Abbvie, Celgene, Janssen, Luitpold, Pfizer, Grant Funding Janssen, Abbive; David Keljo: research support from Genentech and Takeda; Anthony Otley: Advisory Board, Janssen, Abbvie, Research support Lilly, Abbvie, Janssen, Takeda, Celgene; Paul Rufo: Consultant, Shire, Leutpold, Speaker, Abbvie, Research support, TechLab; Cary Sauer: Consultant, Abbvie; Subra Kugathasan: Consultant, Janssen, UCB; Melvin Heyman: Research grants Genentech, Abbvie, Shire, Takeda, Mallinkrodt, Janssen, Gilead; Curtis Huttenhower: Seres Scientific Advisory Board.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ananthakrishnan AN, Luo C, Yajnik V, Khalili H, Garber JJ, Stevens BW, Cleland T & Xavier RJ (2017). Gut Microbiome Function Predicts Response to Anti-integrin Biologic Therapy in Inflammatory Bowel Diseases. Cell Host Microbe, 21, 603–610 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Suda W, Luo C, Kawaguchi T, Motoo I, Narushima S, Kiguchi Y, Yasuma K, Watanabe E, Tanoue T, et al. (2017). Ectopic colonization of oral bacteria in the intestine drives TH1 cell induction and inflammation. Science, 358, 359–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, Fukuda S, Saito T, Narushima S, Hase K, et al. (2013). Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature, 500, 232–6. [DOI] [PubMed] [Google Scholar]
- Bousvaros A, Antonioli DA, Colletti RB, Dubinsky MC, Glickman JN, Gold BD, Griffiths AM, Jevon GP, Higuchi LM, Hyams JS, et al. (2007). Differentiating ulcerative colitis from Crohn disease in children and young adults: report of a working group of the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition and the Crohn's and Colitis Foundation of America. J Pediatr Gastroenterol Nutr, 44, 653–74. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. (2012). Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. Isme j, 6, 1621–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CC, Ching YH, Wang YC, Liu JY, Li YP, Huang YT & Chuang HL (2014). Monocolonization of germ-free mice with Bacteroides fragilis protects against dextran sulfate sodium-induced acute colitis. Biomed Res Int, 2014, 675786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohavy O, Bruckner D, Gordon LK, Misra R, Wei B, Eggena ME, Targan SR & Braun J (2000). Colonic bacteria express an ulcerative colitis pANCA-related protein epitope. Infect Immun, 68, 1542–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron E, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. (2015). Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature, 517, 165–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vries JJ, Arents NL & Manson WL (2008). Campylobacter species isolated from extra-oro-intestinal abscesses: a report of four cases and literature review. Eur J Clin Microbiol Infect Dis, 27, 1119–23. [DOI] [PubMed] [Google Scholar]
- Dubinsky MC, Kugathasan S, Mei L, Picornell Y, Nebel J, Wrobel I, Quiros A, Silber G, Wahbeh G, Katzir L, et al. (2008). Increased immune reactivity predicts aggressive complicating Crohn’s disease in children. Clin Gastroenterol Hepatol, 6, 1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C, Gibbons SM, Gurry T, Irizarry RA & Alm EJ (2017). Meta-analysis of gut microbiome studies identifies disease-specific and shared responses. Nat Commun, 8, 1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevers D, Kugathasan S, Denson LA, Vazquez-Baeza Y, Van Treuren W, Ren B, Schwager E, Knights D, Song SJ, Yassour M, et al. (2014). The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe, 15, 382–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goverse G, Molenaar R, Macia L, Tan J, Erkelens MN, Konijn T, Knippenberg M, Cook EC, Hanekamp D, Veldhoen M, et al. (2017). Diet-Derived Short Chain Fatty Acids Stimulate Intestinal Epithelial Cells To Induce Mucosal Tolerogenic Dendritic Cells. J Immunol, 198, 2172–2181. [DOI] [PubMed] [Google Scholar]
- Haiser HJ, Gootenberg DB, Chatman K, Sirasani G, Balskus EP & Turnbaugh PJ (2013). Predicting and manipulating cardiac drug inactivation by the human gut bacterium Eggerthella lenta. Science, 341, 295–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halfvarson J, Brislawn CJ, Lamendella R, Vazquez-Baeza Y, Walters WA, Bramer LM, D'amato M, Bonfiglio F, Mcdonald D, Gonzalez A, et al. (2017). Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol, 2, 17004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AB, Yassour M, Sauk J, Garner A, Jiang X, Arthur T, Lagoudas GK, Vatanen T, Fornelos N, Wilson R, et al. (2017). A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med, 9, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyams JS, Davis S, Mack DR, Boyle B, Griffiths AM, Leleiko NS, Sauer CG, Keljo DJ, Markowitz J, Baker SS, et al. (2017). Factors associated with early outcomes following standardised therapy in children with ulcerative colitis (PROTECT): a multicentre inception cohort study. Lancet Gastroenterol Hepatol, 2, 855–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail Y, Lee H, Riordan SM, Grimm MC & Zhang L (2013). The effects of oral and enteric Campylobacter concisus strains on expression of TLR4, MD-2, TLR2, TLR5 and COX-2 in HT-29 cells. PLoS One, 8, e56888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JP, Goudarzi M, Singh N, Tong M, Mchardy IH, Ruegger P, Asadourian M, Moon BH, Ayson A, Borneman J, et al. (2016). A Disease-Associated Microbial and Metabolomics State in Relatives of Pediatric Inflammatory Bowel Disease Patients. Cell Mol Gastroenterol Hepatol, 2, 750–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, Ghishan FK, Carvalho FA, Gewirtz AT, Sjovall H, et al. (2014). Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut, 63, 281–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM & Stappenbeck TS (2016). The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell, 165, 1708–1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, et al. (2015). Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe, 17, 662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugathasan S, Denson LA, Walters TD, Kim MO, Marigorta UM, Schirmer M, Mondal K, Liu C, Griffiths A, Noe JD, et al. (2017). Prediction of complicated disease course for children newly diagnosed with Crohn's disease: a multicentre inception cohort study. Lancet, 389, 1710–1718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers CJ, Cohavy O, Misra R, Yang H, Lin YC, Braun J & Targan SR (2002). Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology, 123, 689–99. [DOI] [PubMed] [Google Scholar]
- Langille MG, Zaneveld J, Caporaso JG, Mcdonald D, Knights D, Reyes JA, Clemente JC, Burkepile DE, Vega Thurber RL, Knight R, et al. (2013). Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol, 31, 814–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen JM, Steen-Jensen DB, Laursen JM, Sondergaard JN, Musavian HS, Butt TM& Brix S (2012). Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One, 7, e31976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Price J, Mahurkar A, Rahnavard G, Crabtree J, Orvis J, Hall AB, Brady A, Creasy HH, Mccracken C, Giglio MG, et al. (2017). Strains, functions and dynamics in the expanded Human Microbiome Project. Nature, 550, 61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mciver LJ, Abu-Ali G, Franzosa EA, Schwager R, Morgan XC, Waldron L, Segata N & Huttenhower C (2017). bioBakery: A meta'omic analysis environment. Bioinformatics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan XC, Tickle TL, Sokol H, Gevers D, Devaney KL, Ward DV, Reyes JA, Shah SA, Leleiko N, Snapper SB, et al. (2012). Dysfunction of the intestinal microbiome in inflammatory bowel disease and treatment. Genome Biol, 13, R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nallabelli N, Patil PP, Pal VK, Singh N, Jain A, Patil PB, Grover V & Korpole S (2016). Biochemical and genome sequence analyses of Megasphaera sp. strain DISK18 from dental plaque of a healthy individual reveals commensal lifestyle. Sci Rep, 6, 33665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peloquin JM, Goel G, Villablanca EJ & Xavier RJ (2016). Mechanisms of Pediatric Inflammatory Bowel Disease. Annu Rev Immunol, 34, 31–64. [DOI] [PubMed] [Google Scholar]
- Peyrin-Biroulet L, Standaert-Vitse A, Branche J & Chamaillard M (2007). IBD serological panels: facts and perspectives. Inflamm Bowel Dis, 13, 1561–6. [DOI] [PubMed] [Google Scholar]
- Png CW, Linden SK, Gilshenan KS, Zoetendal EG, Mcsweeney CS, Sly LI, Mcguckin MA & Florin TH (2010). Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol, 105, 2420–8. [DOI] [PubMed] [Google Scholar]
- Pryde SE, Duncan SH, Hold GL, Stewart CS & Flint HJ (2002). The microbiology of butyrate formation in the human colon. FEMS Microbiol Lett, 217, 133–9. [DOI] [PubMed] [Google Scholar]
- Ridlon JM, Kang DJ, Hylemon PB & Bajaj JS (2014). Bile acids and the gut microbiome. Curr Opin Gastroenterol, 30, 332–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruemmele FM, Targan SR, Levy G, Dubinsky M, Braun J & Seidman EG (1998). Diagnostic accuracy of serological assays in pediatric inflammatory bowel disease. Gastroenterology, 115, 822–9. [DOI] [PubMed] [Google Scholar]
- Sartor RB & Wu GD (2017). Roles for Intestinal Bacteria, Viruses, and Fungi in Pathogenesis of Inflammatory Bowel Diseases and Therapeutic Approaches. Gastroenterology, 152, 327–339.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer M, Smeekens SP, Vlamakis H, Jaeger M, Oosting M, Franzosa EA, Ter Horst R, Jansen T, Jacobs L, Bonder MJ, et al. (2016). Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell, 167, 1125–1136 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder BO, Birchenough GMH, Stahlman M, Arike L, Johansson MEV, Hansson GC & Backhed F (2018). Bifidobacteria or Fiber Protects against Diet-Induced Microbiota-Mediated Colonic Mucus Deterioration. Cell Host Microbe, 23, 27–40.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segata N, Haake SK, Mannon P, Lemon KP, Waldron L, Gevers D, Huttenhower C & Izard J (2012). Composition of the adult digestive tract bacterial microbiome based on seven mouth surfaces, tonsils, throat and stool samples. Genome Biol, 13, R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw KA, Bertha M, Hofmekler T, Chopra P, Vatanen T, Srivatsa A, Prince J, Kumar A, Sauer C, Zwick ME, et al. (2016). Dysbiosis, inflammation, and response to treatment: a longitudinal study of pediatric subjects with newly diagnosed inflammatory bowel disease. Genome Med, 8, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha R, Abu-Ali G, Vogtmann E, Fodor AA, Ren B, Amir A, Schwager E, Crabtree J, Ma S, Microbiome Quality Control Project, C., et al. (2017). Assessment of variation in microbial community amplicon sequencing by the Microbiome Quality Control (MBQC) project consortium. Nat Biotechnol, 35, 1077–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivan A, Corrales L, Hubert N, Williams JB, Aquino-Michaels K, Earley ZM, Benyamin FW, Lei YM, Jabri B, Alegre ML, et al. (2015). Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science, 350, 1084–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A, Loening-Baucke V, Lochs H & Hale LP (2005). Spatial organization of bacterial flora in normal and inflamed intestine: a fluorescence in situ hybridization study in mice. World J Gastroenterol, 11, 1131–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tailford LE, Crost EH, Kavanaugh D & Juge N (2015). Mucin glycan foraging in the human gut microbiome. Front Genet, 6, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner D, Otley AR, Mack D, Hyams J, De Bruijne J, Uusoue K, Walters TD, Zachos M, Mamula P, Beaton DE, et al. (2007). Development, validation, and evaluation of a pediatric ulcerative colitis activity index: a prospective multicenter study. Gastroenterology, 133, 423–32. [DOI] [PubMed] [Google Scholar]
- Vetizou M, Pitt JM, Daillere R, Lepage P, Waldschmitt N, Flament C, Rusakiewicz S, Routy B, Roberti MP, Duong CP, et al. (2015). Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science, 350, 1079–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl C, Liptay S, Adler G & Schmid RM (1998). Sulfasalazine: a potent and specific inhibitor of nuclear factor kappa B. J Clin Invest, 101, 1163–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M, Kostic AD & Xavier RJ (2015). An integrative view of microbiome-host interactions in inflammatory bowel diseases. Cell Host Microbe, 17, 577–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier RJ & Podolsky DK (2007). Unravelling the pathogenesis of inflammatory bowel disease. Nature, 448, 427–34. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Differential metabolic potential between mild and severe disease at baseline. Related to Figure 3.
Differentially abundant organisms in the remission versus no remission groups of patients stratified by initial treatment. Related to Figure 4. See also Fig. S4.
Mean abundances of severity-associated OTUs within samples from patients with inactive/mild/moderate/severe disease. Related to Figure 4. See also Fig. S4.
Microbial associations with disease severity in baseline samples. Related to Figure 3. See also Fig. S3
Sample-specific metadata and final microbial OTU profiles. Related to Figure 1. See also Methods and Fig. S1.