Abstract
Background
Farnesol has potential antifungal activity against Candida albicans biofilms, but the molecular mechanism of this activity is still unclear. Farnesol inhibits hyphal growth by regulating the cyclic AMP (cAMP) signalling pathway in C. albicans, and CYR1 and PDE2 regulate a pair of enzymes that are directly responsible for cAMP synthesis and degradation. Here, we hypothesize that farnesol enhances the antifungal susceptibility of C. albicans biofilms by regulating CYR1 and PDE2.
Results
The resistance of the CYR1- and PDE2-overexpressing strains to caspofungin, itraconazole and terbinafine was increased in planktonic cells, and that to amphotericin B was increased in biofilms. Meanwhile, the biofilms of the CYR1- and PDE2-overexpressing strains were thicker (all p < 0.05) and consisted of more hyphae than that of the wild strain. The intracellular cAMP levels were higher in the biofilms of the CYR1-overexpressing strain than that in the biofilms of the wild strain (all p < 0.01), while no changes were found in the PDE2-overexpressing strain. Exogenous farnesol decreased the resistance of the CYR1- and PDE2-overexpressing strains to these four antifungals, repressed the hyphal and biofilm formation of the strains, and decreased the intracellular cAMP level in the biofilms (all p < 0.05) compared to the untreated controls. In addition, farnesol decreased the expression of the gene CYR1 and the protein CYR1 in biofilms of the CYR1-overexpressing strain (all p < 0.05) but increased the expression of the gene PDE2 and the protein PDE2 in biofilms of the PDE2-overexpressing strain (all p < 0.01).
Conclusions
The results indicate that CYR1 and PDE2 regulate the resistance of C. albicans biofilms to antifungals. Farnesol suppresses the resistance of C. albicans biofilms to antifungals by regulating the expression of the gene CYR1 and PDE2, while PDE2 regulation was subordinate to CYR1 regulation.
Keywords: Candida albicans biofilms, Resistance, Farnesol, CYR1, PDE2
Background
Candida albicans, existing as a commensal fungus in healthy individuals, is the major fungal pathogen in humans that can cause systemic candidiasis in immunocompromised patients, especially in organ transplant recipients, HIV-positive patients or those with autoimmune diseases [1]. In clinical infections of C. albicans, the fungi commonly form biofilms, which are associated with chronic infections. Biofilms are protected niches for microorganisms, where they are safer from antibiotic treatment than they are in their planktonic form, and can provide a source of persistent infection. Biofilm-related infections caused by C. albicans can become a major threat to public health, as these infections are refractory and result in a reservoir for continued infection [2–4]. Therefore, it is crucial to find the resistance mechanisms of C. albicans biofilms and to explore novel drugs and compounds that may enhance the efficacy of traditional antifungals in the therapeutic armamentarium against biofilm-related infections.
Farnesol, an exogenous chemical molecule, has a broad range of effects on C. albicans, but the most prominent of these are its ability to influence C. albicans morphology by inhibiting the yeast-to-hypha transition [5, 6] and its antifungal activities against C. albicans biofilms [7, 8]. Our previous research indicated that farnesol inhibited the development of C. albicans biofilms of resistant strains, and there were synergistic effects of farnesol in combination with antifungals [9, 10]. Further studies suggested that farnesol-dependent inhibition of hyphal formation involved the cAMP pathway [5, 11, 12]. Farnesol inhibited hyphal growth as an adenylate cyclase (CYR1) inhibitor and exerted a direct effect on intracellular cAMP levels [1, 11]. Another study found that farnesol upregulated PDE2 expression in C. albicans biofilms grown for 24 h [13]. Those studies did not report any mechanism of the antifungal activities of farnesol on the C. albicans biofilms to be associated with CYR1 or PDE2, and the mechanism needs to be explored.
The yeast-to-hypha transition is correlated with the antifungal resistance of C. albicans. As previously reported, cAMP is the key element in triggering hyphal formation [1, 14, 15]. CYR1 and PDE2 regulate a pair of enzymes that are directly responsible for cAMP synthesis and degradation, respectively [16]. The activation of cAMP signalling is due to the loss of cAMP phosphodiesterase, PDE2, or an increase in cAMP adenylate cyclase, CYR1 [17–19]. Further studies found that CYR1 and PDE2 were associated with the resistance of planktonic C. albicans to antifungals, and the mutants of C. albicans with a deletion of CYR1 or PDE2 had increased sensitivity to azole antifungals [20, 21]. However, the role of overexpressing CYR1 and PDE2 in the resistance of C. albicans biofilms and the role played by farnesol in this process need to be elucidated.
Therefore, we hypothesized that CYR1 and PDE2 regulate the resistance of C. albicans biofilms to antifungals and that farnesol increases the susceptibility of C. albicans biofilms to antifungals by regulating the gene expression of CYR1 and PDE2 in the cAMP pathway. The antifungal resistance of C. albicans biofilms formed by CYR1- and PDE2-overexpressing strains was examined in the presence or absence of farnesol and was compared to that in biofilms formed by the wild strain. Additionally, morphological changes were examined using CLSM and SEM. The intracellular cAMP level of C. albicans biofilms was detected by an ELISA. Furthermore, the expression of genes and proteins was analysed using RT-qPCR and western blotting, respectively.
Results
CYR1 and PDE2 are involved in the resistance of C. albicans to antifungals
For the XTT assay, biofilms formed from the CYR1-overexpressing strain (CYR1OE) and the PDE2-overexpressing strain (PDE2OE) had higher values for the sessile minimum inhibitory concentration (SMIC) that caused a ≥ 50% reduction in the metabolic activity of the biofilms (SMIC50) of amphotericin B than did the wild strain (CAI4-pCaEXP) at all studied phases (Table 1). There were no significant differences in the SMIC50 values of caspofungin, itraconazole and terbinafine among the three strains (CYR1OE, PDE2OE and CAI4-pCaEXP). For the spot assay, the planktonic form of C. albicans of the CYR1OE and PDE2OE strains was more resistant to caspofungin, itraconazole and terbinafine than was that of the wild strain (Fig. 1).
Table 1.
In vitro susceptibility of C. albicans biofilms to antifungals
| Strains | SMIC50 of antifungals (μg/ml) | |||
|---|---|---|---|---|
| 6 h | 12 h | 24 h | 36 h | |
| amphotericin B | ||||
| CAI4-pCaEXP | 1 | 0.5 | 1 | 0.25 |
| CYR1OE | 4 | 8 | > 16 | 16 |
| PDE2OE | 4 | 4 | 1 | 16 |
| caspofungin | ||||
| CAI4-pCaEXP | 0.125 | > 8 | > 8 | > 8 |
| CYR1OE | 0.125 | > 8 | > 8 | > 8 |
| PDE2OE | 8 | > 8 | > 8 | > 8 |
| itraconazole | ||||
| CAI4-pCaEXP | > 16 | > 16 | > 16 | > 16 |
| CYR1OE | > 16 | > 16 | > 16 | > 16 |
| PDE2OE | > 16 | > 16 | > 16 | > 16 |
| terbinafine | ||||
| CAI4-pCaEXP | > 256 | > 256 | > 256 | > 256 |
| CYR1OE | > 256 | > 256 | > 256 | > 256 |
| PDE2OE | > 256 | > 256 | > 256 | > 256 |
Fig. 1.
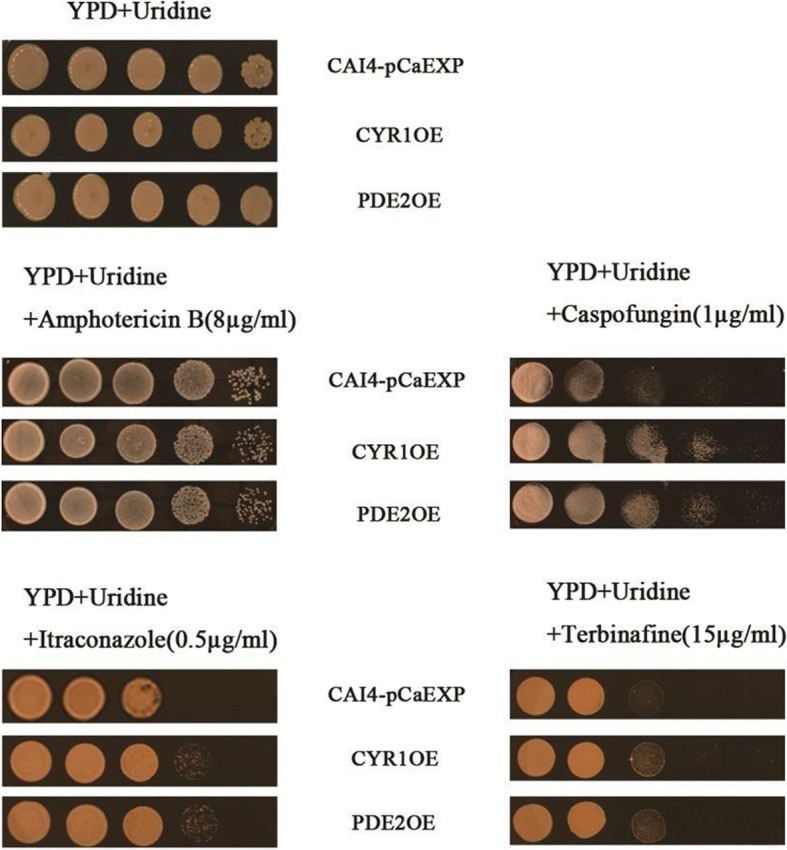
The susceptibility of C. albicans to antifungals. The concentrations of the drugs used above: Fluconazole: 4 μg/ml; Amphotericin B: 8 μg/ml; Caspofungin: 1 μg/ml; Itraconazole: 0.5 μg/ml; Terbinafine: 15 μg/ml. The planktonic form of C. albicans of the CYR1OE and PDE2OE strains was more resistant to caspofungin, itraconazole and terbinafine than was that of the wild strain
Farnesol increased the activities of antifungals against C. albicans biofilms of the CYR1OE and PDE2OE strains
The biofilms of the CYR1OE strain exposed to farnesol showed lower SMICs of amphotericin B (biofilm phase at 6, 12, 24 and 36 h) (Table 2), caspofungin (biofilm phase at 6, 12, 24 and 36 h) (Table 2), itraconazole (biofilm phase at 12 and 24 h) (Table 3) and terbinafine (biofilm phase at 6, 12 and 24 h) (Table 3) than did those that were not exposed to farnesol. Moreover, biofilms of the PDE2OE strain exposed to farnesol showed lower SMICs of amphotericin B (biofilm phase at 6, 12 and 36 h) (Table 2), caspofungin (biofilm phase at 6, 24 and 36 h) (Table 2), itraconazole (biofilm phase at 12, 24 and 36 h) (Table 3) and terbinafine (biofilm phase at 6, 12 and 24 h) (Table 3) than did those that were not exposed to farnesol. However, the farnesol-treated biofilms of the CYR1OE and PDE2OE strains showed higher SMICs of itraconazole (biofilm phase at 6 h) than those that were not treated with farnesol (Table 3).
Table 2.
The SMICs of amphotericin B and caspofungin in C. albicans biofilms
| Farnesol (μM) | amphotericin B(μg/ml) | caspofungin(μg/ml) | ||||
|---|---|---|---|---|---|---|
| CAI4-pCaEXP | CYR1OE | PDE2OE | CAI4-pCaEXP | CYR1OE | PDE2OE | |
| 6 h | ||||||
| 0 | 1 | 4 | 4 | 0.125 | 0.125 | 8 |
| 50 | 1 | 4 | 4 | 0.125 | 0.125 | 8 |
| 100 | 0.5 | 2 | 4 | 0.0625 | 0.0625 | 8 |
| 200 | 1 | 4 | 2 | 0.0625 | 0.0625 | 0.625 |
| 300 | 1 | 4 | 1 | 0.0625 | 0.0625 | 0.25 |
| 600 | 1 | 4 | 1 | 0.0625 | 0.0625 | 0.25 |
| 1200 | 1 | 2 | 1 | 0.0625 | 0.125 | 0.125 |
| 12 h | ||||||
| 0 | 0.5 | 8 | 4 | > 8 | > 8 | 0.0625 |
| 50 | 0.5 | 4 | 4 | > 8 | 8 | 0.0625 |
| 100 | 0.5 | 4 | 2 | > 8 | > 8 | 0.0625 |
| 200 | 0.5 | 4 | 2 | 0.25 | 8 | 0.0625 |
| 300 | 1 | 4 | 2 | 0.25 | 0.0625 | 0.0625 |
| 600 | 1 | 8 | 1 | 0.25 | 0.0625 | 0.0625 |
| 1200 | 16 | 8 | 2 | 0.0625 | 0.125 | 0.0625 |
| 24 h | ||||||
| 0 | 1 | > 16 | 1 | > 8 | > 8 | > 8 |
| 50 | 1 | 16 | 1 | > 8 | 8 | > 8 |
| 100 | 1 | 16 | 1 | > 8 | 0.125 | > 8 |
| 200 | 0.5 | 8 | 1 | > 8 | 0.0625 | 0.25 |
| 300 | 1 | 8 | 1 | > 8 | 0.0625 | 0.0625 |
| 600 | 0.125 | 8 | 1 | 2 | 0.0625 | 0.125 |
| 1200 | 0.125 | 8 | 1 | > 8 | 0.0625 | 0.125 |
| 36 h | ||||||
| 0 | 0.25 | 16 | 16 | > 8 | > 8 | > 8 |
| 50 | 0.25 | 16 | 16 | 4 | 1 | > 8 |
| 100 | 2 | 8 | 8 | 1 | 0.5 | > 8 |
| 200 | 0.125 | 4 | 4 | 0.25 | 0.5 | 0.125 |
| 300 | 0.125 | 4 | 4 | 0.125 | 0.5 | 0.125 |
| 600 | 0.25 | 4 | 2 | 0.0625 | 0.5 | 0.25 |
| 1200 | 0.125 | 4 | 2 | 4 | 0.5 | 0.5 |
Table 3.
The SMICs of itraconazole and terbinafine in C. albicans biofilms
| Farnesol (μM) | itraconazole(μg/ml) | terbinafine(μg/ml) | ||||
|---|---|---|---|---|---|---|
| CAI4-pCaEXP | CYR1OE | PDE2OE | CAI4-pCaEXP | CYR1OE | PDE2OE | |
| 6 h | ||||||
| 0 | > 16 | 0.0625 | 0.3125 | > 256 | > 256 | > 256 |
| 50 | > 16 | 0.0625 | 0.0625 | > 256 | > 256 | > 256 |
| 100 | > 16 | 0.125 | 0.0625 | > 256 | > 256 | > 256 |
| 200 | > 16 | 0.125 | > 16 | > 256 | 2 | 4 |
| 300 | > 16 | 0.125 | > 16 | > 256 | 2 | 2 |
| 600 | > 16 | 0.125 | > 16 | > 256 | 4 | 8 |
| 1200 | > 16 | 0.125 | > 16 | > 256 | 16 | 4 |
| 12 h | ||||||
| 0 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 50 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 100 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 200 | > 16 | 0.5 | 0.125 | > 256 | > 256 | > 256 |
| 300 | > 16 | 0.5 | 0.125 | > 256 | 2 | 4 |
| 600 | > 16 | 0.5 | 0.125 | > 256 | 2 | 4 |
| 1200 | 1 | 1 | 1 | > 256 | 4 | 32 |
| 24 h | ||||||
| 0 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 50 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 100 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 200 | > 16 | 16 | 2 | > 256 | 1 | 1 |
| 300 | > 16 | 0.25 | 0.125 | > 256 | 2 | 2 |
| 600 | > 16 | 0.25 | 0.25 | > 256 | > 256 | > 256 |
| 1200 | > 16 | 0.5 | 0.5 | > 256 | > 256 | > 256 |
| 36 h | ||||||
| 0 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 50 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 100 | > 16 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 200 | 0.125 | > 16 | > 16 | > 256 | > 256 | > 256 |
| 300 | 0.5 | > 16 | 16 | > 256 | > 256 | > 256 |
| 600 | 1 | > 16 | 2 | > 256 | > 256 | > 256 |
| 1200 | 2 | > 16 | 4 | > 256 | > 256 | > 256 |
Under treatment with the same concentration of farnesol, the biofilms of the CYR1OE strain showed higher SMICs of amphotericin B (biofilm phase at 6, 12, 24 and 36 h) (Table 2) and SMICs of itraconazole (biofilm phase at 36 h) (Table 3) than did those of the wild strain (CAI4-pCaEXP). Further, the biofilms of the PDE2OE strain showed higher SMICs of amphotericin B (biofilm phase at 6, 12 and 36 h) (Table 2), caspofungin (biofilm phase at 6 h) (Table 2) and itraconazole (biofilm phase at 36 h) (Table 3) than did those of the wild strain. In addition, compared to the biofilms of the PDE2OE strain, the biofilms of the CYR1OE strain showed higher SMICs of amphotericin B (biofilm phase at 6, 12, 24 and 36 h) (Table 2) under treatment with the same concentration of farnesol.
Farnesol changed the morphology of C. albicans biofilms formed form the CYR1OE and PDE2OE strains
The results of CLSM and SEM all showed that the biofilms of the farnesol-treated groups consisted of fewer hyphae but more pseudohyphae and yeast than did the controls without farnesol treatment (Figs. 2A and 3A). Moreover, the biofilms of CYR1OE and PDE2OE strains consisted of more hyphae and pseudohyphae than did those of the wild strain (Figs. 2A and 3A). Furthermore, SEM showed that the cell surfaces in the biofilms of the wild strain were uneven (Fig. 3A a1–4 and d1–4), while the cell surfaces in the biofilms of the CYR1OE and PDE2OE strains were smooth with spores (Fig. 3A b1–4, c1–4, e1–4 and f1–4). No obvious difference was observed between the cell surfaces of the farnesol-treated biofilms and the untreated controls. The quantitative assessment for the CLSM images showed that the biofilms exposed to farnesol were thinner than that without farnesol (all p < 0.01). The biofilms of the CYR1OE and PDE2OE strains were much thicker than that of the wild strain (all p < 0.05) (Fig. 2B). What’s more, the quantitative assessment for the SEM images showed that the biofilms of the CYR1OE and PDE2OE strains exposed to farnesol consisted of shorter hyphae than that without farnesol (all p < 0.01). The hyphae extended much longer in the later phases of the biofilms than did that in the early phases (all p < 0.01) (Fig. 3B).
Fig. 2.
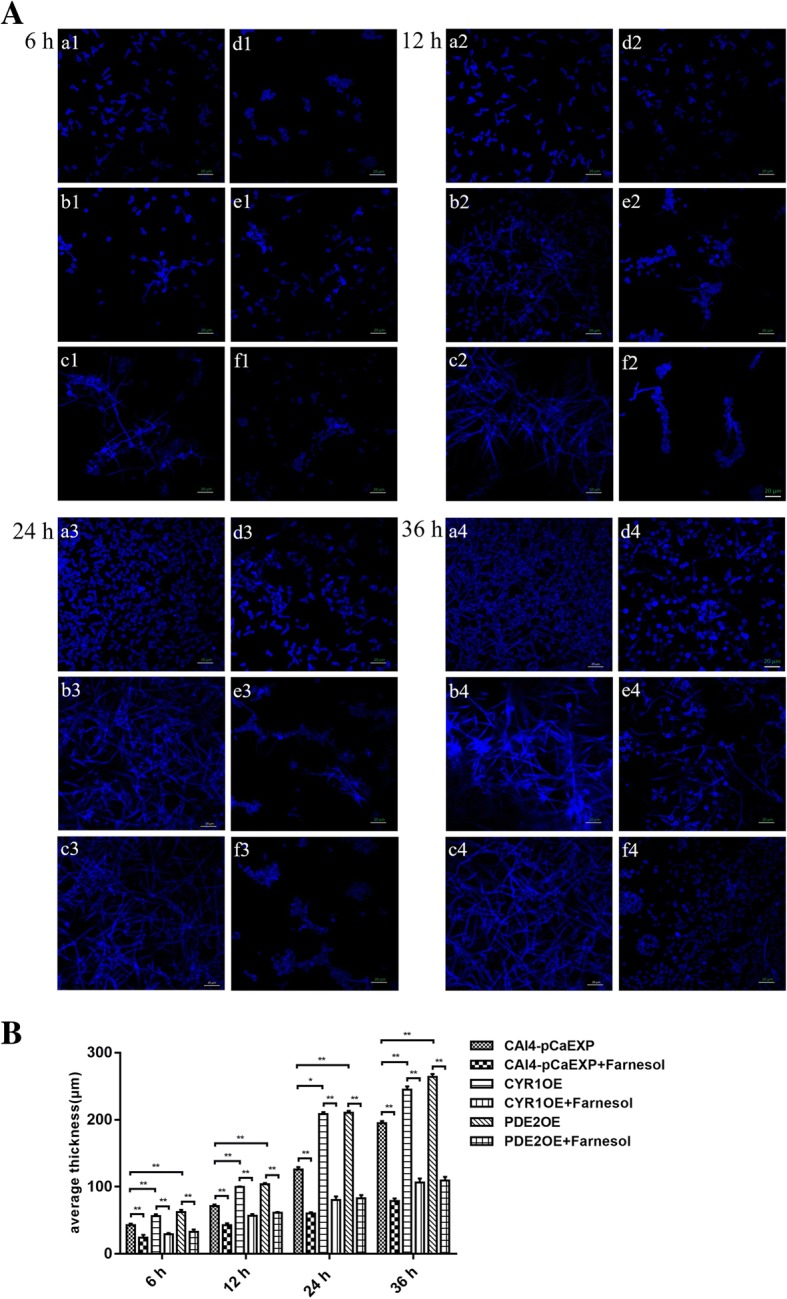
Confocal Laser Scanning Microscopy (CLSM) images of C. albicans biofilms formed from the CYR1OE and PDE2OE strains. A: a1–4: Farnesol untreated wild strain. b1–4: Farnesol untreated CYR1OE strain. c1–4: Farnesol untreated PDE2OE strain. d1–4: Farnesol treated wild strain. e1–4: Farnesol treated CYR1OE strain. f1–4: Farnesol treated PDE2OE strain. Scale bars represent 20 μm for 400× magnifications. The biofilms of d1–4, e1–4 and f1–4 consisted of fewer hyphae, but more pseudohyphae and yeast than did those of a1–4, b1–4 and c1–4. Additionally, the biofilms of b1–4 and c1–4 consisted of more hyphae and pseudohyphae than did that of a1–4. B: The biofilms exposed to farnesol were thinner than that without farnesol and the biofilms of the CYR1OE and PDE2OE strains were much thicker than that of the wild strain. Significance was calculated using one-way ANOVA with post ad-hoc Dunnett’s multiple comparison test. *: p < 0.05. **: p < 0.01
Fig. 3.
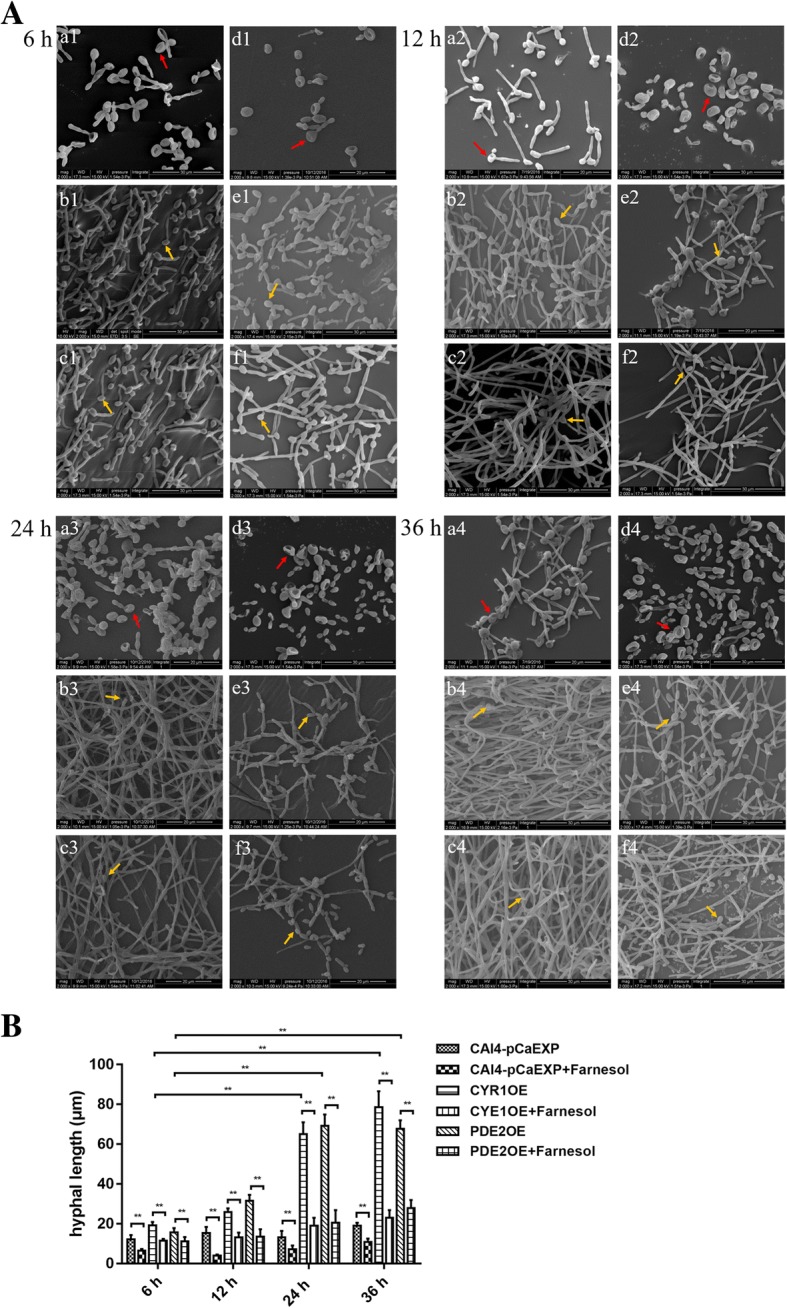
Scanning Electron Microscopy (SEM) images of C. albicans biofilms formed from the CYR1OE and PDE2 strains. A: a1–4: Farnesol untreated wild strain. b1–4: Farnesol untreated CYR1OE strain. c1–4: Farnesol untreated PDE2OE strain. d1–4: Farnesol treated wild strain. e1–4: Farnesol treated CYR1OE strain. f1–4: Farnesol treated PDE2OE strain. Magnification: 2000×. The surface of the cells in a1–4, d1–4 was uneven (red arrows), while the surface of cells in b1–4, c1–4, e1–4, f1–4 was smooth with spores (yellow arrows). The other results were the same as Fig. 2A. B: The biofilms formed from the CYR1OE and PDE2OE strains exposed to farnesol consisted of shorter hyphae than that without farnesol. The hyphae extended much longer in the later phases (24 and 36 h) of the biofilms than did that in the early phases (6 h). Significance was calculated using one-way ANOVA with post ad-hoc Dunnett’s multiple comparison test. *: p < 0.05. **: p < 0.01
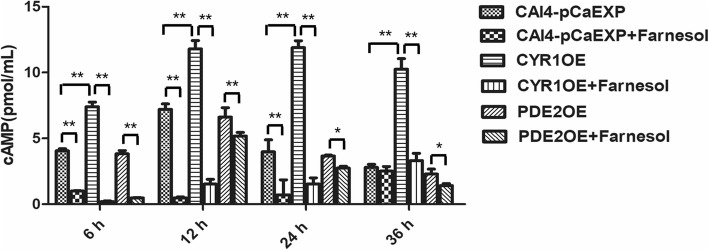
Farnesol decreased the cAMP levels in the C. albicans biofilms of the CYR1OE and PDE2OE strains
The results of the ELISA showed that farnesol decreased the intracellular cAMP levels in the biofilms of the CYR1OE strain (all p < 0.01), the PDE2OE strain (all p < 0.05), and the wild strain (p < 0.01) (Fig. 4), compared to those of the respective untreated controls. In addition, compared to the cAMP levels in the wild strain, the cAMP levels in the CYR1OE strain were significantly higher in all studied phases (all p < 0.01), while those of the PDE2OE strain did not change in any studied phase (all p > 0.05) (Fig. 4).
Fig. 4.
The level of cAMP from C. albicans biofilms in the presence of farnesol. Farnesol decreased the intracellular cAMP levels in the biofilms of the CYR1OE strain, the PDE2OE strain, and the wild strain compared to those of the respective untreated controls. In addition, compared to the cAMP levels in the wild strain, the cAMP levels in the CYR1OE strain were significantly higher in all studied phases. One-way analysis of variance (ANOVA) was employed to assess the statistical significance of differences in matched groups, while paired t-tests were performed for intra-group comparisons. *: p < 0.05. **: p < 0.01
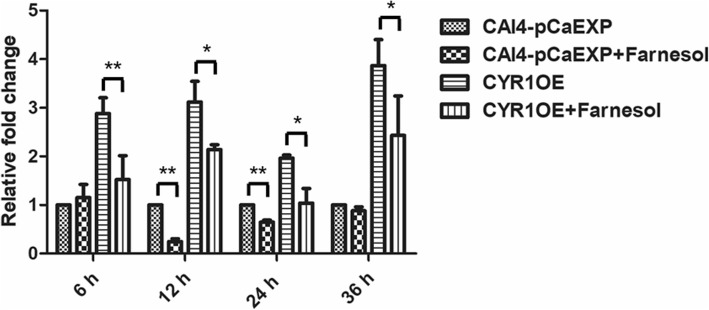
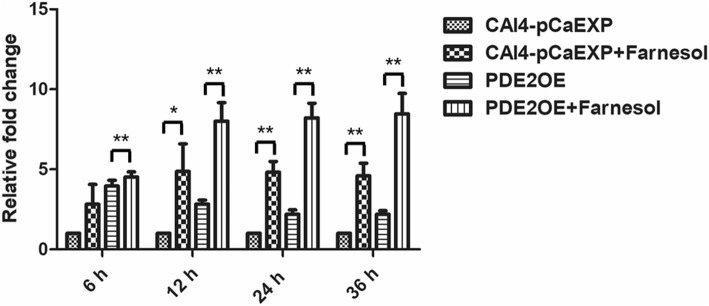
Farnesol decreased CYR1 expression but increased PDE2 expression
The results of RT-qPCR and western blotting showed that farnesol decreased the expression of the gene CYR1 and the protein CYR1 in the biofilms formed from the wild strain (p < 0.01) and the CYR1OE strain (all p < 0.05), compared to the respective untreated controls (Figs. 5 and 6). Farnesol increased the expression of the gene PDE2 and the protein PDE2 in the biofilms formed from the wild strain (p < 0.05) and the PDE2OE strain (all p < 0.01), compared to the respective controls (Figs. 7 and 8).
Fig. 5.
The expression of gene CYR1 in the presence of farnesol. Farnesol decreased the expression of CYR1 in the biofilms of the CAI4-pCaEXP strain at 12 and 24 h biofilm phases and decreased the expression of CYR1 in the biofilms of the CYR1OE strain at 6, 12, 24 and 36 h biofilm phases compared to the respective untreated controls. One-way analysis of variance (ANOVA) was employed to assess the statistical significance of differences in matched groups, while paired t-tests were performed for intra-group comparisons. *: p < 0.05. **: p < 0.01
Fig. 6.
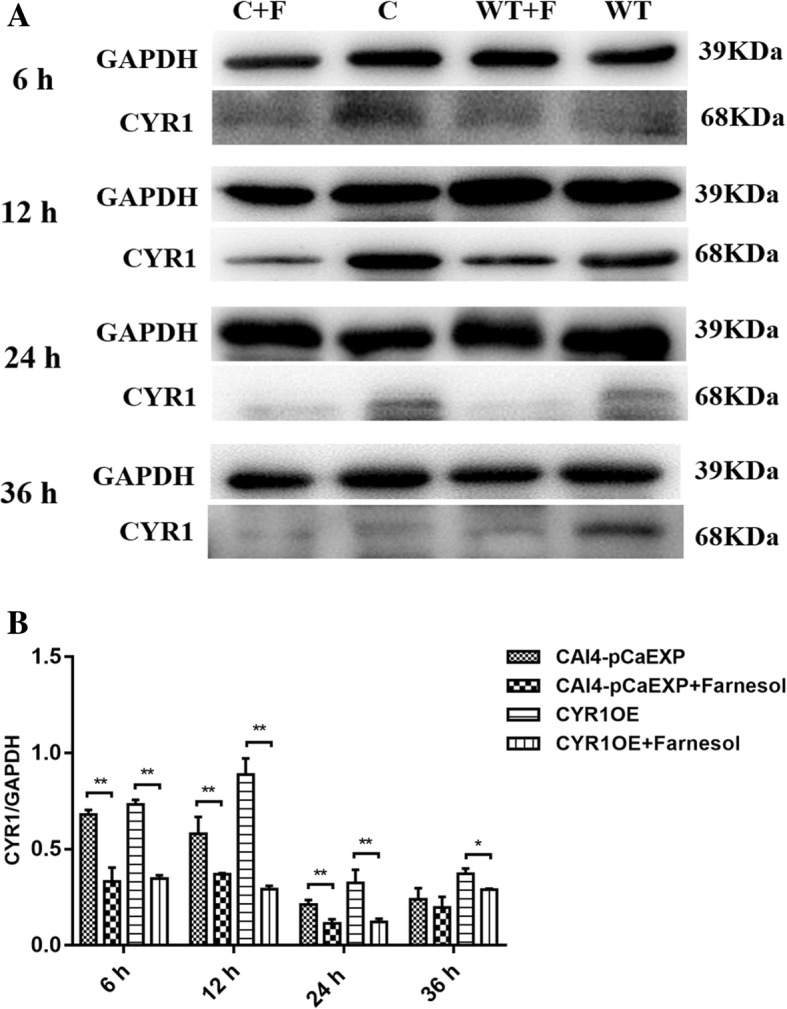
The expression of CYR1 protein in C. albicans biofilms. A: WT: Farnesol untreated wild strain (CAI4-pCaEXP); WT + F: Farnesol treated wild strain (CAI4-pCaEXP); C: Farnesol untreated CYR1-overexpressing strain (CYR1OE); C + F: Farnesol treated CYR1-overexpressing strain (CYR1OE). a and b: Farnesol decreased the expression of CYR1 of the CAI4-pCaEXP strain at 12, 24 and 36 h biofilm phases and decreased the expression of CYR1 of the CYR1OE strain at 6, 12 and 24 h biofilm phases compared to the respective untreated controls. Shown are means ± standard deviation of three independent experiments performed in duplicate. One-way analysis of variance (ANOVA) was employed to assess the statistical significance of differences in matched groups, while paired t-tests were performed for intra-group comparisons. *: p < 0.05. **: p < 0.01
Fig. 7.
The expression of gene PDE2 in the presence of farnesol. Farnesol increased the expression of PDE2 in the biofilms of the CAI4-pCaEXP strain at 12, 24 and 36 h biofilm phases and increased the expression of PDE2 in the biofilms of the PDE2OE strain at 6, 12, 24 and 36 h biofilm phases compared to the respective untreated controls. One-way analysis of variance (ANOVA) was employed to assess the statistical significance of differences in matched groups, while paired t-tests were performed for intra-group comparisons. *: p < 0.05. **: p < 0.01
Fig. 8.
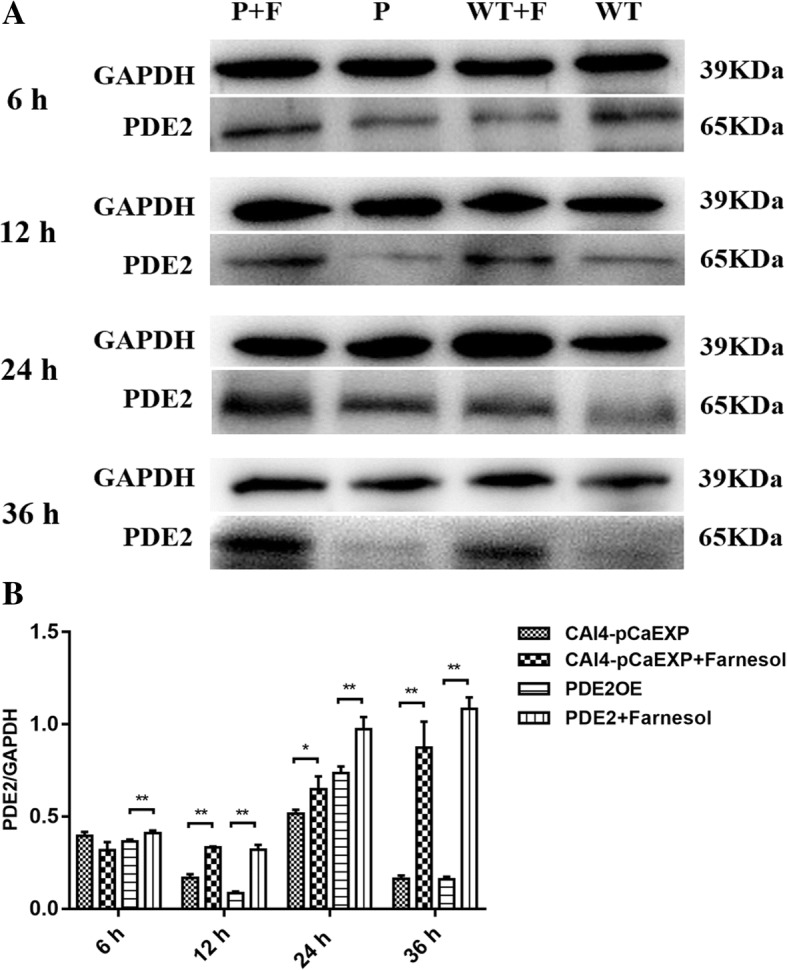
The expression of PDE2 protein in C. albicans biofilms. a: WT: Farnesol untreated wild strain (CAI4-pCaEXP); WT + F: Farnesol treated wild strain (CAI4-pCaEXP); P: Farnesol untreated PDE2-overexpressing strain (PDE2OE); P + F: Farnesol treated PDE2-overexpressing strain (PDE2OE). a and b: Farnesol increased the expression of PDE2 of the CAI4-pCaEXP strain at 12, 24 and 36 h biofilm phases and increased the expression of PDE2 of the PDE2OE strain at 6, 12, 24 and 36 h biofilm phases compared to the respective untreated controls. Shown are means ± standard deviation of three independent experiments performed in duplicate. One-way analysis of variance (ANOVA) was employed to assess the statistical significance of differences in matched groups, while paired t-tests were performed for intra-group comparisons. *: p < 0.05. **: p < 0.01
Discussion
The cAMP pathway regulates the morphological interconversions in C. albicans biofilms and is determined by the processes of synthesis and hydrolysis [16]: adenylyl cyclases (CYR1) catalyse the conversion of ATP to cAMP, while phosphodiesterases (PDE1 and PDE2) hydrolyse cAMP to AMP [22]. The C. albicans CYR1 regulated three developmental programmes, namely, invasive filamentous growth, phenotypic switch, and biofilm formation, while the deletion of CYR1 prevented the growth of hyphae, and block the formation of mature biofilms [23–25]. In addition, previous studies also provided evidence that a C. albicans adenylate cyclase mutant (homozygous deletion) was hyper-susceptible to various antifungals, such as fluconazole, itraconazole, miconazole, terbinafine, fenpropimorph, amphotericin B, and caspofungin [21, 26]. In this study, the data revealed that a CYR1-overexpressing strain had a higher cAMP level and more hyphal growth and biofilm formation than did the wild strain. CYR1 overexpression also increased the resistance of planktonic C. albicans to caspofungin, itraconazole and terbinafine and of sessile C. albicans to amphotericin B. The results confirmed that CYR1 contributed to the resistance and formation of the biofilms, which suggested that CYR1 is associated with the susceptibility of C. albicans biofilms to antifungals.
PDE2, a high-affinity phosphodiesterase, was required for hyphal development and cell wall integrity in C. albicans [16, 27]. The deletion of PDE2 causes elevation of cAMP levels, prohibits normal hyphal development in hypha-inducing liquid medium and the biofilms development [27, 28]. In this study, the PDE2-overexpressing strain had increased hyphal growth and biofilm formation, which is consistent with the finding of a previous study by Jung WH et al. [28] that normal hyphal growth was prohibited in a PDE2-deletion strain. Moreover, a previous study showed that a strain with a PDE2 deletion had decreased resistance of planktonic cells to amphotericin B, flucytosine and fluconazole [20]. Our data further found that PDE2 overexpression increased the resistance of planktonic C. albicans to most antifungals, namely, caspofungin, itraconazole and terbinafine, and of sessile C. albicans to amphotericin B, which suggested that PDE2 is associated with the resistance of C. albicans biofilms to antifungals.
Previous studies showed that farnesol specifically affected the activity of the cAMP signalling pathway and was able to bind to the cyclase domain of the adenylyl cyclase CYR1, exerting a direct effect on intracellular cAMP levels [1, 11, 12]. This study confirmed that farnesol directly inhibited CYR1 activity and decreased the level of intracellular cAMP and hypha formation in the biofilms of the CYR1-overexpressing and wild strains. In addition, farnesol decreased the SMICs of amphotericin B, caspofungin, itraconazole and terbinafine in C. albicans biofilms formed from the CYR1-overexpressing and wild strains. Further, farnesol decreased the expression of the gene CYR1 and the protein CYR1 in those biofilms. The results suggested that farnesol decreased the antifungal resistance of C. albicans by reducing the expression of CYR1.
The study also found that farnesol inhibited hypha formation and decreased the level of intracellular cAMP in the PDE2-overexpressing and wild strains. In addition, farnesol decreased the SMICs of amphotericin B, caspofungin, itraconazole and terbinafine in the biofilms formed from the same strains. In addition, farnesol increased the expression of PDE2 in the biofilm formed from those strains. The results suggested the overexpression of PDE2 regulated by farnesol would not change the inhibitory effect of farnesol on the hyphal growth or the antifungal resistance in the biofilms. Farnesol regulation could decrease the expression of an upstream gene, CYR1, which encodes adenylate cyclase and reduces cAMP level, and even if farnesol increases the expression of a downstream gene, PDE2, and enhances the phosphodiesterase level, there is not enough cAMP to be degraded by the extra phosphodiesterase. The results suggested that PDE2 regulation was subordinate to CYR1 regulation by farnesol. Consistent with this finding, a previous study indicated that the high-affinity cAMP phosphodiesterase PDE2 was not required for repression of hypha formation by farnesol [11]. These data suggested that PDE2 might have a functionality auxiliary to the antifungal specificity of farnesol, which needs to be explored in the future.
It has been generally accepted that hyphal development in C. albicans requires elevated cAMP levels. Intracellular cAMP levels increased during the yeast-to-hypha transition, and the maximum level coincided with maximum germ tube formation [29]. This study showed that the overexpression of CYR1 increases the cAMP levels and the hypha formation of the biofilms, while the overexpression of PDE2 increases the hypha formation without changing the cAMP levels. Jung WH et al. found that Pde2p-catalysed cAMP hydrolysis is required for normal growth and development of hyphae in C. albicans [28], and the deletion of PDE2 causes elevated cAMP levels, severe growth defects and greatly reduced levels of the transcription of an important hypha-associated gene, EFG1 [16, 30]. The results were in accordance with those of a previous study, which suggested that cAMP level regulation by PDE2 may be subordinate to that by CYR1, EFG1 or other downstream genes associated with hyphal growth in C. albicans biofilms.
C. albicans biofilm formation proceeds in three distinct developmental phases: early (0 to 11 h), intermediate (12 to 30 h), and maturation (30 to 72 h) [31]. Farnesol, which can regulate the formation of C. albicans biofilms, decreased the resistance of biofilms grown for 12 h, 24 h and 36 h from CYR1-overexpressing and PDE2-overexpressing strains to itraconazole, while it increased the resistance of biofilms grown for 6 h to itraconazole. The results suggested that phase-specific mechanisms might be involved in the resistance of the biofilms.
Conclusion
In summary, our study demonstrated that CYR1 and PDE2 are associated with the resistance of C. albicans biofilms to antifungals, and farnesol decreased the antifungal resistance of C. albicans biofilms by regulating the expression of the gene CYR1 and PDE2, while PDE2 regulation by farnesol was subordinate to CYR1 regulation by farnesol.
Methods
Strains and media
The strains and plasmids used in this study were provided by the State Key Laboratory of Pharmacy Genetic Engineering, Second Military Medical University, Shanghai, China and are listed in Table 4. The CYR1OE and PDE2OE strains were transformed from CAI4 using CYR1-pCaEXP and PDE2-pCaEXP plasmids, respectively [32–34]. The wild strain CAI4-pCaEXP was transformed from CAI4 using the pCaEXP plasmid. A rich (yeast extract peptone dextrose (YPD) + uridine) (2% peptone, 1% yeast extract, 2% glucose and 0.01% uridine) medium was prepared for wild strain incubation, and a selective (SD-ura-met-cys) medium was prepared for CYR1OE and PDE2OE incubation.
Table 4.
Strains and plasmids used in these studies
| Genotype and Description | reference | |
|---|---|---|
| C. albicans strains | ||
| CAI4 | ura3:: λimm434/ura3:: λimm434 | Fonzi and Irwin (1993) |
| URA3 auxotrophic strain | ||
| CAI4-pCaEXP | ura3:: λimm434/ura3:: λimm434-(pCaEXP URA3) | In this study |
| Wild strain transformed with pCaEXP used as a control of overexpression experiment | ||
| CYR1OE | ura3:: λimm434/ura3:: λimm434-(CYR1-pCaEXP CYR1, URA3) | In this study |
| CYR1-overexpressing strain | ||
| PDE2OE | ura3:: λimm434/ura3:: λimm434-(PDE2-pCaEXP PDE2, URA3) | In this study |
| PDE2-overexpressing strain | ||
| Plasmids | ||
| pCaEXP | URA3 and MET3 promoter integrating plasmid | R. S. Care et al. (1999) |
| CYR1-pCaEXP | Constructed by integration of CYR1 | In this study |
| PDE2-pCaEXP | Constructed by integration of PDE2 | In this study |
Biofilm formation
The strains were grown overnight in 10 ml of YPD + uridine medium at 30 °C in a roller drum [35]. Then, the cells were harvested and diluted into RPMI-1640 medium (Gibco Ltd., Paisley, UK) at an initial concentration of 1 × 106 cells/ml. The suspensions were inoculated into the polystyrene surface of 96-well plates or culture discs (Corning Inc., N.Y., USA) [31]. After 2 h, non-adherent cells were removed by washing twice with sterile phosphate-buffered saline (PBS). The plates were then incubated for 6, 12, 24, or 36 h for the development of biofilms. The media were replenished once every 2 h, and farnesol (Sigma Chemical Co., St. Louis, MO) was added subsequently, depending on the experimental groups.
Susceptibility tests for C. albicans
For the XTT reduction assay for sessile C. albicans, stock solutions of amphotericin B (2.5 mg/ml), caspofungin (10 mg/ml), itraconazole (20 mg/ml) (Sigma-Aldrich, St Louis, MO, USA) and terbinafine (50 mg/ml) (Selleckchem, Houston, TX, USA) were prepared in sterile distilled water (for amphotericin B) or dimethyl sulfoxide (DMSO; for caspofungin, itraconazole and terbinafine). Farnesol was dissolved in 100% (vol/vol) methanol. According to the method M27-A3 from the Clinical and Laboratory Standards Institute (CLSI) (2008), drugs were prepared in serial 2-fold dilutions, and their final concentrations ranged from 0.125 to 16 μg/ml for amphotericin B, 0.0625 to 8 μg/ml for caspofungin, 0.03125 to 16 μg/ml for itraconazole and 0.5 to 256 μg/ml for terbinafine. After incubation with the antifungal agents for 24 h, biofilm SMICs were determined using the XTT (0.25 mg/ml) and menadione (25 μM) assays [36]. The SMIC50 was determined as the minimum antifungal concentrations that caused a ≥ 50% reduction in the metabolic activity of the biofilms compared to that of the control. The tests were repeated in triplicate for each assay, and each group was tested in triplicate on different days.
For the spot assay of planktonic C. albicans [37], the strains were incubated in YPD + uridine medium and grown for 16 h in an orbital shaker at 30 °C. Antifungals were added to the medium at a concentration of 8 μg/ml for amphotericin B, 1 μg/ml for caspofungin, 0.5 μg/ml for itraconazole and 15 μg/ml for terbinafine. For the spot assay, 5 μl of five-fold serial dilutions of yeast suspensions (cells suspended in normal saline to an OD600 nm of 0.1) was spotted onto YPD + uridine plates in the absence or presence of the antifungal drugs [34]. Growth was not affected by the presence of solvent used in the examination. The assay was repeated three times and growth differences were measured after incubation at 30 °C for 24–48 h.
Morphological observation by CLSM and SEM
Biofilms were washed with PBS and fixed with 4% paraformaldehyde. For CLSM observation, biofilms were stained with 500 μl calcofluor white stain [38] (0.0025 g/ml; Sigma Chemical Co. St. Louis, MO) for 30 min at 37 °C in the dark. Then, the biofilms were observed and images were taken using a Zeiss LSM700 microscope (Carl Zeiss, Inc., Oberkochen, Germany) with a 495 nm argon ion laser. The three-dimensional image analysis software ZEN (Carl Zeiss, Inc., Oberkochen, Germany) was used to analyse the thickness of biofilms (depending on the height of the biofilm) formed from different groups. For SEM analysis, biofilms were placed in 2.5% (vol/vol) glutaraldehyde overnight at 4 °C. Samples were dehydrated in a series of washes with increasing concentrations of ethanol (70% for 10 min, 95% for 10 min and 100% for 20 min), dried in a desiccator and coated with gold powder [35, 39]; then, the biofilms were observed using a scanning electron microscope (1530VP, LEO, Oberkochen, Germany). The hyphal length of each germinated spore was measured using the software Image J (1.51, National Institutes of Health, Bethesda, MD, USA). All assays were performed in triplicate on three different occasions to ensure reproducibility and representative images were taken.
Assay of intracellular cAMP
The levels of intracellular cAMP of the biofilms were examined using an ELISA according to the method of Yun et al., with slight modifications [23]. Briefly, biofilms were washed once with water and once with MES buffer (10 mM MES containing 0.1 mM EDTA; pH = 6) and then dried in a desiccator and supplemented with MES buffer with 10% trichloroacetic acid up to the same concentration. The cells were frozen in liquid nitrogen and thawed on ice twice and sonicated under chilled conditions (twice at 130 W for 2 min) using an ultrasonic cell crusher (UH-500B, Celton, Tianjin, China). After centrifugation, trichloroacetic acid was extracted four times with water-saturated ether. The cAMP content was measured with a cAMP enzyme immunoassay system (Amersham, Sigma, USA) following the manufacturer’s instructions. The tests were repeated in triplicate for each assay, and each group was tested in triplicate on different days.
RT-qPCR
Isolation of the total RNA of C. albicans biofilms was carried out using a modified hot phenol method as described previously [40]. Reverse transcription was performed on 1 mg of total RNA using a cDNA synthesis kit (TaKaRa Bio Co., Ltd., Dalian, China) as specified in the manufacturer’s instructions, and the synthesized cDNA was stored at − 80 °C until needed. The synthetic cDNA described above was used for real-time PCR analysis, which was performed on an ABI 7300 Fast Real-time PCR machine (Applied Biosystems, Rotkreuz, Switzerland) using Absolute qPCR SYBR Green Mix (Thermo Scientific, Waltham, MA, USA). Amplification was achieved using the following cycle settings: 10 min at 95 °C followed by 40 cycles of 95 °C for 15 s, 55 °C for 60 s, and 72 °C for 20 s. After the amplification, a melting curve was analysed to ensure the absence of primer dimers. Expression of genes was calculated using the 2-ΔΔCt method [10]. ACT1 was used as a reference gene. The primers were all designed by Shanghai Generay BioTech Co., Ltd. (Table 5). The test was repeated three times.
Table 5.
Primer sequences used in this study
| Primer name | Sequence (5′ → 3′) |
|---|---|
| CYR1-F | AAGACGATGAAACAGCCACA |
| CYR1-R | AGTTGGTAAGCCAGTAGTCGG |
| PDE2-F | ATGCTGTGGGACATTGGAGT |
| PDE2-R | TAAAGTAGTGCCGTTGGTGC |
| ACT1-F | GCCGGTGACGACGCTCCAAGAGCTG |
| ACT1-R | CCGTGTTCAATTGGGTATCTCAAGGTC |
Western blotting
Total protein extracts were prepared from an immunoprecipitation protocol as previously described [41]. Protein samples were mixed with a one-fifth volume of 5 × sample buffer for SDS-PAGE. Samples were boiled for 5 min and separated by SDS-PAGE using an 8–10% acrylamide gel. Proteins were electro-transferred to PVDF membranes (Bio-Rad Laboratories, Inc.) and blocked with 5% skimmed milk in PBS with 0.1% Tween-20. Blots were hybridized with antibodies against C. albicans CYR1 or PDE2 (1:1000), followed by incubation in PBST containing the secondary antibody (1:10000 dilution) for 1 h. GAPDH (1:1000 dilution, Bioworld, Minneapolis, MN, USA) was used as a reference protein. Protein bands were detected by using a chemiluminescence system (Merck & Co., Inc., Kenilworth, NJ, USA). All operations were repeated three times and the gray value were measured with Gel-Pro Analyzer Software (Media Cybernetics, Rockville, MD, USA). The CYR1 and PDE2 level was standardized by the gray value ratio of CYR1/GAPDH and PDE2/GAPDH, respectively.
Statistical analysis
One-way analysis of variance (ANOVA) was employed to assess the statistical significance of differences in matched groups, while paired t-tests were performed for intra-group comparisons. Differences were considered statistically significant at p < 0.05 or p < 0.01. The analyses were performed with SPSS statistics 17.0 software (SPSS Inc., Chicago, IL).
Acknowledgements
Not Applicable.
Funding
This work was supported by the National Natural Sciences Foundation of China under Grant No. 81271151 and No. 81371156 and the Foundation of the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD, 2014–37).
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- cAMP
cyclic AMP
- cDNA
complementary DNA
- CLSI
Clinical and Laboratory Standards Institute
- CLSM
confocal laser scanning electron microscopy
- CYR1OE
CYR1-overexpressing
- DMSO
dimethyl sulfoxide
- ELISA
enzyme-linked immunosorbent assay
- MES
morpholineethanesulfonic acid
- PBS
phosphate-buffered saline
- PBST
phosphate-buffered saline with Tween-20
- PDE2OE
PDE2-overexpressing
- PVDF membrane
microporous membrane of polyvinylidene fluoride
- RT-qPCR
reverse transcription quantitative real-time polymerase chain reaction
- SEM
scanning electron microscopy
- SMIC
sessile minimum inhibitory concentration
- XTT
2,3-bis(2-methoxy-4-nitro-5-sulfophenyl)-5-[(phenylamino)carbonyl]-2H-tetrazolium hydroxide
- YPD
yeast extract peptone dextrose
Authors’ contributions
XW conceived the study, participated in study design and data analysis and was responsible for writing and submitting the final manuscript. SYC carried out the experimental studies relaxed to the XTT assay, spot assay and intracellular cAMP assay; performed statistical analysis and drafted the manuscript. JPX participated in RT-qPCR and western blot analysis and manuscript writing. CSL participated in biofilm formation. LLZ participated in morphological observations by CLSM and SEM. All authors read and approved the manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shengyan Chen, Email: m18851726981@163.com.
Jinping Xia, Email: 17621732585@163.com.
Chengxi Li, Email: 1584494188@qq.com.
Lulu Zuo, Email: 1209847978@qq.com.
Xin Wei, Email: weixinart@163.com.
References
- 1.Lindsay AK, Deveau A, Piispanen AE, Hogan DA. Farnesol and cyclic AMP signaling effects on the hypha-to-yeast transition in Candida albicans. Eukaryot Cell. 2012;11(10):1219–1225. doi: 10.1128/EC.00144-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donlan RM. Biofilm formation: a clinically relevant microbiological process. Clin Infect Dis. 2001;33(8):1387–1392. doi: 10.1086/322972. [DOI] [PubMed] [Google Scholar]
- 3.Wang YC, Lan CY, Hsieh WP, Murillo LA, Agabian N, Chen BS. Global screening of potential Candida albicans biofilm-related transcription factors via network comparison. BMC Bioinformatics. 2010;26(11):53. doi: 10.1186/1471-2105-11-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rajendran R, Sherry L, Nile CJ, Sherriff A, Johnson EM, Hanson MF, Williams C, Munro CA, Jones BJ, Ramage G. Biofilm formation is a risk factor for mortality in patients with Candida albicans bloodstream infection-Scotland, 2012-2013. Clin Microbiol Infect. 2016;22(1):87–93. doi: 10.1016/j.cmi.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ramage G, Saville SP, Wickes BL, Lopez-Ribot JL. Inhibition of Candida albicans biofilm formation by farnesol, a quorum-sensing molecule. Appl Environ Microbiol. 2002;68(11):5459–5463. doi: 10.1128/AEM.68.11.5459-5463.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dižová S, Bujdáková H. Properties and role of the quorum sensing molecule farnesol in relation to the yeast can-dida albicans. Pharmazie. 2017;72(6):307–312. doi: 10.1691/ph.2017.6174. [DOI] [PubMed] [Google Scholar]
- 7.Cordeiro RA, Teixeira CE, Brilhante RS, Castelo-Branco DS, Paiva MA, Giffoni LJ, Lima DT, Monteiro AJ, Sidrim JJ, Rocha MF. Minimum inhibitory concentrations of amphotericin B, azoles and caspofungin against Candida species are reduced by farnesol. Med Mycol. 2013;51(1):53–59. doi: 10.3109/13693786.2012.692489. [DOI] [PubMed] [Google Scholar]
- 8.Katragkou A, McCarthy M, Alexander EL, Antachopoulos C, Meletiadis J, Jabra-Rizk MA, Petraitis V, Roilides E, Walsh TJ. In vitro interactions between farnesol and fluconazole, amphotericin B or micafungin against Candida albicans biofilms. J Antimicrob Chemother. 2015;70(2):470–478. doi: 10.1093/jac/dku374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xia J, Qian F, Xu W, Zhang Z, Wei X. In vitro inhibitory effects of farnesol and interactions between farnesol and antifungals against biofilms of Candida albicans resistant strains. Biofouling. 2017;33(4):283–293. doi: 10.1080/08927014.2017.1295304. [DOI] [PubMed] [Google Scholar]
- 10.Yu LH, Wei X, Ma M, Chen XJ, Xu SB. Possible inhibitory molecular mechanism of farnesol on the development of fluconazole resistance in Candida albicans biofilm. Antimicrob Agents Chemother. 2012;56(2):770–775. doi: 10.1128/AAC.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis-Hanna A, Piispanen AE, Stateva LI, Hogan DA. Farnesol and dodecanol effects on the Candida albicans Ras1-cAMP signalling pathway and the regulation of morphogenesis. Mol Microbiol. 2008;67(1):47–62. doi: 10.1111/j.1365-2958.2007.06013.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall RA, Turner KJ, Chaloupka J, Cottier F, De Sordi L, Sanglard D, Levin LR, Buck J, Muhlschlegel FA. The quorum-sensing molecules farnesol/homoserine lactone and dodecanol operate via distinct modes of action in Candida albicans. Eukaryot Cell. 2011;10(8):1034–1042. doi: 10.1128/EC.05060-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cao YY, Cao YB, Xu Z, Ying K, Li Y, Xie Y, Zhu ZY, Chen WS, Jiang YY. cDNA microarray analysis of differential gene expression in Candida albicans biofilm exposed to farnesol. Antimicrob Agents Chemother. 2005;49(2):584–589. doi: 10.1128/AAC.49.2.584-589.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H. Transcriptional control of dimorphism in Candida albicans. Curr Opin Microbiol. 2001;4(6):728–735. doi: 10.1016/S1369-5274(01)00275-2. [DOI] [PubMed] [Google Scholar]
- 15.Nigg M, Bernier L. From yeast to hypha: defining transcriptomic signatures of the morphological switch in the dimorphic fungal pathogen Ophiostoma novo-ulmi. BMC Genomics. 2016;17(1):920. doi: 10.1186/s12864-016-3251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Park JI, Grant CM, Dawes IW. The high-affinity cAMP phosphodiesterase of Saccharomyces cerevisiae is the major determinant of cAMP levels in stationary phase: involvement of different branches of the Ras-cyclic AMP pathway in stress responses. Biochem Biophys Res Commun. 2005;327(1):311–319. doi: 10.1016/j.bbrc.2004.12.019. [DOI] [PubMed] [Google Scholar]
- 17.Bahn YS, Staab J, Sundstrom P. Increased high-affinity phosphodiesterase PDE2 gene expression in germ tubes counteracts CAP1-dependent synthesis of cyclic AMP, limits hypha production and promotes virulence of Candida albicans. Mol Microbiol. 2003;50(2):391–409. doi: 10.1046/j.1365-2958.2003.03692.x. [DOI] [PubMed] [Google Scholar]
- 18.Bai C, Xu XL, Wang HS, Wang YM, Chan FY, Wang Y. Characterization of a hyperactive Cyr1 mutant reveals new regulatory mechanisms for cellular cAMP levels in Candida albicans. Mol Microbiol. 2011;82(4):879–893. doi: 10.1111/j.1365-2958.2011.07859.x. [DOI] [PubMed] [Google Scholar]
- 19.Xie JL, O'Meara TR, Polvi EJ, Robbins N, Cowen LE. Staurosporine induces Filamentation in the human fungal pathogen Candida albicans via signaling through Cyr1 and protein kinase a. mSphere. 2017;1(2):2. doi: 10.1128/mSphere.00056-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jung WH, Warn P, Ragni E, Popolo L, Nunn CD, Turner MP, Stateva L. Deletion of PDE2, the gene encoding the high-affinity cAMP phosphodiesterase, results in changes of the cell wall and membrane in Candida albicans. Yeast. 2005;22:285–294. doi: 10.1002/yea.1199. [DOI] [PubMed] [Google Scholar]
- 21.Jain P, Akula I, Edlind T. Cyclic AMP signaling pathway modulates susceptibility of candida species and Saccharomyces cerevisiae to antifungal azoles and other sterol biosynthesis inhibitors. Antimicrob Agents Chemother. 2003;47(10):3195–3201. doi: 10.1128/AAC.47.10.3195-3201.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hogan DA, Muhlschlegel FA. Candida albicans developmental regulation: adenylyl cyclase as a coincidence detector of parallel signals. Curr Opin Microbiol. 2011;14(6):682–686. doi: 10.1016/j.mib.2011.09.014. [DOI] [PubMed] [Google Scholar]
- 23.Yun CW, Tamaki H, Nakayama R, Yamamoto K, Kumagai H. Gpr1p, a putative G-protein coupled receptor, regulates glucose-dependent cellular cAMP level in yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1998;252(1):29–33. doi: 10.1006/bbrc.1998.9600. [DOI] [PubMed] [Google Scholar]
- 24.Böttcher B, Pöllath C, Staib P, Hube B, Brunke S. Candida species rewired hyphae developmental programs for Chlamydospore formation. Front Microbiol. 2016;27(7):1697. doi: 10.3389/fmicb.2016.01697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao C, Wu M, Bing J, Tao L, Ding X, Liu X, Huang G. Global regulatory roles of the cAMP/PKA pathway revealed by phenotypic, transcriptomic and phosphoproteomic anal-yses in a null mutant of the PKA catalytic subunit in Candida albicans. Mol Microbiol. 2017;105(1):46–64. doi: 10.1111/mmi.13681. [DOI] [PubMed] [Google Scholar]
- 26.Wenwen S, Liuping Z, Xiaoyan L, Lei F, Sun S. The synergistic antifungal effects of sodium phenylbutyrate combined with azoles against Candida albicans via regulating Ras/cAMP/PKA signalling pathway and virulence. Can J Microbiol. 2018. 10.1139/cjm-2018-0337. [DOI] [PubMed]
- 27.Wilson D, Tutulan-Cunita A, Jung W, Hauser NC, Hernandez R, Williamson T, Piekarska K, Rupp S, Young T, Stateva L. Deletion of the high-affinity cAMP phosphodiesterase encoded by PDE2 affects stress responses and virulence in Candida albicans. Mol Microbiol. 2007;65(4):841–856. doi: 10.1111/j.1365-2958.2007.05788.x. [DOI] [PubMed] [Google Scholar]
- 28.Jung WH, Stateva LI. The cAMP phosphodiesterase encoded by CaPDE2 is required for hyphal development in Candida albicans. Microbiology. 2003;149(Pt 10):2961–2976. doi: 10.1099/mic.0.26517-0. [DOI] [PubMed] [Google Scholar]
- 29.Sabie FT, Gadd GM. Effect of nucleosides and nucleotides and the relationship between cellular adenosine 3′:5′-cyclic monophosphate (cyclic AMP) and germ tube formation in Candida albicans. Mycopathologia. 1992;119(3):147–156. doi: 10.1007/BF00448812. [DOI] [PubMed] [Google Scholar]
- 30.Schaekel A, Desai PR, Ernst JF. Morphogenesis-regulated localization of protein kinase a to genomic sites in Candida albicans. BMC Genomics. 2013;14(1):842. doi: 10.1186/1471-2164-14-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra J, Kuhn DM, Mukherjee PK, Hoyer LL, McCormick T, Ghannoum MA. Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J Bacteriol. 2001;183(18):5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leberer E, Harcus D, Dignard D, Johnson L, Ushinsky S, Thomas DY, Schroppel K. Ras links cellular morphogenesis to virulence by regulation of the MAP kinase and cAMP signalling pathways in the pathogenic fungus Candida albicans. Mol Microbiol. 2001;42(3):673–687. doi: 10.1046/j.1365-2958.2001.02672.x. [DOI] [PubMed] [Google Scholar]
- 33.Care RS, Trevethick J, Binley KM, Sudbery PE. The MET3 promoter: a new tool for Candida albicans molecular genetics. Mol Microbiol. 1999;34(4):792–798. doi: 10.1046/j.1365-2958.1999.01641.x. [DOI] [PubMed] [Google Scholar]
- 34.Fu MS, De Sordi L, Muhlschlegel FA. Functional characterization of the small heat shock protein Hsp12p from Candida albicans. PLoS One. 2012;7(8):e42894. doi: 10.1371/journal.pone.0042894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reuss O, Vik A, Kolter R, Morschhauser J. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene. 2004;341:119–127. doi: 10.1016/j.gene.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 36.Kuhn DM, George T, Chandra J, Mukherjee PK, Ghannoum MA. Antifungal susceptibility of Candida biofilms: unique efficacy of amphotericin B lipid formulations and echinocandins. Antimicrob Agents Chemother. 2002;46(6):1773–1780. doi: 10.1128/AAC.46.6.1773-1780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ansari M. A., Z. Fatima, and S. Hameed. Antifungal action of methylene blue involves mitochondrial dysfunction and disruption of redox and membrane homeostasis in C. albicans. Open Microbiol J. 2016;10:12–22. doi: 10.2174/1874285801610010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nett J, Lincoln L, Marchillo K, Andes D. Beta −1,3 glucan as a test for central venous catheter biofilm infection. J Infect Dis. 2007;195:1705–1712. doi: 10.1086/517522. [DOI] [PubMed] [Google Scholar]
- 39.Atshan SS, Shamsudin MN, Lung LT, Sekawi Z, Ghaznavi-Rad E, Pei CP. Comparative characterisation of genotypically different clones of MRSA in the production of biofilms. J Biomed Biotechnol. 2012;2012:417247. doi: 10.1155/2012/417247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taskova RM, Zorn H, Krings U, Bouws H, Berger RG. A comparison of cell wall disruption techniques for the isolation of intracellular metabolites from Pleurotus and Lepista sp. Z Naturforsch C. 2006;61(5–6):347–350. doi: 10.1515/znc-2006-5-608. [DOI] [PubMed] [Google Scholar]
- 41.Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS Pathog. 2009;5(7):e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and analysed during the current study are available from the corresponding author upon reasonable request.