Abstract
Background
Imprinted genes are regulated by DNA methylation at imprinting-associated differentially methylated regions (iDMRs). Abnormal expression of imprinted genes is implicated in imprinting disorders and tumors. In colorectal cancer (CRC), methylation and imprinting status of the IGF2/H19 domain have been studied. However, no comprehensive methylation analysis of iDMRs in CRC has been reported. Furthermore, the relationship between iDMR methylation status and other methylation-related issues, such as CpG island methylator phenotype (CIMP) and long interspersed element-1 (LINE-1) methylation, remains unclear.
Results
We analyzed the methylation status of 38 iDMRs in 106 CRC patients. We also investigated CIMP, LINE-1 methylation, KRAS and BRAF gene mutations, and loss of imprinting (LOI) of IGF2. We further examined the relationship between these factors and clinicopathological factors. The overall trend in iDMR methylation was towards hypermethylation, and iDMRs could be grouped into three categories: susceptible, resistant, and intermediate-to-aberrant methylation. The susceptible and resistant iDMRs consisted of all types of iDMR (gametic and somatic, maternally and paternally methylated). Hypermethylation of multiple iDMRs (HyMiD)-positive status was statistically associated with CIMP-positive status, but not associated with mutations in the BRAF and KRAS genes. HyMiD-positive status was inversely associated with LINE-1 methylation. Among four iDMRs within the IGF2/H19 domain, IGF2-DMR0 hypomethylation occurred most frequently, but was not associated with IGF2 LOI. Finally, we statistically calculated predictive prognostic scores based on aberrant methylation status of three iDMRs.
Conclusion
In CRC tissues, some iDMRs were susceptible to hypermethylation independent of the type of iDMR and genomic sequence. Although HyMiD-positive status was associated with CIMP-positive status, this was independent of the BRAF and KRAS pathways, which are responsible for CIMP. Since IGF2-DMR0 hypomethylation and aberrant methylation of other iDMRs within the IGF2/H19 domain were not associated with IGF2 LOI, dysfunction of any of the molecular components related to imprinting regulation may be involved in IGF2 LOI. The prognostic score calculated based on aberrant methylation of three iDMRs has potential clinical applications as a prognostic predictor in patients. Further study is required to understand the biological significance of, and mechanisms behind, aberrant methylation of iDMRs and IGF2 LOI in CRCs.
Electronic supplementary material
The online version of this article (10.1186/s13148-018-0578-9) contains supplementary material, which is available to authorized users.
Keywords: Genomic imprinting, DNA methylation, iDMR, CIMP, BRAF mutation, KRAS mutation, LINE-1, IGF2-DMR0, IGF2 LOI
Background
Genomic imprinting is an epigenetic phenomenon resulting in differential expression of a subset of mammalian genes in a parent-specific manner. Generally, imprinted genes form clusters in certain chromosomal regions called imprinting domains. The expression of imprinted genes is regulated by epigenetic mechanisms, especially DNA methylation, at imprinting-associated differentially methylated regions (iDMRs) within these domains [1, 2]. iDMRs are characterized by DNA methylation on one of the two parental alleles. iDMRs are classified as maternally methylated and paternally methylated, as well as gametic or somatic. In gametic iDMRs, DNA methylation is acquired during gametogenesis and maintained from zygote to somatic cell. Most gametic iDMRs are present in imprinting control regions (ICRs), while somatic iDMRs are established during early embryogenesis after fertilization under the control of nearby ICRs [1]. Many imprinted genes play important roles in cell growth and differentiation, embryogenesis, fetal development, placental formation, and metabolism [3]. Loss of imprinting (LOI), or loss of monoallelic expression due to aberrant DNA methylation at iDMRs, is implicated in imprinting disorders such as Beckwith-Wiedemann syndrome, Angelman syndrome, and Prader-Willi syndrome [2, 4, 5].
Abnormal expression of imprinted genes is also implicated in the pathogenesis of cancers, because many imprinted genes regulate cell growth and differentiation [6]. IGF2 LOI occurs in childhood tumors such as Wilms tumor, hepatoblastoma, and rhabdomyosarcoma, as well as in a majority of adult tumors, such as prostate, breast, lung, colorectal, and liver cancer [7]. IGF2 LOI in Wilms tumor and hepatoblastoma is reportedly correlated with hypermethylation of H19-DMR, which is the ICR in the IGF2/H19 domain [8, 9]. Disrupted expression of other imprinted genes has also been reported in various other cancers. For instance, in esophageal cancer cell lines, silencing of CDKN1C was correlated with hypomethylation of KvDMR1, which is the ICR in the KCNQ1OT1 domain [10]. Hypermethylation of ARHI and PEG3 iDMRs, which are imprinted tumor suppressor genes, is associated with downregulation of these genes in ovarian cancer [11]. It has also been proposed that the expression and methylation status of several imprinted genes are associated with clinicopathological factors. For example, IGF2-DMR0 hypomethylation is associated with shorter survival in colorectal cancer (CRC) patients [12]. ARHI expression is associated with prolonged disease-free survival in epithelial ovarian cancer and pancreatic endocrine tumor patients [13, 14].
CRC is the third most common cancer globally in men and the second most common in women [15]. Among imprinted domains, the IGF2/H19 domain has been well studied in CRC. It was reported that IGF2 LOI was associated with IGF2-DMR0 hypomethylation, but not H19-DMR hypermethylation [16]. IGF2 LOI in lymphocytes has been suggested as a potential risk predictor in CRC [17], while a different study contested the association between IGF2-DMR0 hypomethylation and IGF2 LOI in CRC [18]. Additionally, the association was not detected in osteosarcoma, ovarian, or breast cancer [19–21]. The relationship between IGF2 LOI and IGF2-DMR0 hypomethylation thus remains unclear. To date, no comprehensive methylation analysis of iDMRs in CRC has been carried out. In this study, we comprehensively analyzed the methylation status of 38 iDMRs in tumors and adjacent normal mucosae from 106 CRC patients. We also investigated CpG island methylator phenotype (CIMP) status, long interspersed nuclear element-1 (LINE-1) methylation, mutations in BRAF and KRAS, and allelic expression of IGF2. Finally, we examined the relationship between these genetic and epigenetic factors and clinicopathological factors in CRC.
Results
CRC patient clinical information
A total of 106 CRC patients were included in this study. Patients with metastases, preoperative chemotherapy, and inflammatory bowel disease were excluded. We analyzed paired frozen tumor tissues and adjacent normal mucosae. Patient clinical information is provided in Additional file 1: Table S1. Sub-division of the groups by site of tumor occurrence (right colon, left colon, or rectum) revealed no statistically significant differences in terms of age at diagnosis, sex, cancer stage, lymph node metastasis, or pathological type (Additional file 1: Table S1).
Methylation status of 38 iDMRs
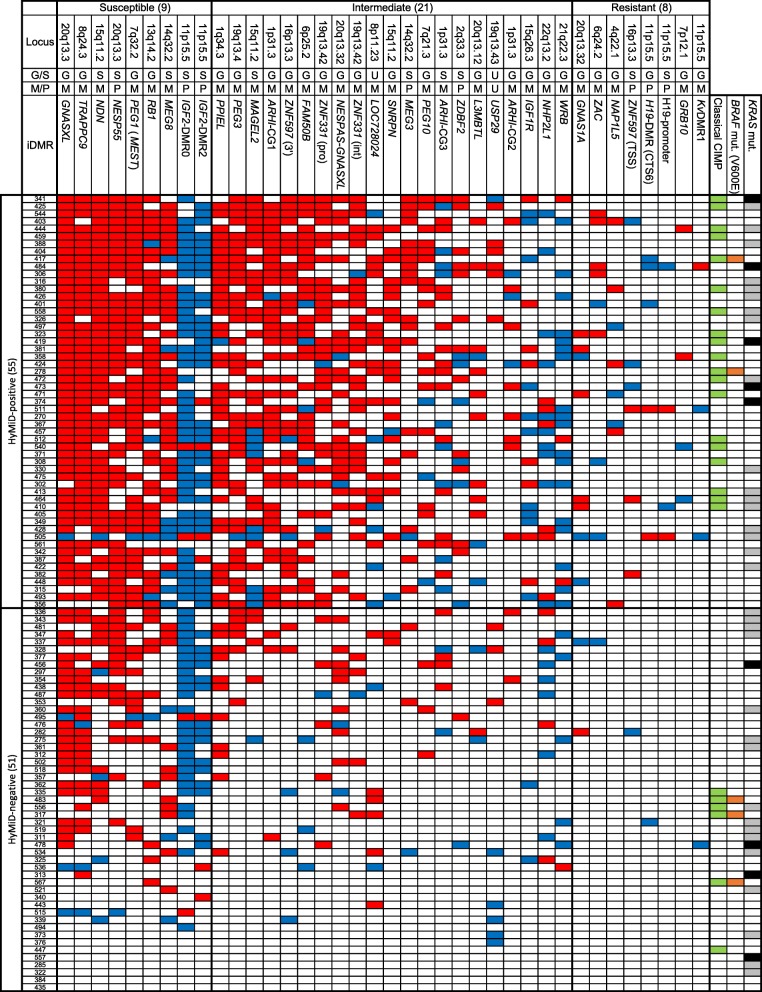
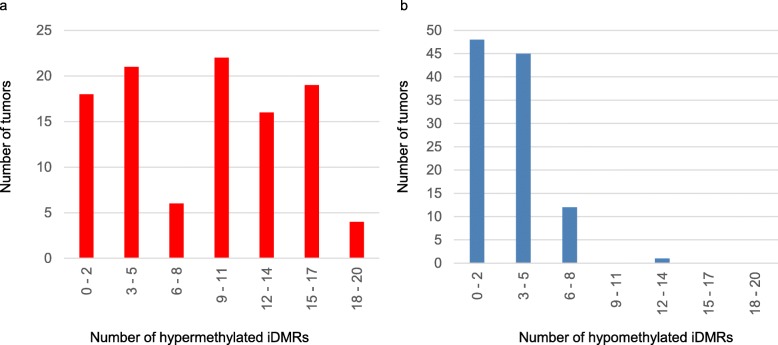
We performed a preliminary assessment of methylation status of 45 iDMRs, including iDMRs identified by Court et al. [22], in all adjacent normal mucosae by bisulfite pyrosequencing. We found that 37 iDMRs were differentially methylated with a high degree of precision (standard deviation < 10% of methylation). IGF2-DMR2, which was hypermethylated (87% methylation) in normal mucosae, was included because of its location within the IGF2 gene. Therefore, we analyzed 38 iDMRs in this study. The methylation statuses of all iDMRs in tumor tissues were summarized into a qualitative chart (Fig. 1) (see the “Methods” section for the definition of aberrant methylation). The median number of hypermethylated iDMRs per tumor was 10 (ranging from 0 to 19), and of hypomethylated iDMRs was 3 (ranging from 0 to 12), indicating that hypermethylation of iDMRs is more likely in CRC tissues (Fig. 2a, b). In addition, we found that iDMRs could be grouped into three categories based on their susceptibility to aberrant methylation (Fig. 1). The first group was susceptible and consisted of nine iDMRs aberrantly methylated in half of all tumors or more (53 or more) (Fig. 1). Seven of these (GNASXL, TRAPPC9, NDN, NESP55, PEG1, RB1, and MEG8) were predominantly hypermethylated, although IGF2-DMR0 and IGF2-DMR2 were predominantly hypomethylated. The second group exhibited resistance to aberrant methylation and consisted of eight iDMRs (GNAS1A, ZAC, NAP1L5, ZNF597 (TSS), H19-DMR, H19-promoter, GRB10, and KvDMR1) that were aberrantly methylated in less than 10% of tumors (under 11) (Fig. 1). The last group was an intermediate group consisting of the remaining 21 iDMRs (Fig. 1). These findings indicate that susceptibility to aberrant methylation varies according to the individual iDMR and that certain iDMRs were highly susceptible to aberrant methylation while others were resistant. The susceptible group as well as the resistant group consisted of both gametic and somatic iDMRs as well as both maternally and paternally methylated iDMRs. In addition, we could not find a consensus sequence among the representative five hypermethylated iDMRs (GNASXL, TRAPPC9, NDN, NESP55, and PEG1). These results suggest that susceptibility to aberrant methylation does not depend on iDMR type or genomic sequence.
Fig. 1.
Methylation statuses of 38 iDMRs, CIMP status, and BRAF and KRAS mutations in 106 CRC patients. The three-digit number in the leftmost column indicates each patient’s ID. Hypermethylation and hypomethylation of iDMR are indicated by red and blue, respectively. CIMP-positive samples are indicated in green in the column on the right. The BRAF mutation (V600E) is indicated in orange. The KRAS (G13D) mutation and other KRAS mutations are indicated in black and gray, respectively. G, gametic iDMR; S, somatic iDMR; M, maternally methylated iDMR; P, paternally methylated iDMR; U, unidentified; ZNF597 (TSS), transcription start site region of ZNF597; ZNF597 (3′), 3′ region of ZNF597; ZNF331 (pro), promoter region of ZNF331; ZNF331 (int), intron of ZNF331. Definitions of aberrant methylation of iDMR and CIMP-positive are provided in the “Methods” section
Fig. 2.
Frequency of iDMR aberrant methylation. a The distribution of hypermethylated iDMRs in tumors by number. b The distribution of hypomethylated iDMRs in tumors by number. The data indicate that iDMRs are more susceptible to hypermethylation than hypomethylation in CRC tissues
Relationship between hypermethylation of multiple iDMRs (HyMiD) and CIMP
Because the median number of hypermethylated iDMRs per tumor detected in our study was 10, we defined tumors with hypermethylation in 10 or more iDMRs as hypermethylation of multiple iDMRs (HyMiD)-positive (55 tumors) (Fig. 1). We further defined tumors with hypermethylation in less than 10 iDMRs as HyMiD-negative (51 tumors). In addition, CpG island methylator phenotype (CIMP) is a well-known indicator of widespread CpG island promoter hypermethylation, first introduced by Toyota et al. [23]. Therefore, we analyzed CIMP status in all tumor tissues with five classical CIMP markers (hMLH1, MINT1, MINT2, MINT31, p16) using bisulfite pyrosequencing. We found that 25 tumors (24%) could be classified as CIMP-positive, which was defined as having at least two methylated markers among the five (Fig. 1). Comparison of HyMiD status with CIMP status revealed that HyMiD-positive status was correlated with CIMP-positive status (p = 0.005) (Table 1).
Table 1.
Relationship between HyMiD and CIMP
| CIMP-positive | CIMP-negative | p value | |
|---|---|---|---|
| HyMiD-positive | 19 | 36 | 0.005 |
| HyMiD-negative | 6 | 45 |
Fisher’s exact test
Relationship between HyMiD status and BRAF or KRAS mutations
It was recently reported that the activated BRAF (V600E)-directed MAFG and activated KRAS (G13D)-directed ZNF304 pathways were responsible for CIMP in CRC [24, 25]. It was also reported that the BRAF and KRAS mutations were associated with CIMP-high and CIMP-low CRC, respectively [26–28]. The correlation between HyMiD-positive and CIMP-positive statuses therefore indicated the possibility of an association between HyMiD-positive status and the above two CIMP pathways. We explored the BRAF and KRAS mutations (codon 12, 13, 59, and 61) in tumor tissues. The BRAF mutation was identified in 5 tumors (4.7%), and KRAS mutations were identified in 42 tumors (39.6%) (Fig. 1 and Additional file 1: Table S2). CIMP-positive status was correlated with the BRAF mutation but not with KRAS mutations, including G13D (Additional file 1: Table S3). The lack of correlation between CIMP-positive status and KRAS mutations was an unanticipated result, but consistent with previous reports [29]. This discrepancy may be due to differing definitions of CIMP-positive and CIMP-high (as opposed to CIMP-low) [27, 28]. On the other hand, HyMiD-positive status was correlated with neither the BRAF mutation nor KRAS mutation (Table 2). Additionally, we investigated MAFG consensus binding sites in all iDMRs and found that few were present (Additional file 1: Table S4), supporting non-correlation of HyMiD-positive status with the BRAF mutation. These results suggest that the BRAF-directed MAFG and KRAS-directed ZNF304 pathways were not responsible for HyMiD-positive status in CRC.
Table 2.
Relationship between HyMiD status and BRAF or KRAS mutation
| HyMiD-positive | HyMiD-negative | p value | |
|---|---|---|---|
| BRAF mut (+) | 2 | 3 | 0.464 |
| BRAF mut (−) | 53 | 48 | |
| KRAS mut (+) | 22 | 20 | 0.546 |
| KRAS mut (−) | 33 | 31 | |
| KRAS G13D (+) | 5 | 4 | 0.548 |
| KRAS G13D (−) | 50 | 47 |
BRAF mut BRAF mutation (V600E), KRAS mut all KRAS mutations
Fisher’s exact test
Relationship between HyMiD status and LINE-1 methylation
The methylation level of LINE-1 retrotransposons, which constitute approximately 20% of the human genome, is a good surrogate marker of genome-wide methylation [30, 31]. LINE-1 methylation levels were investigated in tumor tissues by bisulfite pyrosequencing, and the relationship with HyMiD status was examined. The LINE-1 methylation level in HyMiD-positive tumors (median 55.1%, 22.1–64.8%) was significantly lower than in HyMiD-negative tumors (median 60.2%, 43.7–72.1%) (p = 0.0001), indicating an inverse relationship between HyMiD status and LINE-1 methylation (Fig. 3). LINE-1 methylation did not differ between CIMP-positive and CIMP-negative CRC (Additional file 1: Figure S1).
Fig. 3.
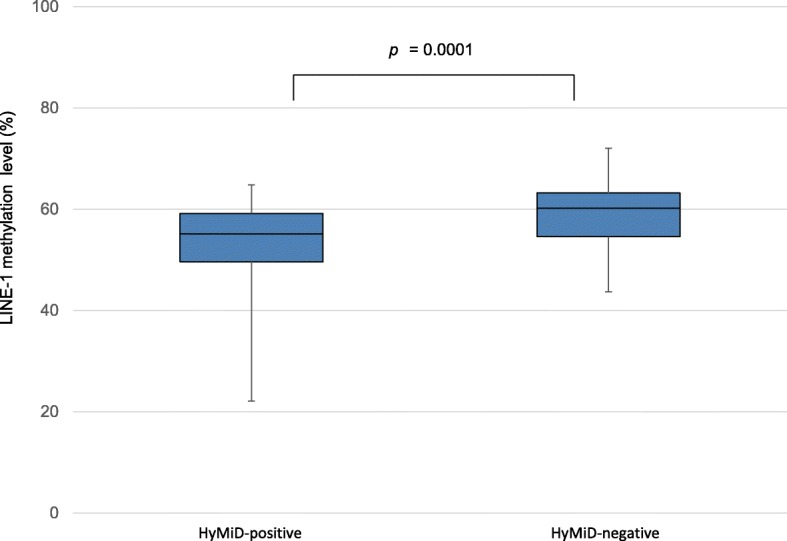
Inverse relationship between HyMiD status and LINE-1 methylation. LINE-1 methylation in HyMiD-positive tumors (median 55.1%, 22.1–64.8%) was significantly lower than in HyMiD-negative tumors (median 60.2%, 43.7–72.1%), indicating that an HyMiD-positive status was inversely correlated with LINE-1 methylation in CRC tissues. (p = 0.0001, Mann-Whitney U test)
Methylation status of iDMRs within the IGF2/H19 domain
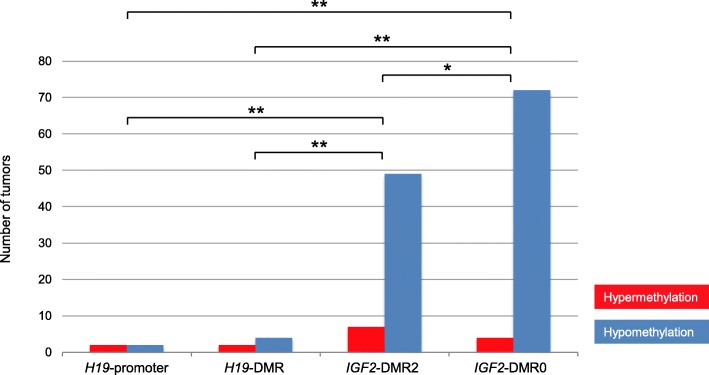
Among imprinting domains, the IGF2/H19 domain has been particularly studied in CRC patients. Although four iDMRs (H19-DMR, H19-promoter, IGF2-DMR0, and IGF2-DMR2) are located within this domain, only a few studies have analyzed all of these iDMRs simultaneously. Therefore, we analyzed the methylation status of all iDMRs within the IGF2/H19 domain. Methylation of H19-DMR and H19-promoter, which belong to the resistant iDMR group, was nearly unchanged (Figs. 1 and 4). Only two tumors were hypermethylated at both H19-DMR and H19-promoter. Four were hypomethylated at H19-DMR, and two were hypomethylated at H19-promoter. Conversely, hypomethylation at IGF2-DMR0 and IGF2-DMR2 was most frequently observed in 72 (67.9%) and 49 tumors (44.2%), respectively, as the two iDMRs belonged to the susceptible iDMR group (Figs. 1 and 4). Only four tumors were hypermethylated at IGF2-DMR0, and seven were hypermethylated at IGF2-DMR2. Hypomethylation of the IGF2-DMRs, especially DMR0, occurred at strikingly high frequency (Fig. 4).
Fig. 4.
Aberrant methylation of four iDMRs within the IGF2/H19 domain in tumors. Hypermethylation of the H19-promoter and H19-DMR were found in only a few samples. However, hypomethylation of IGF2-DMRs, especially DMR0, was comparatively overrepresented among iDMRs (*p = 0.0061, **p < 0.0001, χ2 test)
Allelic expression of IGF2 and H19
We next analyzed the allelic expression of IGF2 and H19. For this purpose, we searched for single nucleotide polymorphisms (SNPs) among the samples. We found that 39 and 40 patients were heterozygous for IGF2 (rs680) and H19 (rs2839702), respectively. Allelic expression was analyzed quantitatively in the heterozygous samples by pyrosequencing. There were four possible patterns of allelic IGF2 expression in tumor tissue compared to matched adjacent normal mucosa: maintenance of imprinting (MOI) in both tissues, LOI in both tissues, LOI in tumor and MOI in normal mucosa, and MOI in tumor and LOI in normal mucosa. We found all patterns, and there was no significant difference in the number of sample pairs in each group (Additional file 1: Table S5). We further examined the relationship between IGF2 LOI and IGF2-DMR0 hypomethylation in tumors and found no correlation (p = 0.901) (Table 3). Likewise, no correlation was found between IGF2 LOI and IGF2-DMR2 hypomethylation (Additional file 1: Table S6). Thus, it is unlikely that hypomethylation of IGF2-DMRs caused IGF2 LOI. As for H19, all tumor tissues except for one (ID 511) showed monoallelic expression, despite variation in iDMR methylation, strongly suggesting an absence of correlation between allelic expression of H19 and aberrant methylation of iDMRs within the IGF2/H19 domain.
Table 3.
Relationship between IGF2-DMR0 hypomethylation and IGF2 LOI in tumor
| Number of MOI cases | Number of LOI cases | p value | ||
|---|---|---|---|---|
| IGF2-DMR0 hypomethylation | (+) | 9 | 21 | 0.901 |
| (−) | 5 | 6 |
MOI maintenance of imprinting, LOI loss of imprinting
Fisher’s exact test
Relationship between aberrant methylation of iDMRs or IGF2 LOI and prognosis
Since an association between IGF2-DMR0 hypomethylation and poor prognosis in CRC patients was previously reported [12], we also examined the relationship between clinicopathological factors predicting outcome and HyMiD status, IGF2-DMR0 hypomethylation, and IGF2 LOI. However, we did not observe any correlations between the epigenetic features and clinicopathological factors (Additional file 1: Tables S7, S8, and S9).
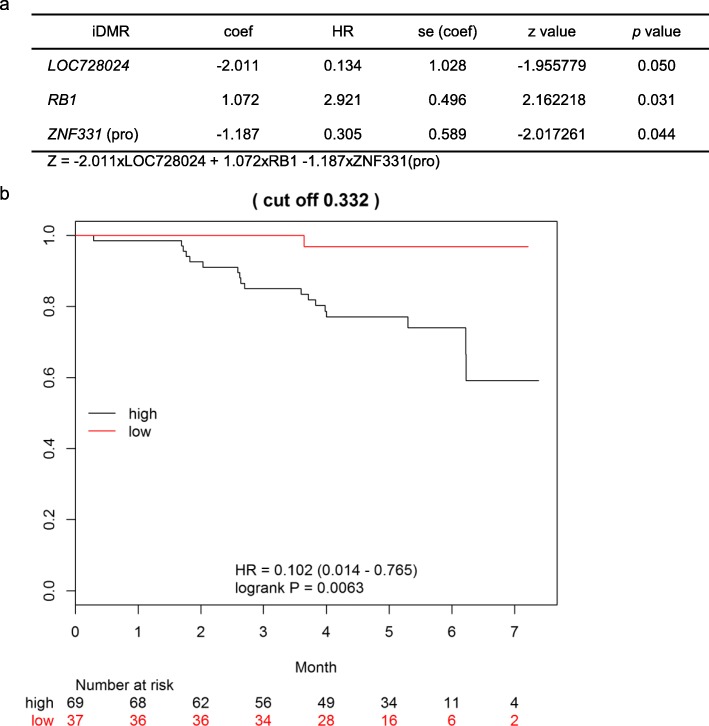
We next analyzed the prognostic value of scores determined using a Cox proportional hazards regression model with stepwise selection. The prognosis score was computed as Z = − 2.011 × LOC728024 + 1.072 × RB1 – 1.187 × ZNF331-promoter. When aberrant methylation (hyper- or hypo-) at LOC728024, RB1, or ZNF331-promoter is found, the corresponding term is assigned a value of 1. The optimal cutoff value is 0.332, and HR = 0.102 (95% CI 0.014–0.765, p = 0.0063) between groups with Z ≥ 0.332 and Z < 0.332 (Fig. 5). The result of this analysis indicated that aberrant methylation of the three iDMRs was correlated with the patients’ prognosis and that the Z score was a potential prognostic marker.
Fig. 5.
Cox proportional hazards regression analysis. a Model fitting summary. The prognosis score was computed as Z = − 2.011 × LOC728024 + 1.072 × RB1 – 1.187 × ZNF331-promoter. When aberrant methylation (hyper- or hypo-) at LOC728024, RB1, or ZNF331-promoter is found, the corresponding term is assigned a value of 1. coef: coefficient; se: standard error. z value = coef/se(coef). b Kaplan-Meier survival estimates. High prognostic scores (Z ≥ 0.332) were significantly associated with poor clinical prognosis compared low scores (Z < 0.332) (p = 0.0063, log-rank test, HR = 0.102 (95% CI 0.014–0.765))
Discussion
In this study, we found that iDMRs were more susceptible to hypermethylation than hypomethylation in CRC tumor tissues and that the seven iDMRs were highly susceptible to aberrant hypermethylation. A recent report by Kim et al., using The Cancer Genome Atlas (TCGA) datasets from 22 cancer types, indicated that the direction of aberrant methylation is skewed towards hypermethylation and that hypermethylation of PEG3, DLK1, PEG1, and GNAS occurred more frequently [32]. The hypermethylated iDMRs identified by Kim et al. were ICRs, and almost entirely gametic iDMRs except for DLK1, whereas the hypermethylated iDMRs identified in the present study consisted of four gametic iDMRs and three somatic iDMRs. Furthermore, we could not find any consensus sequences among the five hypermethylated iDMRs. These results suggest that not only gametic DMRs but also somatic DMRs are aberrantly hypermethylated independent of genomic sequences in cancer, or at least in CRC. In addition, PEG1 was a common iDMR in both studies, suggesting that PEG1 hypermethylation is implicated in the development or progression of various cancer types.
HyMiD-positive status, which was defined as having at least ten hypermethylated iDMRs, was correlated with CIMP-positive status. Although the BRAF (V600E)-directed MAFG and KRAS (G13D)-directed ZNF304 pathways were responsible for CIMP in CRCs [24, 25], there was no correlation between HyMiD-positive status and mutations in BRAF or KRAS. While the number of tumors with BRAF mutation was low, the scarcity of MAFG-binding sites in iDMRs supported non-correlation between a HyMiD-positive status and BRAF mutation. These results suggest that the two pathways are not responsible for HyMiD-positive status in CRC, which may therefore be driven by other distinct molecular processes.
Genome-wide hypomethylation is believed to contribute to tumorigenesis by activating proto-oncogenes and inducing chromosomal instability (CIN) [33–35]. LINE-1 methylation is often used as a good surrogate marker for genome-wide methylation [31]. The results of analyses of the relationship between hypermethylation, such as CIMP, and genome-wide hypomethylation, such as LINE-1 hypomethylation, have been conflicting. CIMP-high tumors had higher LINE-1 methylation than non-CIMP-high tumors [36]; however, other studies including our present one showed that LINE-1 hypomethylation was independent from classical CIMP status [37, 38]. The discrepancy might be attributable to the array of CIMP markers and variations in the definition of CIMP. In this study, we identified a novel inverse association between HyMiD-positive with LINE-1 methylation. This difference in relationship to LINE-1 methylation between HyMiD and CIMP supports the existence of BRAF/KRAS-independent molecular processes in HyMiD.
The IGF2/H19 domain has been well investigated because IGF2 LOI is a common event in many cancers, including CRC. In this domain, H19-DMR plays a pivotal role in imprinting regulation in normal cells [2]. H19-DMR hypermethylation leads to IGF2 LOI and H19 silencing in Wilms tumor and hepatoblastoma [8, 9]. However, in this study, almost all tumors showed normal methylation of H19-DMR, strongly suggesting that the cause of IGF2 LOI is not H19-DMR hypermethylation in CRCs. Some studies have proposed that IGF2-DMR0 hypomethylation could be the cause of IGF2 LOI because of the association between IGF2-DMR0 hypomethylation and IGF2 LOI [16, 39, 40]. However, another study contested this theory based on the lack of a tight link between IGF2-DMR0 hypomethylation and IGF2 LOI, and it is improbable that the hypomethylation of paternally methylated IGF2-DMR0 could lead to reactivation of the silent maternal IGF2 allele [18]. Because there was no correlation between IGF2 LOI and IGF2-DMR0 hypomethylation, our results support the latter theory. We also could not find a correlation between IGF2 LOI and IGF2-DMR2 hypomethylation. It is therefore clear that hypomethylation of IGF2-DMRs was not a cause of IGF2 LOI. The cause of IGF2 LOI in CRC might not be an alteration of DNA methylation within the IGF2/H19 domain, but rather the dysfunction of any of the components related to imprinting regulation, such as the intrachromosomal loop, sequence alteration of H19-DMR, or expression of the CTCF or PRC2 complexes [41].
The association between IGF2-DMR0 hypomethylation and poor prognosis has been previously documented in CRC patients [12]. This raised the possibility that the methylation status of iDMRs and imprinting status might be associated with clinicopathological factors. Although we could not find any association between HyMiD, IGF2-DMR0 hypomethylation, or IGF2 LOI and various clinicopathological factors, we were able to determine a prognostic score by including the status of aberrant methylation of the three iDMRs in a hazard regression model. This score would be useful to assess prognosis in CRC, although the mechanism for the role of aberrant methylation of iDMRs in mediating outcome is still unknown.
Conclusions
We performed the first comprehensive methylation analysis of iDMRs in CRC patients. The iDMRs were more susceptible to hypermethylation than hypomethylation, but individual iDMRs varied in their susceptibility to aberrant methylation. We found that HyMiD-positive status was associated with CIMP-positive status and LINE-1 hypomethylation. We also confirmed that IGF2-DMR0 hypomethylation was not an appropriate surrogate marker for IGF2 LOI. Moreover, we calculated a clinically relevant prognostic score based on aberrant methylation of the three iDMRs (LOC728024, RB1, and ZNF331-promoter). Further studies are required to understand the mechanisms behind aberrant methylation of iDMRs and IGF2 LOI, and their significance in CRC patients.
Methods
Tissue samples
CRC patients with metastases were excluded from this study due to the very small number in this subgroup. Patients with preoperative chemotherapy or inflammatory bowel disease were also excluded to eliminate the influence of chemotherapy and chronic inflammation on DNA methylation [42–44]. Paired frozen tumor tissue and adjacent normal mucosa were obtained from 106 CRC patients who were admitted to Saga University Hospital. Dissected tissues were stored at − 80 °C followed by DNA and RNA extraction.
DNA extraction and bisulfite conversion
Genomic DNA was extracted from each sample using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Five hundred nanograms of genomic DNA was subjected to bisulfite conversion using an EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA). The converted DNA was then eluted in 100 μl of water. The Human WGA Methylated & Non-Methylated DNA Sets (Zymo Research) were used as fully methylated and unmethylated DNA controls, respectively.
Methylation analysis by bisulfite pyrosequencing
The methylation status of iDMRs, CIMP markers, and LINE-1 was analyzed by bisulfite pyrosequencing using a PyroMark Q24 pyrosequencing instrument (Qiagen) according to the manufacturer’s instructions. We designed primers for bisulfite-mediated PCR and pyrosequencing, using PyroMark Assay Design 2.0 software (Qiagen) in the majority of cases. The methylation percentage of each CpG site was calculated by PyroMark Q24 software (Qiagen). To validate the quantitative capability of bisulfite-pyrosequencing methylation analysis, all primer sets were evaluated using varying mixtures of the unmethylated control and the fully methylated control DNA: 0%, 25%, 50%, 75%, and 100% methylated DNA. All primers used in this study are indicated in Additional file 1: Table S10.
We compared the average methylation of CpG sites within each iDMR between tumor tissues and matched adjacent normal mucosa. Aberrant methylation of each iDMR was defined as the situation of tissue pairs in which the methylation difference between tumor and normal mucosa exceeded 15%. The methylation status of five classical CIMP markers (hMLH1, MINT1, MINT2, MINT31, p16) in tumor tissue was evaluated to determine CIMP status. Each marker was classified as positive (average methylation of CpG sites within a marker > 15%) or negative (< 15%). The tumor sample was classified as CIMP-positive if it was positive in two or more of the five markers and as CIMP-negative if it was positive in less than two markers. LINE-1 methylation level in tumor tissue was determined by the average methylation of CpG sites.
KRAS and BRAF mutation analysis
Detection of the BRAF (V600E) mutation was performed by pyrosequencing, according to a previously published method [45]. A sample was considered mutation-positive if the mutant allele frequency was greater than 15%. Exons 2 and 3 of KRAS were amplified individually by PCR. The two PCR products were combined for each individual sample. Asymmetrical “Y-shaped” adaptors were ligated to PCR products after the end repair and A-tailing using KAPA Hyper Prep Kit (KAPA Biosystems, Wilmington, MA, USA). After adapter ligation, samples were amplified by PCR with indexed primers using the KAPA HiFi HotStart Ready Mix (KAPA Biosystems). The prepared amplicon libraries were sequenced using the MiSeq system (Illumina, San Diego, CA, USA). Mean read depth was > 1000 for all samples. A sample was considered mutation-positive if a mutation was present in at least 1% of the consensus reads [46].
Allelic expression analysis of IGF2 and H19
The genotyping of tumor tissue was performed by PCR-RFLP utilizing the two SNPs, HaeIII (rs680) and AluI (rs2839702) polymorphisms, located at IGF2 exon 9 and at H19 exon 5, respectively. In heterozygous samples, total RNA was extracted from both tumor tissue and matched adjacent normal mucosa using the ISOGEN II kit (Nippon Gene, Tokyo, Japan) following the manufacturer’s instructions. Total RNA was treated with RNase-free DNase I (Takara, Tokyo, Japan), and reverse transcription was performed using random primers and ReverTra Ace reagent mix (Toyobo, Osaka, Japan). To assess allelic expression of IGF2 and H19, the RT-PCR products containing the SNPs were sequenced on a PyroMark Q24 pyrosequencing instrument (Qiagen), following the manufacturer’s instructions. When more than 85% of transcripts were recognized to be derived from a single allele, the expression mode was defined as monoallelic expression, or MOI. When less than 85% of transcripts were from one single allele, and more than 15% of transcripts were from another allele, the expression mode was defined as biallelic expression, or LOI.
Statistical analysis
Correlations among HyMiD status, CIMP status, and KRAS and BRAF mutations were analyzed using Fisher’s exact test. Differences in LINE-1 methylation levels between HyMiD-positive and HyMiD-negative samples were analyzed using the Mann-Whitney U test. Frequencies of aberrant methylation of H19-promoter, H19-DMR, and IGF2-DMRs were analyzed with the χ2 test. Correlations between IGF2 LOI and IGF2-DMR0 hypomethylation or IGF2-DMR2 hypomethylation were investigated by Fisher’s exact test. p values below 0.01 were considered to be statistically significant.
Correlations between clinicopathological factors and HyMiD status, IGF2-DMR0 hypomethylation, or IGF2 LOI were statistically analyzed using the Kruskal-Wallis test, Fisher’s exact test, the Mann-Whitney U test, or the Log-rank test. In addition, all results were corrected for multiple hypotheses testing using the Bonferroni correction, and p values below 0.005 (0.05/10 = 0.005) were deemed statistically significant.
The prognosis score was computed based on a Cox proportional hazards regression model with stepwise selection. The optimal cutoff value for the score was computed by maximizing the log-rank test statistic. The Kaplan-Meier method was used to estimate survival distributions for each group. The log-rank test was used to compare groups, and the hazard ratio (HR) and 95% confidence interval (CI) were calculated. p values below 0.01 were considered statistically significant.
Additional file
Table S1. Information of patients analyzed in this study. Table S2 KRAS mutations identified in 106 CRC tissues. Table S3 Relationship between CIMP and BRAF or KRAS mutations. Table S4 MAFG consensus binding sites in the iDMRs. Table S5 IGF2 imprinting status in paired tumors and normal mucosae. Table S6 Relationship between IGF2-DMR2 hypomethylation and IGF2 LOI in tumors. Table S7 Relationship between clinicopathological factors and HyMiD status. Table S8 Relationship between clinicopathological factors and IGF2-DMR0 hypomethylation. Table S9 Relationship between clinicopathological factors and IGF2 LOI. Table S10 Primers used in this study. Figure S1 Non-relationship between CIMP status and LINE-1 methylation. (PDF 478 kb)
Acknowledgements
Not applicable.
Funding
This study was supported in part by: grants from the Grant-in-Aid for Scientific Research (C) program of the Japan Society for the Promotion of Science [16K09970 to K.H., 17K08687 to H.S., 17K09307 to K.F.]; grants for Practical Research Projects for Rare/Intractable Diseases from the Japan Agency for Medical Research and Development (AMED) [17ek0109280h0001, 17ek0109234h0001, and 17ek0109205h0001 to H.S.]; a grant for Child Health and Development research from the National Center for Child Health and Development [26-13 to H.S.]; a grant for Research on Intractable Diseases from the Ministry of Health, Labor, and Welfare [H29-nanchitou(nan)-ippan-025 to H.S.]; the Joint Research Program of the Institute for Molecular and Cellular Regulation at Gunma University [16029 to K.H.]; and AstraZeneca Research Support (201500315 to R.I.)
Availability of data and materials
All data generated or analyzed throughout this study are included in this published article and its supplementary information files.
Abbreviations
- CIMP
CpG island methylator phenotype
- CIN
Chromosomal instability
- CRC
Colorectal cancer
- HyMiD
Hypermethylation of multiple iDMRs
- iDMR
Imprinting-associated differentially methylated region
- LINE-1
Long interspersed nuclear element-1
- LOI
Loss of imprinting
- MOI
Maintenance of imprinting
- TCGA
The Cancer Genome Atlas
Authors’ contributions
KH and HS designed and supervised the study; YK and HN provided the tissue samples and clinical data; HH, SA, TM, HY, and KH validated the quantitative capability of bisulfite-pyrosequencing; HH, KH, HM, CH, KY performed the experiments; HH and AK performed statistical analyses; KH and HS drafted the manuscript. KJ, RI, and KF provided comments on the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee for Human Genome and Gene Analyses of the Faculty of Medicine at Saga University. Written informed consent was obtained from the patients.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Hidenori Hidaka, Email: h_hida8@yahoo.co.jp.
Ken Higashimoto, Email: higashim@cc.saga-u.ac.jp.
Saori Aoki, Email: bluerose_leo@yahoo.co.jp.
Hiroyuki Mishima, Email: hmishima@nagasaki-u.ac.jp.
Chisa Hayashida, Email: c-haya@nagasaki-u.ac.jp.
Toshiyuki Maeda, Email: topgun.mae@gmail.com.
Yasuo Koga, Email: kogayas@cc.saga-u.ac.jp.
Hitomi Yatsuki, Email: yatsuki@cc.saga-u.ac.jp.
Keiichiro Joh, Email: joh@cc.saga-u.ac.jp.
Hirokazu Noshiro, Email: noshiro@cc.saga-u.ac.jp.
Ryuichi Iwakiri, Email: iwaryu9451.iwa@gmail.com.
Atsushi Kawaguchi, Email: akawa@cc.saga-u.ac.jp.
Koh-ichiro Yoshiura, Email: kyoshi@nagasaki-u.ac.jp.
Kazuma Fujimoto, Email: fujimotk@cc.saga-u.ac.jp.
Hidenobu Soejima, Email: soejimah@cc.saga-u.ac.jp.
References
- 1.Tomizawa S, Sasaki H. Genomic imprinting and its relevance to congenital disease, infertility, molar pregnancy and induced pluripotent stem cell. J Hum Genet. 2012;57:84–91. doi: 10.1038/jhg.2011.151. [DOI] [PubMed] [Google Scholar]
- 2.Soejima H, Higashimoto K. Epigenetic and genetic alterations of the imprinting disorder Beckwith-Wiedemann syndrome and related disorders. J Hum Genet. 2013;58:402–409. doi: 10.1038/jhg.2013.51. [DOI] [PubMed] [Google Scholar]
- 3.Piedrahita JA. The role of imprinted genes in fetal growth abnormalities. Birth Defects Res A Clin Mol Teratol. 2011;91:682–692. doi: 10.1002/bdra.20795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cerrato F, Sparago A, Verde G, De Crescenzo A, Citro V, Cubellis MV, Rinaldi MM, Boccuto L, Neri G, Magnani C, et al. Different mechanisms cause imprinting defects at the IGF2/H19 locus in Beckwith-Wiedemann syndrome and Wilms’ tumour. Hum Mol Genet. 2008;17:1427–1435. doi: 10.1093/hmg/ddn031. [DOI] [PubMed] [Google Scholar]
- 5.Rabinovitz S, Kaufman Y, Ludwig G, Razin A, Shemer R. Mechanisms of activation of the paternally expressed genes by the Prader-Willi imprinting center in the Prader-Willi/Angelman syndromes domains. Proc Natl Acad Sci U S A. 2012;109:7403–7408. doi: 10.1073/pnas.1116661109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murrell A. Genomic imprinting and cancer: from primordial germ cells to somatic cells. ScientificWorldJournal. 2006;6:1888–1910. doi: 10.1100/tsw.2006.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Higashimoto K, Joh K, Soejima H. Genomic imprinting syndromes and cancer. In: Kaneda A, Tsukada Y, editors. DNA and histone methylation as cancer targets. New York: Humana Press; 2017. pp. 297–344. [Google Scholar]
- 8.Bjornsson HT, Brown LJ, Fallin MD, Rongione MA, Bibikova M, Wickham E, Fan JB, Feinberg AP. Epigenetic specificity of loss of imprinting of the IGF2 gene in Wilms tumors. J Natl Cancer Inst. 2007;99:1270–1273. doi: 10.1093/jnci/djm069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Honda S, Arai Y, Haruta M, Sasaki F, Ohira M, Yamaoka H, Horie H, Nakagawara A, Hiyama E, Todo S, Kaneko Y. Loss of imprinting of IGF2 correlates with hypermethylation of the H19 differentially methylated region in hepatoblastoma. Br J Cancer. 2008;99:1891–1899. doi: 10.1038/sj.bjc.6604754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soejima H, Nakagawachi T, Zhao W, Higashimoto K, Urano T, Matsukura S, Kitajima Y, Takeuchi M, Nakayama M, Oshimura M, et al. Silencing of imprinted CDKN1C gene expression is associated with loss of CpG and histone H3 lysine 9 methylation at DMR-LIT1 in esophageal cancer. Oncogene. 2004;23:4380–4388. doi: 10.1038/sj.onc.1207576. [DOI] [PubMed] [Google Scholar]
- 11.Feng W, Marquez RT, Lu Z, Liu J, Lu KH, Issa JP, Fishman DM, Yu Y, Bast RC., Jr Imprinted tumor suppressor genes ARHI and PEG3 are the most frequently down-regulated in human ovarian cancers by loss of heterozygosity and promoter methylation. Cancer. 2008;112:1489–1502. doi: 10.1002/cncr.23323. [DOI] [PubMed] [Google Scholar]
- 12.Baba Y, Nosho K, Shima K, Huttenhower C, Tanaka N, Hazra A, Giovannucci EL, Fuchs CS, Ogino S. Hypomethylation of the IGF2 DMR in colorectal tumors, detected by bisulfite pyrosequencing, is associated with poor prognosis. Gastroenterology. 2010;139:1855–1864. doi: 10.1053/j.gastro.2010.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosen DG, Wang L, Jain AN, Lu KH, Luo RZ, Yu Y, Liu J, Bast RC., Jr Expression of the tumor suppressor gene ARHI in epithelial ovarian cancer is associated with increased expression of p21WAF1/CIP1 and prolonged progression-free survival. Clin Cancer Res. 2004;10:6559–6566. doi: 10.1158/1078-0432.CCR-04-0698. [DOI] [PubMed] [Google Scholar]
- 14.Dalai I, Missiaglia E, Barbi S, Butturini G, Doglioni C, Falconi M, Scarpa A. Low expression of ARHI is associated with shorter progression-free survival in pancreatic endocrine tumors. Neoplasia. 2007;9:181–IN182. doi: 10.1593/neo.06838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 16.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP. Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res. 2002;62:6442–6446. [PubMed] [Google Scholar]
- 17.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, Wu Y, He X, Powe NR, Feinberg AP. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science. 2003;299:1753–1755. doi: 10.1126/science.1080902. [DOI] [PubMed] [Google Scholar]
- 18.Ito Y, Koessler T, Ibrahim AE, Rai S, Vowler SL, Abu-Amero S, Silva AL, Maia AT, Huddleston JE, Uribe-Lewis S, et al. Somatically acquired hypomethylation of IGF2 in breast and colorectal cancer. Hum Mol Genet. 2008;17:2633–2643. doi: 10.1093/hmg/ddn163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murphy SK, Huang Z, Wen Y, Spillman MA, Whitaker RS, Simel LR, Nichols TD, Marks JR, Berchuck A. Frequent IGF2/H19 domain epigenetic alterations and elevated IGF2 expression in epithelial ovarian cancer. Mol Cancer Res. 2006;4:283–292. doi: 10.1158/1541-7786.MCR-05-0138. [DOI] [PubMed] [Google Scholar]
- 20.Ulaner GA. Loss of imprinting of IGF2 and H19 in osteosarcoma is accompanied by reciprocal methylation changes of a CTCF-binding site. Hum Mol Genet. 2003;12:535–549. doi: 10.1093/hmg/ddg034. [DOI] [PubMed] [Google Scholar]
- 21.Barrow TM, Barault L, Ellsworth RE, Harris HR, Binder AM, Valente AL, Shriver CD, Michels KB. Aberrant methylation of imprinted genes is associated with negative hormone receptor status in invasive breast cancer. Int J Cancer. 2015;137:537–547. doi: 10.1002/ijc.29419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Court F, Tayama C, Romanelli V, Martin-Trujillo A, Iglesias-Platas I, Okamura K, Sugahara N, Simon C, Moore H, Harness JV, et al. Genome-wide parent-of-origin DNA methylation analysis reveals the intricacies of human imprinting and suggests a germline methylation-independent mechanism of establishment. Genome Res. 2014;24:554–569. doi: 10.1101/gr.164913.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyota M, Ahuja N, Ohe-Toyota M, Herman JG, Baylin SB, Issa JP. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci U S A. 1999;96:8681–8686. doi: 10.1073/pnas.96.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. doi: 10.7554/eLife.02313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang M, Ou J, Hutchinson L, Green MR. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Mol Cell. 2014;55:904–915. doi: 10.1016/j.molcel.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kambara T, Simms LA, Whitehall VL, Spring KJ, Wynter CV, Walsh MD, Barker MA, Arnold S, McGivern A, Matsubara N, et al. BRAF mutation is associated with DNA methylation in serrated polyps and cancers of the colorectum. Gut. 2004;53:1137–1144. doi: 10.1136/gut.2003.037671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ogino S, Kawasaki T, Kirkner GJ, Loda M, Fuchs CS. CpG island methylator phenotype-low (CIMP-low) in colorectal cancer: possible associations with male sex and KRAS mutations. J Mol Diagn. 2006;8:582–588. doi: 10.2353/jmoldx.2006.060082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ogino S, Kawasaki T, Kirkner GJ, Kraft P, Loda M, Fuchs CS. Evaluation of markers for CpG island methylator phenotype (CIMP) in colorectal cancer by a large population-based sample. J Mol Diagn. 2007;9:305–314. doi: 10.2353/jmoldx.2007.060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iacopetta B, Grieu F, Phillips M, Ruszkiewicz A, Moore J, Minamoto T, Kawakami K. Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci. 2007;98:1454–1460. doi: 10.1111/j.1349-7006.2007.00548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miousse IR, Koturbash I. The fine LINE: methylation drawing the cancer landscape. Biomed Res Int. 2015;2015:131547. doi: 10.1155/2015/131547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang AS, Estecio MR, Doshi K, Kondo Y, Tajara EH, Issa JP. A simple method for estimating global DNA methylation using bisulfite PCR of repetitive DNA elements. Nucleic Acids Res. 2004;32:e38. doi: 10.1093/nar/gnh032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Bretz CL, Lee S. Epigenetic instability of imprinted genes in human cancers. Nucleic Acids Res. 2015;43:10689–10699. doi: 10.1093/nar/gkv867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–5413. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 34.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science. 2003;300:455. doi: 10.1126/science.1083557. [DOI] [PubMed] [Google Scholar]
- 35.Matsuzaki K, Deng G, Tanaka H, Kakar S, Miura S, Kim YS. The relationship between global methylation level, loss of heterozygosity, and microsatellite instability in sporadic colorectal cancer. Clin Cancer Res. 2005;11:8564–8569. doi: 10.1158/1078-0432.CCR-05-0859. [DOI] [PubMed] [Google Scholar]
- 36.Ogino S, Kawasaki T, Nosho K, Ohnishi M, Suemoto Y, Kirkner GJ, Fuchs CS. LINE-1 hypomethylation is inversely associated with microsatellite instability and CpG island methylator phenotype in colorectal cancer. Int J Cancer. 2008;122:2767–2773. doi: 10.1002/ijc.23470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Estecio MR, Gharibyan V, Shen L, Ibrahim AE, Doshi K, He R, Jelinek J, Yang AS, Yan PS, Huang TH, et al. LINE-1 hypomethylation in cancer is highly variable and inversely correlated with microsatellite instability. PLoS One. 2007;2:e399. doi: 10.1371/journal.pone.0000399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bariol C, Suter C, Cheong K, Ku S-L, Meagher A, Hawkins N, Ward R. The relationship between hypomethylation and CpG island methylation in colorectal neoplasia. Am J Pathol. 2003;162:1361–1371. doi: 10.1016/S0002-9440(10)63932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng YW, Idrees K, Shattock R, Khan SA, Zeng Z, Brennan CW, Paty P, Barany F. Loss of imprinting and marked gene elevation are 2 forms of aberrant IGF2 expression in colorectal cancer. Int J Cancer. 2010;127:568–577. doi: 10.1002/ijc.25086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miroglio A, Jammes H, Tost J, Ponger L, Gut IG, El Abdalaoui H, Coste J, Chaussade S, Arimondo PB, Lamarque D, Dandolo L. Specific hypomethylated CpGs at the IGF2 locus act as an epigenetic biomarker for familial adenomatous polyposis colorectal cancer. Epigenomics. 2010;2:365–375. doi: 10.2217/epi.10.24. [DOI] [PubMed] [Google Scholar]
- 41.Zhang H, Niu B, Hu JF, Ge S, Wang H, Li T, Ling J, Steelman BN, Qian G, Hoffman AR. Interruption of intrachromosomal looping by CCCTC binding factor decoy proteins abrogates genomic imprinting of human insulin-like growth factor II. J Cell Biol. 2011;193:475–487. doi: 10.1083/jcb.201101021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartnett L, Egan LJ. Inflammation, DNA methylation and colitis-associated cancer. Carcinogenesis. 2012;33:723–731. doi: 10.1093/carcin/bgs006. [DOI] [PubMed] [Google Scholar]
- 43.Abu-Remaileh M, Bender S, Raddatz G, Ansari I, Cohen D, Gutekunst J, Musch T, Linhart H, Breiling A, Pikarsky E, et al. Chronic inflammation induces a novel epigenetic program that is conserved in intestinal adenomas and in colorectal cancer. Cancer Res. 2015;75:2120–2130. doi: 10.1158/0008-5472.CAN-14-3295. [DOI] [PubMed] [Google Scholar]
- 44.Baharudin R, Ab Mutalib NS, Othman SN, Sagap I, Rose IM, Mohd Mokhtar N, Jamal R. Identification of predictive DNA methylation biomarkers for chemotherapy response in colorectal cancer. Front Pharmacol. 2017;8:47. doi: 10.3389/fphar.2017.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spittle C, Ward MR, Nathanson KL, Gimotty PA, Rappaport E, Brose MS, Medina A, Letrero R, Herlyn M, Edwards RH. Application of a BRAF pyrosequencing assay for mutation detection and copy number analysis in malignant melanoma. J Mol Diagn. 2007;9:464–471. doi: 10.2353/jmoldx.2007.060191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.de Biase D, Visani M, Baccarini P, Polifemo AM, Maimone A, Fornelli A, Giuliani A, Zanini N, Fabbri C, Pession A, Tallini G. Next generation sequencing improves the accuracy of KRAS mutation analysis in endoscopic ultrasound fine needle aspiration pancreatic lesions. PLoS One. 2014;9:e87651. doi: 10.1371/journal.pone.0087651. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Information of patients analyzed in this study. Table S2 KRAS mutations identified in 106 CRC tissues. Table S3 Relationship between CIMP and BRAF or KRAS mutations. Table S4 MAFG consensus binding sites in the iDMRs. Table S5 IGF2 imprinting status in paired tumors and normal mucosae. Table S6 Relationship between IGF2-DMR2 hypomethylation and IGF2 LOI in tumors. Table S7 Relationship between clinicopathological factors and HyMiD status. Table S8 Relationship between clinicopathological factors and IGF2-DMR0 hypomethylation. Table S9 Relationship between clinicopathological factors and IGF2 LOI. Table S10 Primers used in this study. Figure S1 Non-relationship between CIMP status and LINE-1 methylation. (PDF 478 kb)
Data Availability Statement
All data generated or analyzed throughout this study are included in this published article and its supplementary information files.