Abstract
Background
Environmental stress induced genetic polymorphisms have been suggested to arbitrate functional modifications influencing adaptations in microbes. The relationship between the genetic processes and concomitant functional adaptation can now be investigated at a genomic scale with the help of next generation sequencing (NGS) technologies. Using a NGS approach we identified genetic variations putatively underlying chromium tolerance in a strain of Aspergillus flavus isolated from a tannery sludge. Correlation of nsSNPs in the candidate genes (n = 493) were investigated for their influence on protein structure and possible function. Whole genome sequencing of chromium tolerant A. flavus strain (TERIBR1) was done (Illumina HiSeq2000). The alignment of quality trimmed data of TERIBR1 with reference NRRL3357 (accession number EQ963472) strain was performed using Bowtie2 version 2.2.8. SNP with a minimum read depth of 5 and not in vicinity (10 bp) of INDEL were filtered. Candidate genes conferring chromium resistance were selected and SNPs were identified. Protein structure modeling and interpretation for protein-ligand (CrO4− 2) docking for selected proteins harbouring non-synonymous substitutions were done using Phyre2 and PatchDock programs.
Results
High rate of nsSNPs (approximately 11/kb) occurred in selected candidate genes for chromium tolerance. Of the 16 candidate genes selected for studying effect of nsSNPs on protein structure and protein-ligand interaction, four proteins belonging to the Major Facilitator Superfamily (MFS) and recG protein families showed significant interaction with chromium ion only in the chromium tolerant A. flavus strain TERIBR1.
Conclusions
Presence of nsSNPs and subsequent amino-acid alterations evidently influenced the 3D structures of the candidate proteins, which could have led to improved interaction with (CrO4− 2) ion. Such structural modifications might have enhanced chromium efflux efficiency of A. flavus (TERIBR1) and thereby offered the adaptation benefits in counteracting chromate stress. Our findings are of fundamental importance to the field of heavy-metal bio-remediation.
Electronic supplementary material
The online version of this article (10.1186/s12864-018-5244-2) contains supplementary material, which is available to authorized users.
Keywords: Non synonymous SNPs (nsSNPs), Mutation, Protein structure and function, Protein-ligand interaction, Adaptation
Background
Bioremediation of heavy metals by microbial cells has been recognized as a potential alternative to the existing physico-chemical technologies for recovery of heavy metals from industrial effluents [1]. Metal uptake in microorganisms takes place either actively (bioaccumulation) or passively (biosorption) [2–6]. Several species of bacteria and fungi have been identified for their bioaccumulation or absorption potentials and reduced cost and toxicity achieved by microbial bioremediation approach are appreciated over the conventional methods [7]. Various bacterial species detoxify chromium by periplasmic absorption, intracellular bioaccumulation and biotransformation through direct enzymatic reaction or indirectly with metabolites. Filamentous fungi have been identified as a potential biomass for removal of heavy metals from solutions and species of Aspergillus, Rhizopus and Penicillium are reported useful in biological treatment of the sludge [8–11], Several reports support the prominent ability of Aspergillus flavus in detoxification of chromium and other heavy metals [12]. However, the molecular mechanisms underlying heavy metal detoxification in fungi are largely unknown. Understanding the genes and pathways involved in metal accumulation/tolerance in fungi has several biotechnological implications for bioremediation of heavy metal-contaminated sites.
The extensive use of chromium in diverse industrial processes has made it a significant environmental contaminant. Chromium is a Class A human carcinogen [13, 14] and exists in eleven valence states (from −IV to +VI), among which Cr (III) and Cr (VI) are the most stable forms in the environment. Due to high water solubility Cr (VI) is 100-folds more toxic over Cr (III). As per the United States Environmental Protection Agency (US EPA) the maximum contaminant level for Cr (VI) and total chromium content in domestic water supplies is 0.05 and 2 mg/l respectively [15]. Cr (VI) actively crosses biological membranes [16] and generates active intermediates Cr (V) and/or Cr (IV), free radicals, and Cr (III). Cellular accumulation of Cr (III) causes damage to DNA and alters the structure and activity of proteins [17, 18]. The existing physico-chemical processes for treating chromium-contaminated water bodies include precipitation, ion exchange, reverse osmosis, evaporation and electro dialysis, which are reported to display poor efficiency [14, 19–24].
For survival in Cr (VI) contaminated environments, microorganisms must develop efficient systems to detoxify the effects of chromium. These mechanisms involve detoxification or repair strategies such as Cr (VI) efflux pumps, Cr (VI) reduction to Cr (III), and activation of enzymes involved in the detoxifying processes, repair of DNA lesions, sulfur metabolism, and iron homeostasis [16, 18, 25]. Additionally, alterations in gene function due to mutation have been suggested to support survival under chromium toxic conditions [26]. Biotransformation and biosorption are suggested as the putative fungal processes that help them transform or adsorb heavy metals [27]. The fungal cell walls predominantly consist of chitins, glucans, mannans and proteins in addition to other polysaccharides, lipids and pigments [28, 29]. The functional groups on these structural components enable binding of metal ions on the fungal cell walls [30]. Uptake and reduction of hexavalent chromium has been suggested as the mechanisms for chromium tolerance in Aspergillus sp. [27, 31].
Information on genes supporting survival under environmental stress in bacterial system has been recently curated in BacMet database (http://bacmet.biomedicine.gu.se) which primarily contains several experimentally verified Chromate ion transporter (CHR) genes [32] responsible for chromium efflux, transport or binding, and other enzymes involved in chromium uptake. However, very less knowledge is available on genetic mechanisms responsible for chromium tolerance in fungi. In the Neurospora crassa strain 74-A, chr-1 gene that encodes a putative CHR-1 protein and belongs to the CHR superfamily was identified [33]. However, contrary to the bacterial ChrA (chromate transport protein) homologues that confer chromate resistance by exporting chromate ions from the cell’s cytoplasm, the experimental data suggested that the N. crassa CHR-1 protein functions as a transporter that takes up chromate [34]. The presence of CHR-1 protein was reported to cause chromate sensitivity and chromium accumulation in N. crassa.
Experimental evidences in a recent study suggested that environmental stress could induce adaptation in a wide range of micro-organisms by extensive positive pleiotropy in a manner that multiple beneficial mutations dramatically enhance numerous fitness components simultaneously [35]. Environmentally induced mutations and polymorphisms in DNA and subsequently the alteration in proteins are hypothesized to offer a significant evolutionary advantage by enabling faster adaptation to toxic conditions [36]. We identified a high chromium tolerant Aspergillus flavus strain (TERIBR1) from a tannery sludge in Kanpur, Uttar Pradesh, India. TERIBR1 showed accumulation of Cr (III) in its biomass while growing in Cr containing media. It showed no toxic effect of Cr (VI) up to 250 mg/l. In order to identify the genetic factors underlying chromium tolerance in TERIBR1, we investigated effects of nonsynonymous variations (nsSNPs) in candidate genes on protein structure and their interaction with chromate ion.
Our study comprises whole genome sequencing of A. flavus strain TERIBR1 followed by single nucleotide polymorphism (SNPs) analysis in candidate genes for chromium-resistance. Protein modeling for candidate genes with nsSNPs was done and interactions between modeled proteins and the ligand (CrO4− 2) were assessed by protein-ligand docking. For all comparative genomics and genetics analyses the A. flavus strain TERIBR1 was considered as the “test” and previously sequenced strain NRRL3357 as the “reference” type.
Materials and methods
Fungal strain and DNA extraction
The protocol followed for isolation and characterization of fungi from a tannery sludge is previously described [37]. Briefly, the Cr-resistant fungi were isolated from a tannery sludge [containing 250 mg/l of Cr (III)] through an enrichment culture technique. The sludge sample was collected from a tannery waste disposal site in Kanpur, India. Pure culture of the isolated A. flavus strain (TERIBR1) was grown in potato dextrose broth (PDB) at 28 °C in a shaking incubator (100 rpm) for 72 h in dark condition. After incubation, culture was centrifuged at 5000 g for 10 min at room temperature. The pellet was washed thrice with sterile distilled water to remove any media components and was further used for DNA extraction. Genomic DNA was extracted using the DNeasy plant mini kit (QIAGEN, USA), according to the manufacturer’s instructions. Genetic characterization of isolated fungi was done using universal fungal ITS (nuclear ribosomal internal transcribed spacer) primer set [ITS1: 5’ TCCGTAGGTGAACCTGCGG, 3′ and ITS4: 5’ TCCTCCGCTTATTGATATGC 3′; [38] that amplified the ITS1, 5.8S and ITS2 regions of the nuclear ribosomal RNA genes.
Growth kinetics and sensitivity to Cr (VI)
The effect of different concentrations of chromium [Cr (VI)], 0 mg/l, 100 mg/l and 250 mg/l, on the growth of A. flavus strains TERIBR1 and NRRL3357 was compared. The strains were grown in PDB and mycelial biomass (dry weight) was measured at different time periods (0, 1, 2, 3, 4 and 5 days).
Genome sequencing and assembly
Genome sequencing was performed at MOgene LC, USA, using next generation sequencing technology Illumina as reported previously [39]. Two paired end libraries (insert sizes 180 bp and 500 bp) and one mate pair library (5 kb) were constructed. DNA libraries were purified using AMPure XP beads. KAPA was done to quantify the libraries, which were then normalized and pooled at 4 nM concentration.
A total of 8 GB raw data was subjected to adaptor- and quality-based trimming. Quality-passed data was assembled using the de novo genome assembler AllpathsLG [40]. Reads with overlaps were first combined to form contigs. The reads were mapped back to contigs. With paired-end reads, contigs from the same transcript, as well as the distances between these contigs, were detected. In order to generate scaffolds, contigs were connected using “N” to represent unknown sequences between two contigs. Mate-pair reads were used for gap filling of scaffolds in order to get sequences with minimal N’s and the longest length. The whole genome project has been deposited at https://submit.ncbi.nlm.nih.gov/subs/wgs/under Bioproject PRJNA362980.
Structural and functional annotation of A. flavus TERIBR1 genome was done using MAKER [41] pipeline, InterProScan [42] and nrBlast [39] as described previously.
Identification of single nucleotide polymorphisms (SNPs)
Genome and protein sequences for reference genome were retrieved from the Aspergillus flavus Database (http://fungidb.org/fungidb/app/record/organism/aflaNRRL3357). The alignment of quality trimmed data of TERIBR1 with NRRL3357 (assembly) was performed using Bowtie2 version 2.2.8 [43]. Samtools [http://samtools.sourceforge.net/] was used for SNP identification.
SNP analysis in candidate genes for chromium resistance
Genes conferring chromium resistance in bacterial system were selected from BacMet database [32]. BacMet is freely available antibacterial biocide and metal resistance genes database for bacteria. InterProScan analysis [42] was performed to identify A. flavus genes harbouring atleast one IPR domains that are present in the chromium resistance genes documented in the BacMet database. SNPs were identified in the selected candidate genes using variant calling format (VCF) file and Blastn tool. SNPs were further annotated as synonymous or non-synonymous (nsSNPs) using an in-house perl script.
Protein structure modeling
Protein modeling was done by fold recognition methods through Phyre2 server [44]. The amino acid sequences of candidate genes in both the reference (NRRL3357) and the test strains were modeled. The top model with highest confidence and coverage was selected for each protein. The predicted confidence score and coverage for all the final structures were recorded. To assess the reliability of all the predicted models, structural analysis and verification was exercised. The selected models were validated using the PROCHECK [45] and ERRAT [46] to estimate the stereo chemical figures, geometry, and hydrogen bonding energy, torsion angles and error rate of the predicted structures. In addition, energy minimization was performed with in vacuo GROMOS96 43B1 parameters set using GROMOS96 implementation in Swiss-Pdb Viewer [47]. The energy optimized protein structures were used for protein-small ligand docking.
Prediction of ligand binding sites
Prior to docking, a web based approach 3DLigandSite [48] was used to predict the ligand binding sites. 3DLigandSite utilizes protein-structure prediction to provide structural models for proteins that have not been solved. Ligands bound to structures similar to the query are superimposed onto the model and used to predict the binding site.
Protein- ligand docking
In order to investigate protein–ligand interactions, proteins were docked with the chromate ion (CrO4− 2) through a rigid docking protocol using PatchDock (http://bioinfo3d.cs.tau.ac.il/PatchDock/) [49, 50] which docks the ligand with the protein based on structure complementarity. Also, binding sites predicted by 3DLigandSite in the receptor/proteins were specified and uploaded in PatchDock analysis. The protein-ligand interactions were interpreted based on Atomic Contact Energy (ACE) and docking score. The pdb file of chromate ion was downloaded from the RCSB PDB (research collaborator fo structural Bioinformatics protein data bank) site [51]. The PDB structures of target proteins and protein-ligand interaction were visualized using the PyMOL [52].
Results
Growth kinetics and sensitivity to Cr (VI)
Dry weight of fungal biomass was recorded at different time periods (from 1 to 5 days) for both the strains under the conditions mentioned above. No significant difference in growth was observed between the two strains under the control condition (Fig. 1). However, stark difference in the mycelial biomass (dry weight) between the reference strain (NRRL3357) and the test strain (TERIBR1) was observed when potato dextrose broth was amended with chromium 100 mg/l and 250 mg/l. Growth kinetics of the TERIBR1 strain at chromium concentration of 100 mg/l were similar to that observed under control condition (no chromium). The reference strain exhibited delayed growth response with concomitant decrease in biomass in comparison to the test strain at different time intervals (between day 1 and day 5) when the growth media was amended with chromium at concentrations of 100 mg/l and 250 mg/l.
Fig. 1.
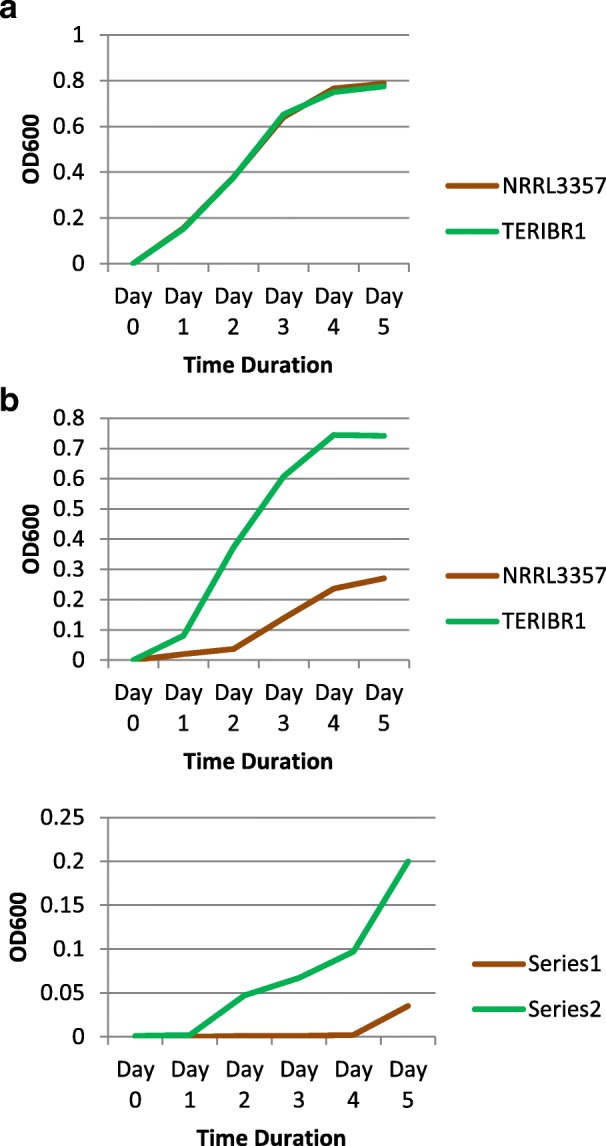
Chromium [Cr (VI)] dose response exhibited by TERIBR1 and NRRL3357 strains of A. flavus. Chromium dose/growth response (measured by dry weight) exhibited by TERIBR1 and NRRL3357 strains of A. flavus grown up to 5 days in potato dextrose broth supplemented with Cr (VI): (a) 0 mg/l, (b) 100 mg/l and 250 mg/l
Global genome structure
The genome of A. flavus strain TERIBR1 was sequenced to 200x coverage and reads were assembled into 322 scaffolds. The sum of the scaffolds length is equal to 38.2 Mb. The three largest scaffolds are 2.76 kb, 2.64 kb, and 2.50 kb in size. The MAKER annotation pipeline predicted 13,587 protein coding genes as compared to 13,659 in NRRL3357. Gain or loss of unique genes, DNA duplication, gene family expansion, and translocation of transposon-like elements are often observed between different isolates of a fungal species [53]. This may suggest that some of the genes present in NRRL3357 could have been lost in TERIBR1, possibly during environmental adaptations.
Identification of candidate genes in A. flavus
No homologue of CHR-1 protein (XP_961667.3) coded by N. crassa was identified in both the A. flavus strains included in this study. A total of 34 InterProScan domains coding for transporter or regulator proteins responsible for chromium bio-accumulation or tolerance in bacteria were reported in the BacMet database. nrBlast was performed to identify genes containing at least one IPR domain associated with chromium tolerance in the genome of A.
Flavus strain TERIBR1, NRRL3357 (http://fungidb.org/fungidb/app/record/organism/aflaNRRL3357) and AF70 (https://www.ncbi.nlm.nih.gov/assembly/GCA_000952835.1). 23/34 bacterial IPR domains were not found in any of the three strains of A. flavus. A total of 493 candidate genes was identified to harbor one or more IPR domains of interest in TERIBR1(Table 1). IPR domains mdrL/yfmO (IPR011701; n = 334), recG (IPR001650; n = 71), ruvB (IPR003959; n = 45) and recG (IPR011545; n = 44) were among the maximally present protein domains related to chromium resistance.
Table 1.
Distribution of IPR domains important in chromium bio-accumulation in A. flavus strains TERIBR1, NRRL3357 & AF70
| Gene Family (BacMet db) | Description | Interproscan Domain | # of Genes containing IPR domains of interest | ||
|---|---|---|---|---|---|
| NRRL3357 | TERIBR1 | AF70 | |||
| Chromate ion transporter (CHR) family (chrA) | Efflux | IPR003370 | 1 | 1 | 2 |
| Rhodanese family (chrE) | Enzyme | IPR001763 | 9 | 6 | 10 |
| NADH_dh2 family (chrR) | Enzyme | IPR005025 | 4 | 4 | 4 |
| IPR000415 | 0 | 3 | 0 | ||
| MFS superfamily (mdrL/yfmO) | Efflux | IPR011701 | 374 | 334 | 394 |
| Contains 1 DEAD/DEAH box helicase domain (recG) | Enzyme | IPR011545 | 43 | 44 | 42 |
| IPR001650 | 74 | 71 | 80 | ||
| IPR004365 | 2 | 5 | 4 | ||
| RuvB family (ruvB) | Enzyme | IPR003959 | 47 | 45 | 48 |
| IPR012301 | 2 | 2 | 2 | ||
Identification of single nucleotide polymorphisms (SNPs)
The read alignment rate of TERIBR1 with NRRL3357 (assembly) was 78.62% (29,001,807 / 36,890,268) of which 78.23% (22,681,743) were uniquely mapped reads. A total of 201,145 SNPs (read depth > 5) was identified at a frequency of ~ 5 SNPs per Kb of the TERIBR1 genome. SNP mapping in n = 493 candidate genes, homologous among A. flavus NRRL3357 and TERIBR1 isolates was done using Samtools. No SNP was identified in 325/493 genes. SNPs identified in the remaining n = 168 genes were annotated as synonymous or non-synonymous (Additional file 1: Table S1). 28/168 candidate genes contained only synonymous polymorphisms whereas 16/168 candidate genes, belonging to MFS (n = 12), recG (n = 3) and chrE (n = 1) protein families, showed higher rate of nsSNP as compared to other candidate genes (Additional file 2: Table S2).
Protein- ligand docking
For studying protein-chromate ion interaction, we predicted tertiary protein structures of homologous pairs of the 16 highly polymorphic proteins (Additional file 2: Table S2) using Phyre2 server (Additional file 3: Table S3). Prediction for Cr binding sites in the target proteins was done by 3DLigandSite (Table 2). Strength of protein-ligand interaction was measured based on the atomic contact energy (ACE) in the PatchDock score (Table 3). Also change in free energy (ΔG) of the amino acid residues present in the predicted binding and ligand docking sites was recorded (Fig. 2). Structures of 8 proteins in both the reference and test strains did not show any possible interaction between the ligand and the target proteins. Ligand docking was observed in both the strains for four proteins (g8975, g685, g6212, g9525; Additional file 4: Figure S1). Binding residues that showed a drop in free energy on chromate docking in PatchDock analysis are depicted on the 3D structures of these four proteins (Additional file 4: Figure S1).
Table 2.
Prediction of binding site and protein – ligand interaction using 3DLigandSite and PatchDock softwares respectively
| Protein ID | nsSNP | Predicted binding sites | Docking status and residues in recognition cavity | # nsSNP | Gene Family |
|||
|---|---|---|---|---|---|---|---|---|
| NRRL3357 | TERI BRI |
NRRL 3357 |
TERI BR1 |
NRRL3357 | TERIBR1 | |||
| AFL2G_00299 | g652 | A346D, D351N, M389 T | A261, G262, I263 | No B.S. | N/A | ≤2 | mdrL/ yfmO | |
| AFL2G_04853 | g9548 | P254L, K261I, K263E, A262D, M34 T | T67, F68, V69, S70, P71, L72, A73, S74, S75, L104, Y107, V108, P111, G161, C164, L165, W188, P192, Y280, L283, Y284, T288, Y393, T416, A417, S420, L421, V422, A424, L425, L426 | Y122, W203, P207, Y319, L322, Y323 | N/A | 2 to 5 | mdrL/ yfmO | |
| AFL2G_04391 | g8975 | P341A, D349E, H356Y, P373L, P374L | Q119, F240, H403, T404, N405, V407, Q408, L454, F477, S481, Y485, V508, L511, Q512, V514, S515, R516, F518, V519, L520, P521, S524 | Q115, F240, N405, A406, Q408, T409, L454, F477, S481, Y485, V508, L511, Q512, V514, S515, R516, F518, V519, L520, P521, S524, R552 | Dock | 2 to 5 | mdrL/ yfmO | |
| AFL2G_02473 | g5755 | K53 N, N59D, K213 M, V293I, K340R | H97, W124, I125, L126, V127, M128, F129, F130, A131, L132, N133, I134, D135, I183, G184, P185, D186, R187, W188, I189, P190, I191, Q192, I193, I194, L195, S197, F226, D229, V253, S257, A288, S291, I292, G295, F296, S298, F299, L302 | W115, I116, L117, V118, M119, F120, A122, I174, G175, P176, D177, R178, W179, I180, P181, I182, Q183, I184, I185, L186, F217, S282, G286, S289, F290, L293, V294 | N/A | 2 to 5 | mdrL/ yfmO | |
| AFL2G_00264 | g685 | S126G, T139A, G179E, Y112F | T334, L335, G400, K401, S402, L403, E461, H465, F680, G681, R711 | T321, L322, M386, G387, K388, S389, L390, E442, H446, F726, G727, R757 | Dock | 2 to 5 | recG | |
| AFL2G_05826 | g6641 | P307L, Q11P, V19G, E102D, V126A, A129V | No B.S. | S270, M273, I274, Q396 | N/A | > 5 | mdrL/ yfmO | |
| AFL2G_09247 | g6212 | R471H, R437Q, S837P, L1229 V, V192I, L233S | K272, L273, L274, Q277, G309, L310, G311, K312, T313, V314, E380, K384, L919, G920, L921, N922, R947, R950, L951 | L544, V546, K547, L548, L549, Q552, G584, L585, G586, K587, T588, V589, E655, K659, L1194, G1195, N1197, R1222, R1225, L1226 | Dock | > 5 | recG | |
| AFL2G_08767 | g9986 | F222I, A244P, Q270P, G340R, A342G, F431 L, S472I | W290, L291, Y292, L294, M295, I353, L354, V355, M356, H357, L358, W359, T360, P362, P363, F401, I404, Y455, M458, N459, L462, T465, R466 | K277, Y278, Q279, V281, E282, A283, T285, I288, A337, V338, M339, V340, G341, G342, A343, S344, P346, P347, F385, I388, N443, L446, L447, R450, L453, I454 | N/A | Dock K277, Q279, T285, L446 |
> 5 | mdrL/ yfmO |
| AFL2G_05032 | g9401 | N373D, S445 N, E503G, S535 L, V572G, F592Y, K610E, I50M | H614, H616, L666, H668, H670 | C97, A98, F100, L101, Y104, I107, M159, A160, I161, I162, Y164, S165, A168, I169, F198, A202, V205, S257, T260, H261, A264, N267, K268 | N/A | > 5 | mdrL/ yfmO | |
| AFL2G_06586 | g3683 | S517 N, Q324E, L899S, S57 L, D63G, T18I, K285R | Q193, L194, K195, Q198, M221, G222, L223, G224, K225, T226, I227, E266, I643 | L145, S147, Q148, L149, G179, K180, T181, I182, E221, K224, W225, E573, G574, R604 | N/A | Dock L149, K180, T181, I182 |
> 5 | recG |
| AFL2G_11779 | g4359 | G294D, K360E, V388I, F393 L, L468P, Q66H, R19K, V637I, A646T | L582, V586, M589, N590, M593, A621, Y623, L631, H632, A635, H636, H640, W647, I659 | R34, T36, A94, V95, Y100, S101, A178, I206, P207, L208, A209, V211 | N/A | > 5 | chrE | |
| AFL2G_09661 | g4104 | R80M, L110 V, V144 L, N150S, F191 L, G198R, Y199C, E407G, G5E, I25N | No B.S. | N/A | Dock V144 L, N150S | > 5 | mdrL/ yfmO | |
| AFL2G_04878 | g9525 | G210S, C363W, I368S, H438Y, M484, P106A, V131I, V146A, N156S, F163 L, L182F, Q59H | S63, I66, F92 | S141, I144, F170 | Dock | > 5 | mdrL/ yfmO | |
| AFL2G_00229 | g712 | R163L, G180C, S215C, S220Y, A226P, V693I, S765 N, F834I, Q854H, C938S, V121A | G570, A571, N572, S573, G574, L575, V595, R596, S597, K600, L624, D625, M626, L627, N652, A653, G654, I655, V673, V704, G705, S706, Y745, K749, P780, G781, P782, T783, S785, G786, L787 | G666, A667, N668, S669, G670, L671, V691, R692, K696, L720, D721, M722, L723, N748, A749, G750, I751, V769, V800, G801, S802, Y841, K845, P876, G877, P878, T879, S881, G882, L883 | N/A | > 5 | mdrL/ yfmO | |
| AFL2G_04255 | g9088 | L52 V, S101G, A212T, F214 L, T217A, S237 L, A250V, P252L, P271S, M280 T, K292 N, S297R, V303I | A104, L105, P108, S110, L138, I139, V141, G142, M165, M169, A226, I256, F338, L341, N342, M367, Y477, G481, L483 | A195, P198 | N/A | > 5 | mdrL/ yfmO | |
| AFL2G_11442 | g4641 | M254I, P321T, I433V, D661G, P675Q, F682 L, D110N, A111V, I114V, K143 T, T3A, H13Q, T24A, C46S, K826R | A120, F121, V122, V123, S124, A125, A126, S127, S128, L156, F159, A160, S163, M187, P216, L217, Y240, S244, Y355, F359, D363, T513, V514, Y517, C518, A519, G521, G522, M523 | S372, A492, V493, L494, P496, F603, F606, W628, V629, A630, M631, Y632, V633, G634, I635, M636, L637, L640, S724 | N/A | Dock Y632, F606, W628, A492 |
> 5 | mdrL/ yfmO |
SNPs marked in bold were predicted binding site present in the predicted recognition cavity of the protein. B.S. stands for binding site
Table 3.
Docking analysis using PatchDock for selected proteins of A. flavus strain TERIBR1
| Protein ID TERIBR1 | Score | Area | ACE (kcal/mol) |
|---|---|---|---|
| g652 | 2764 | 330.5 | −13.58 |
| g9548 | 2496 | 304.5 | −58.67 |
| bg8975 | 2806 | 333.4 | −29.90 |
| g5755 | 2746 | 329.6 | 31.22 |
| bg685 | 2576 | 321.4 | −1.40 |
| g641 | 2846 | 322.9 | 19.42 |
| bg6212 | 2924 | 326.8 | −62.95 |
| a g9986 | 2644 | 296.6 | −46.56 |
| g9401 | 2594 | 324.6 | −77.47 |
| a g3683 | 2788 | 306.3 | −30.21 |
| g4359 | 2664 | 285.8 | −60.63 |
| a g4104 | 3034 | 335.4 | −72.70 |
| bg9525 | 2966 | 362.2 | −83.82 |
| g712 | 2454 | 299.4 | −7.63 |
| g9088 | 2368 | 258 | −62.84 |
| a g4641 | 2772 | 302.9 | −66.10 |
aProtein - ligand interaction observed only in A. flavus strain TERIBR1
bProtein - ligand interaction observed in both the strains of A. flavus
The entries marked in bold indicate significant interaction of ligand with the protein
Fig. 2.
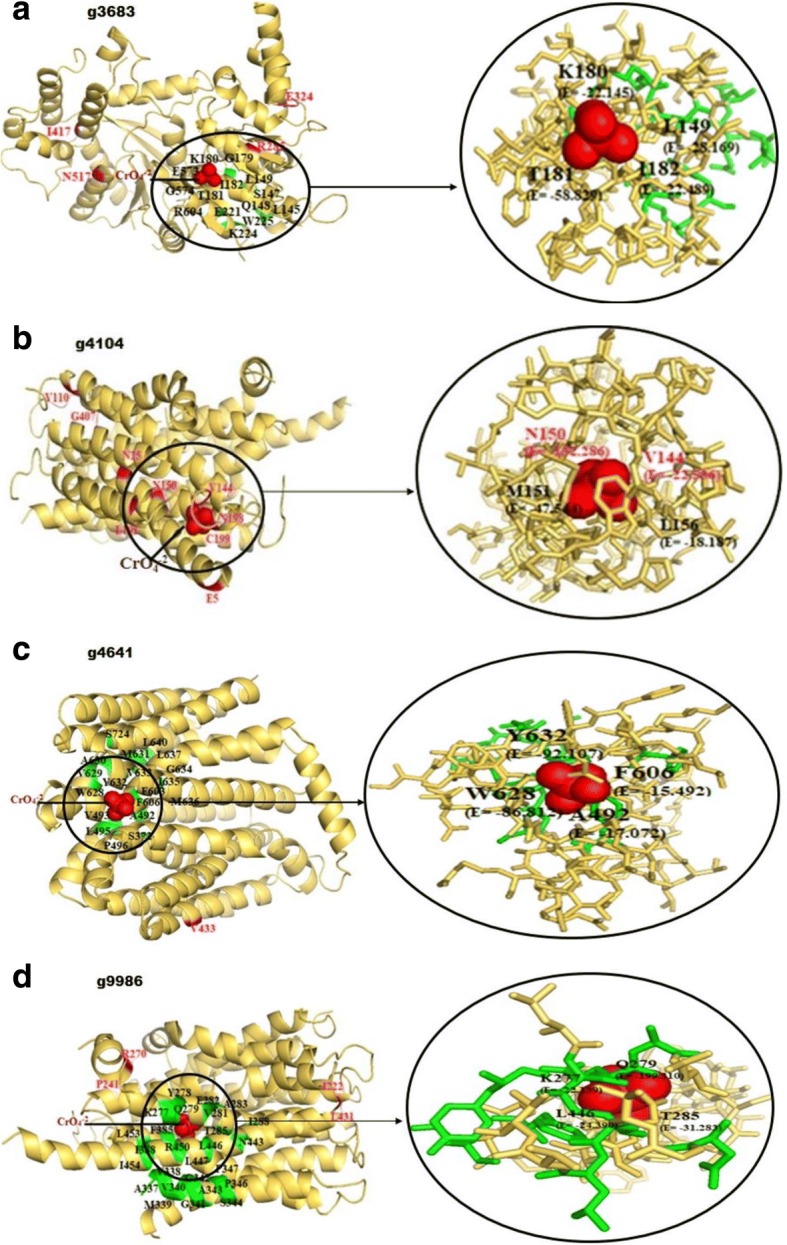
Protein-chromate ion interaction observed with four MFS transporter proteins of A. flavus strain TERIBR1. Docking of chromate ion with MFS transporter proteins in occluded conformation. The chromate ion is depicted as a sphere model. The amino acids of the interacting protein showing negative energy are depicted as bright orange sticks and the interacting binding sites as green sticks. Presence of nsSNPs in the protein sequence is shown in magenta. Amino-acids present in the close vicinity of the binding sites are marked in black (sSNP) and magenta (nsSNP). Figure was produced using the PyMOL Molecular Graphics System
Interestingly, the presence of non-synonymous mutations correlated with change in bioactive conformation and drop in free energy (ΔG) of four proteins (g9986, g3683, g4104, g4641) belonging to three MFS and one recG (helicase) superfamilies in the test strain only (Fig. 2). The structural changes in these proteins lead to successful protein-ligand interactions.
Discussion
As expected for functional conservation, majority of candidate genes in the TERIBR1 genome showed the presence of a large number of sSNPs and a few nsSNPs. Notably, 28/168 candidate genes contained only synonymous polymorphisms. Synonymous codon positions, though do not alter amino acid sequences of the encoded proteins, they may determine secondary structure, stability and translation rate of the mRNA [54]. Presence of sSNPs in the chromium tolerance candidate genes in the test strain could have affected folding and post-translational modifications of the nascent polypeptides which could in turn affect candidate protein expression and function towards Cr tolerance.
The polymorphism rate in 16 candidate genes that showed a high frequency of nsSNPs as compared to synonymous changes (Table 2) was ~ 16 SNPs/Kb with a frequency of ~ 11 nsSNPs/Kb. The observed high rate of nsSNPs in chromium-tolerance candidate genes of TERIBR1 as compared to the housekeeping genes (0.4 nsSNPs/kb; Table 4) could mirror environmental stress induced DNA variations and might provide an advantage in counteracting chromate stress. These included genes from mdrL/yfmO (12), recG (3) and chrE (1) families. The mdrL/yfmO genes belonged to the major facilitator superfamily (MFS), which codes for a metal ion-specific efflux protein [55]. High frequency of nsSNPs observed in the mdrL/yfmO genes in TERIBR1 could have led to altered protein structure and subsequent chromium efflux efficacy under extreme environmental condition, which we discussed in detail under the protein-ligand docking section. recG is a conserved enzyme present in bacteria, archaea, and eukaryota. recG encodes for the ATP-dependent recG DNA helicase which plays a critical role in DNA recombination and repair [56]. In vivo experiments conducted in E. coli showed that chromium salt stimulates several stress promoters associated with different types of DNA damage, indicating that DNA is one of the main targets for Cr (III) inside the cell [57]. After being internalized in cells Cr (VI) is reduced to Cr (III); recG eliminates polymerase arresting lesions (PALs), caused by Cr (III). The observed high frequency of nsSNPs in recG genes observed in our study might have resulted in higher efficiency of the enzyme to remove PAL lesions, thus mediating chromium stress tolerance in the fungal strain. In congruence, a study in Pseudomonas corrugata suggested that recG helicase played a crucial role in chromium tolerance by dismissing PAL lesions caused by Cr (VI)/Cr (III) [58]. The chrE gene encodes a rhodanese type enzyme [59]. Rhodanese protein subfamilies are suggested to be involved in different biological functions including cyanide detoxification, arsenic resistance and chromate responsive DNA-binding regulator. In addition, UniProt database defines ChrE as proteins involved in the processing of chromium-glutathione-complexes. An abundance of nsSNPs in these candidate genes for chromium tolerance could be the result of environment induced variations, perhaps for achieving functional relevance in TERIBR1. Environmentally guided changes in DNA and subsequently the proteins could be advantageous and may enable functional adaptation to extreme environmental influences [36].
Table 4.
SNP frequency in housekeeping genes in A. flavus
| Gene ID NRRL3357 | Gene ID TERIBR1 | Annotation | Gene length (nucl) | change in nucl | change in aa | status of SNPs |
|---|---|---|---|---|---|---|
| AFL2T_10032 | g899 | Calmodulin | 4047 | 0 | 0 | 0 |
| AFL2T_10117 | g962 | RPL5 (ribosomal protein) | 1071 | 0 | 0 | 0 |
| AFL2T_03358 | g2143 | Polyketide Synthase Acetate | 1692 | 0 | 0 | 0 |
| AFL2T_03019 | g2421 | Chitin Synthase 1 | 2655 | 0 | 0 | 0 |
| AFL2T_08232 | g3003 | cyclophilin | 522 | 0 | 0 | 0 |
| AFL2T_08160 | g3065 | Ubiquitin-conjugating enzyme | 450 | 0 | 0 | 0 |
| AFL2T_01340 | g5399 | Vacuolar protein sorting association protein | 324 | 0 | 0 | 0 |
| AFL2T_01191 | g5533 | Cytochrome oxidase | 348 | 0 | 0 | 0 |
| AFL2T_12005 | g6415 | Ubiquitin-conjugating enzyme | 510 | 0 | 0 | 0 |
| AFL2T_02547 | g6955 | Ubiquitin-conjugating enzyme | 513 | 0 | 0 | 0 |
| AFL2T_02762 | g7132 | L- Asparaginase | 690 | 0 | 0 | 0 |
| AFL2T_09767 | g7323 | ATP_D (ATP synthase subunit beta) | 1821 | 0 | 0 | 0 |
| AFL2T_06390 | g8072 | Polyketide Synthase Acetate | 2529 | 0 | 0 | 0 |
| AFL2T_09983 | g10276 | Ubiquitin-conjugating enzyme | 501 | 0 | 0 | 0 |
| AFL2T_09876 | g10370 | L- Asparaginase | 1677 | 0 | 0 | 0 |
| AFL2T_07389 | g10698 | Elongation Factor Alpha like protein | 1185 | 0 | 0 | 0 |
| AFL2T_06969 | g3347 | ATP_D (Atp synthase subunit beta) | 1539 | 0 | 0 | 0 |
| AFL2T_06937 | g3377 | Chitin Synthase 1 | 5283 | 0 | 0 | 0 |
| AFL2T_05991 | g6789 | GAPDH/Glyceraldehyde 3-phosphate dehydrogenase | 1524 | 0 | 0 | 0 |
| AFL2T_02677 | g7061 | Vacuolar protein sorting association protein | 1563 | 0 | 0 | 0 |
| AFL2T_05240 | g3927 | cyclophilin | 1122 | 0 | 0 | 0 |
| AFL2T_02454 | g5775 | TBPI (tata box binding protein) | 690 | 0 | 0 | 0 |
| AFL2T_03769 | g1786 | Actin interacting protein 3 | 2931 | 0 | 0 | 0 |
| AFL2T_05664 | g6503 | Histone | 807 | 0 | 0 | 0 |
| AFL2T_11201 | g8361 | Ubiquitin-conjugating enzyme | 333 | 0 | 0 | 0 |
| AFL2T_12447 | g9127 | Ubiquitin-conjugating enzyme | 837 | 0 | 0 | 0 |
| AFL2T_12048 | g11077 | DNA Topoisomerase II | 1032 | 0 | 0 | 0 |
| AFL2T_04711 | g9681 | Ubiquitin-conjugating enzyme | 450 | 0 | 0 | 0 |
| AFL2T_07021 | g3301 | Ubiquitin-conjugating enzyme | 474 | 0 | 0 | 0 |
| AFL2T_07052 | g3271 | Lactate Dehydrogenase A | 1065 | 0 | 0 | 0 |
| AFL2T_11998 | g6422 | Ubiquitin-conjugating enzyme | 456 | 0 | 0 | 0 |
| AFL2T_05713 | g6542 | Vacuolar protein sorting association protein | 387 | 0 | 0 | 0 |
| AFL2T_03033 | g2409 | Chitin Synthase 1 | 1671 | 0 | 0 | 0 |
| AFL2T_08388 | g2865 | Vacuolar protein sorting association protein | 2022 | 0 | 0 | 0 |
| AFL2T_08078 | g3130 | Histone | 429 | 0 | 0 | 0 |
| AFL2T_05673 | g6508 | 28 s rRNA | 450 | 0 | 0 | 0 |
| AFL2T_04621 | g9757 | cyclophilin | 630 | 0 | 0 | 0 |
| AFL2T_07907 | g4829 | Vacuolar protein sorting association protein | 351 | 0 | 0 | 0 |
| AFL2T_01105 | g5607 | Ubiquitin-conjugating enzyme | 1167 | 0 | 0 | 0 |
| AFL2T_09240 | g6218 | Ubiquitin-conjugating enzyme | 558 | 0 | 0 | 0 |
| AFL2T_09015 | g9269 | Polyketide Synthase Acetate | 6828 | 0 | 0 | 0 |
| AFL2T_10236 | g1076 | Vacuolar protein sorting association protein | 2313 | 0 | 0 | 0 |
| AFL2T_05795 | g6613 | 28 s rRNA | 1815 | 0 | 0 | 0 |
| AFL2T_03329 | g2169 | Ubiquitin-conjugating enzyme | 2631 | 0 | 0 | 0 |
| AFL2T_09350 | g6131 | 18 s rRNA | 2382 | 0 | 0 | 0 |
| AFL2T_00575 | g419 | Chitin Synthase 1 | 3588 | C150T | H50H | sSNPs |
| AFL2T_00433 | g530 | Vacuolar protein sorting association protein | 2562 | C1483G | P496A | nsSNPs |
| AFL2T_02076 | g8000 | Elongation Factor Alpha like protein | 3222 | A890C | K297Q | nsSNPs |
| AFL2T_09781 | g7310 | Vacuolar protein sorting association protein | 2925 | T1645C | S548S | sSNPs |
| AFL2T_09150 | g8774 | Polyketide Synthase Acetate | 7425 | T5820C | L1938 L | sSNPs |
| AFL2T_06936 | g3378 | Chitin Synthase 1 | 5574 | A315G, C2100T, A2841T | G105G, D700D, I947I | sSNPs |
| AFL2T_06204 | g8236 | Chitin Synthase 1 | 3315 | C1668T | L556 L | sSNPs |
| AFL2T_02195 | g6007 | Vacuolar protein sorting association protein | 4545 | A2925G, C4044T | E975E, F1348F | sSNPs |
| AFL2T_08239 | g2996 | Calmodulin | 5103 | A260G, T2232C, C2757G | D87G, T744 T, L919 L | nsSNPs, sSNPs, sSNPs |
| AFL2T_07518 | g5174 | Polyketide Synthase Acetate | 6366 | G1575A, G3053A | S525 N, S1018 N | nsSNPs |
| AFL2T_00612 | g388 | ATP_D (Atp synthase subunit beta) | 1671 | A1515C | A505A | sSNPs |
| AFL2T_05167 | g3861 | Vacuolar protein sorting association protein | 3528 | G456 T, G807A | G152G, T269 T | sSNPs |
| AFL2T_04317 | g9038 | Vacuolar protein sorting association protein | 1920 | T1383C | I461I | sSNPs |
| AFL2T_12048 | g6382 | DNA Topoisomerase II | 3183 | T1871C | V624A | nsSNPs |
| AFL2T_12403 | g9165 | Polyketide Synthase Acetate | 4944 | T1265C, C2231T, Y2271G | M422 T, T744I, X757K | nsSNPs |
| AFL2T_08114 | g3103 | Elongation Factor Alpha like protein | 1383 | T273A | I91I | sSNPs |
| AFL2T_02416 | g5810 | Vacuolar protein sorting association protein | 2124 | C1606T | L536 L | sSNPs |
| AFL2T_06144 | g8287 | aflatoxin regulatory protein | 945 | C46T | L16F | nsSNPs |
| AFL2T_07648 | g5058 | Ubiquitin-conjugating enzyme | 1278 | T570C | G190G | sSNPs |
| AFL2T_03037 | g2405 | secretory lipase | 1353 | W1271A | X424N | nsSNPs |
| AFL2T_05603 | g4255 | Vacuolar protein sorting association protein | 2757 | C1164T, G1491 T, C1884T | I388I, T497 T, I628I | sSNPs |
| AFL2T_09157 | g8767 | Ras protein | 6435 | C1509T, A2193G, G2304A, T2358C, G3018C, G4056A | S503S, S731S, L768 L, N786 N, T1006 T, P1352P | sSNPs |
| AFL2T_12399 | g9169 | Chitin Synthase 1 | 3222 | C1317G | V439 V | sSNPs |
| AFL2T_06989 | g3329 | Elongation Factor Alpha like protein | 2583 | T1461C | T487 T | sSNPs |
| AFL2T_11104 | g8446 | Polyketide Synthase Acetate | 7482 | T3962C, A5970G, A5981G | M1320 T, I1990M, D1994G | nsSNPs |
| AFL2T_01971 | g7909 | Vacuolar protein sorting association protein | 5862 | G281A, A2246T, A2316G, C2766T, T3135A | R94K, N749I, S772S, I922I, D1045E | nsSNPs, nsSNPs, sSNPs, sSNPs, nsSNPs |
| AFL2T_01302 | g5433 | Vacuolar protein sorting association protein | 2457 | C704T, T828C | A235V, G276G | nsSNPs |
| AFL2T_11645 | g4481 | ATP_D (Atp synthase subunit beta) | 1116 | T972C | I324I | sSNPs |
| AFL2T_04569 | g9796 | Elongation Factor Alpha like protein | 2400 | C546T, T1785C, T2253C | D182D, L595 L, F751F | sSNPs |
| AFL2T_05904 | g6711 | Elongation Factor Alpha like protein | 2730 | T146C, C1836T, G2002A, A2435G | F49S, P612P, V668I, D812G | nsSNPs, sSNPs, nsSNPs, nsSNPs |
| AFL2T_02030 | g7958 | Ubiquitin-conjugating enzyme | 1176 | T641C | V214A | nsSNPs |
| AFL2T_09952 | g10303 | TBPI (tata box binding protein) | 1338 | T789G | Y263Y | sSNPs |
| AFL2T_02696 | g7079 | Elongation Factor Alpha like protein | 2169 | T576C, A825T, G1332A, T1557C | D192D, I275I, E444E, F519F | sSNPs |
| AFL2T_12346 | g8605 | Elongation Factor Alpha like protein | 2874 | A1914G | E638E | sSNPs |
| AFL2T_10814 | g1537 | Ubiquitin-conjugating enzyme | 708 | C507T | D169D | sSNPs |
| AFL2T_07094 | g3237 | Polyketide Synthase Acetate | 6606 | T3478A, T4780C, G4927A, A5446G, G5677A, T5862C,T6264C | C1160S, Y1594H, E1643K, N1816D, A1893T, S1954S, C2088C | nsSNPs, nsSNPs, nsSNPs, nsSNPs, nsSNPs, sSNPs, sSNPs |
| AFL2T_02027 | g7956 | Vacuolar protein sorting association protein | 1044 | T663C | Y221Y | sSNPs |
| AFL2T_06635 | g3639 | L- Asparaginase | 1074 | A257G, C663A | D86G, N221 K | nsSNPs |
| AFL2T_09646 | g7430 | Ubiquitin-conjugating enzyme | 921 | T6C, G36C | S2S, A12A | sSNPs |
| AFL2T_01296 | g5440 | Vacuolar protein sorting association protein | 876 | C93T, G369A | T31 T, L123 L | sSNPs |
| AFL2T_05777 | g6597 | Vacuolar protein sorting association protein | 2058 | T538C, G1317A, G1438A, G1938C | Y180H, S439S, E480K, K646 N | nsSNPs, sSNPs, nsSNPs, nsSNPs |
| AFL2T_08606 | g2683 | Chitin Synthase 1 | 5172 | G3339A, G3618A, A3831G, G4764C, G4952A | T1113 T, L1206 L, L1277 L, P1588P, R1651Q | sSNPs, sSNPs, sSNPs, sSNPs, nsSNPs |
| AFL2T_09101 | g9340 | Elongation Factor Alpha like protein | 2859 | C1173T, G1275A, T1895C, G1947A, A2172G, C2670T | D391D, L425 L, V632A, R649R, V724 V | sSNPs, sSNPs, nsSNPs, sSNPs, sSNPs |
| AFL2T_08131 | g3088 | cyclophilin | 486 | C158T | A53V | nsSNPs |
| AFL2T_00781 | g238 | Polyketide Synthase Acetate | 6951 | G2689A, C3072T, A3121G, A3864G | V897I, F1024F, F1041A, K1288 K | nsSNPs, sSNPs, nsSNPs, nsSNPs |
| AFL2T_01027 | g12 | Vacuolar protein sorting association protein | 732 | K401C | X134A | nsSNPs |
| AFL2T_00198 | g739 | Ubiquitin-conjugating enzyme | 3240 | C849A, T852A, G1475C, T1905C, A2459G, G2659A | F283 L, I284I, R492P, G635G, N820S, A887T | nsSNPs, sSNPs, nsSNPs, sSNPs, nsSNPs, nsSNPs |
| AFL2T_11313 | g4750 | aflatoxin regulatory protein | 1164 | T208C, C889A, G922A | S70P, E297E, G308R | nsSNPs, sSNPs, nsSNPs |
| AFL2T_08488 | g2771 | Elongation Factor Alpha like protein | 3249 | G1398A, A1683A, G2161A, G2328A | E466E, V561 V, A721T, Q776Q | sSNPs, sSNPs, nsSNPs, sSNPs |
| AFL2T_07094 | g10842 | Polyketide Synthase Acetate | 1455 | G526A, T711C, T1113C, A295G | A176T, S237S, L371 L | nsSNPs, sSNPs, sSNPs |
| AFL2T_06011 | g6804 | Ubiquitin-conjugating enzyme | 504 | A210G | P70P | sSNPs |
| AFL2T_01283 | g5448 | Chitin Synthase 1 | 2589 | T1072C, A1536G | N358 N, K512 K | sSNPs |
| AFL2T_02416 | g10910 | Vacuolar protein sorting association protein | 370 | A55T | L18 L | sSNPs |
| AFL2T_05917 | g6723 | Ubiquitin-conjugating enzyme | 741 | A567G | K189 K | sSNPs |
| AFL2T_11034 | g8506 | GAPDH/Glyceraldehyde 3-phosphate dehydrogenase | 1077 | C237T, C357A | H79H, G119G | sSNPs |
| AFL2T_02787 | g7154 | Cytochrome oxidase | 1482 | A849G, C857T, A1003T, | E283E, T286I, T335F | sSNPs, nsSNPs, nsSNPs |
| AFL2T_03260 | g2222 | secretory lipase | 1365 | C936T, G985A, T990C, G1286A, T1291C | N312 N, G329R, T330 T, G429D, L431 L | sSNPs, nsSNPs, sSNPs, nsSNPs, sSNPs |
| AFL2T_07361 | g10719 | Lactate Dehydrogenase A | 933 | G702C, C753T, G879A | G234G, F251F, V293 V | sSNPs |
| AFL2T_09556 | g7507 | Ras protein | 1458 | C565T, G681A, T771C, T1047A | L189 L, T227 T, T257 T, P349P | sSNPs |
| AFL2T_03516 | g2002 | Vacuolar protein sorting association protein | 2853 | A1176C, C1180T | G392G, L394 L | sSNPs |
| AFL2T_04629 | g9750 | Elongation Factor Alpha like protein | 1443 | T475G, C543T, G1185C, T1302C | T181 T, V395 V, A434A | sSNPs |
| AFL2T_01738 | g8814 | cyclophilin | 1638 | A501C, A513G, A522G, A624G, T1011C, A1043G, T1176C | V167 V, E171E, V174 V, E208E, A337A, K348R, L392 L | sSNPs, sSNPs, sSNPs, sSNPs, sSNPs, nsSNPs, sSNPs |
| AFL2T_04801 | g9604 | Cytochrome oxidase | 555 | T207A, T463G | G69G, F155C | sSNPs, nsSNPs |
| AFL2T_12397 | g9171 | Vacuolar protein sorting association protein | 1758 | T1659C | G553G | sSNPs |
| AFL2T_08911 | g9862 | Polyketide Synthase Acetate | 7170 | T3097C, A3519C, G3689A, A3761T | W1033R, T1173 T, R1230Q, Y1254F | nsSNPs, sSNPs, nsSNPs, nsSNPs |
| AFL2T_00897 | g134 | cyclophilin | 1893 | G616 T, C855T, T1191C, A1222G | V206 L, F285F, Y397Y, T408A | nsSNPs, sSNPs, sSNPs, nsSNPs |
| AFL2T_04106 | g10177 | cyclophilin | 642 | A72T | T24 T | sSNPs |
| AFL2T_06925 | g3387 | Cytochrome oxidase | 348 | C63A | V21 V | sSNPs |
| AFL2T_07038 | g3286 | Chitin Synthase 1 | 2076 | G1794A | T598 T | sSNPs |
| AFL2T_08473 | g2782 | cyclophilin | 498 | C320G | T107 T | sSNPs |
| AFL2T_01646 | g2579 | secretory lipase | 909 | C10T, T309C, G509C, C646T | L4L, H103H, R170P, L216 L | sSNPs, sSNPs, nsSNPs, sSNPs |
| AFL2T_03998 | g10190 | Histone | 768 | C96T, A255G, C391T | F32F, S85S, P131S | sSNPs, sSNPs, nsSNPs |
| AFL2T_07224 | g5682 | aflatoxin regulatory protein | 1218 | C318G, G408C, G552 T, C581T, A794G, A979G, G1075A, C1137T | T106 T, P136P, S184S, A194V, Y265C, S327G, V359 M, S379S | sSNPs, sSNPs, sSNPs, nsSNPs, nsSNPs, nsSNPs, nsSNPs, sSNPs |
| AFL2T_00797 | g223 | L- Asparaginase | 1137 | T426C, T693C, G831C, C855T, T858C | G142G, G231G, Q277H, I285I, D286D | sSNPs, sSNPs, nsSNPs, sSNPs, sSNPs |
| AFL2T_08030 | g3169 | secretory lipase | 1269 | A642G, A795G, T903C, A904G, A913G, T927C, T933C, T963C, C1140T | A214A, L265 L, Y301Y, N302D, I305V, D309D, F311F, N321 N, G380G | sSNPs, sSNPs, sSNPs, sSNPs, nsSNPs, sSNPs, sSNPs, sSNPs, sSNPs |
| AFL2T_08467 | g2788 | cyclophilin | 1641 | T83C, C408A, G610A, G655A, T666C, C690T, T750A, A789G | V28A, L136 L, A204T, A219T, F222F, Y230Y, T250 T, E263E | nsSNPs, sSNPs, nsSNPs, nsSNPs, sSNPs, sSNPs, sSNPs, sSNPs |
| AFL2T_04948 | g9453 | Polyketide Synthase Acetate | 1977 | C1314G, T1444C, G1519A, A1590G, C1767A, C1854T, G1962A | R438R, W482R, V507I, L530 L, I589I, N618 N, R654R | sSNPs, nsSNPs, nsSNPs, sSNPs, sSNPs, sSNPs, sSNPs |
| AFL2T_07791 | g4925 | Vacuolar protein sorting association protein | 3813 | G39A, C213G, G234A, A291G, C354T | L13 L, S71S, Q78Q, E97E, H118H | sSNPs |
| AFL2T_12205 | g8728 | secretory lipase | 939 | T25C, G182A, T380C, C435T, T699C, T714G, G768A, G828A | L8L, S61 N, I127T, S145S, I233I, L238 L, P256P, A276A | sSNPs, nsSNPs, nsSNPs, sSNPs, sSNPs, sSNPs, sSNPs, sSNPs |
| AFL2T_01987 | g7921 | cyclophilin | 537 | A504G | ,K168 K | sSNPs |
| AFL2T_05263 | g3947 | Vacuolar protein sorting association protein | 891 | G301A, G401A, C558T, C654T, T749G, A750G, A775G | A101T, G134D, D186D, I218I, I250R, I250R, M259 V | nsSNPs, nsSNPs, sSNPs, sSNPs, nsSNPs, nsSNPs, nsSNPs |
| AFL2T_01745 | g8808 | GAPDH/Glyceraldehyde 3-phosphate dehydrogenase | 1041 | C192G, T222C, C345T, C348T, C393T, T474C | D64E, I74I,G115G, A116A, F113F, A158A | nsSNPs, sSNPs, sSNPs, sSNPs, sSNPs, sSNPs |
| AFL2T_04609 | g9767 | RPL5 (ribosomal protein) | 531 | C153T | Y51Y | sSNPs |
Frequency of SNPs = 0.9 SNPs/kb
Frequency of sSNPs = 0.7 SNPs/kb
Frequency of nsSNP = 0.4 SNPs/kb
Several studies have shown that non-synonymous substitutions are likely to affect protein structure [60]. Mapping of nsSNPs to a known 3D structure reveals whether the replacement is likely to destroy the hydrophobic property of a protein, electrostatic interactions or interactions with ligands. Many nsSNPs have been found near or inside the protein-protein interaction interfaces that alter the protein function [61]. Sequence-based structure predictions help in identifying the positions of a protein that are located in the active site. Protein – ligand docking analysis further helps in identifying crucial amino-acids that are involved in ligand binding.
Non-synonymous mutations mediated change in free energy (ΔG) and concomitant bioactive conformation of four proteins (g9986, g3683, g4104, g4641) belonging to the MFS and recG helicase super families were noteworthy. A decrease in free energy and atomic contact energy (ACE) putatively resulted in target-ligand interaction with a significant PatchDock score in the case of the proteins coded by the chromium tolerant strain, TERIBR1 (Table 2); whereas no ligand interaction was observed in the corresponding proteins coded by reference strain. Figure 2 shows the results of the molecular docking studies of the four proteins (g9986, g4104, g4641, g3683) coded by TERIBR1 strain. Ligand binding free energy estimates (ACE) indicated a significant decrease in free energy of these proteins (Table 3). The nsSNPs in the candidate genes of the chromium tolerant A. flavus strain TERIBR1 seemed to have influenced protein structure that could have mediated protein and chromium interaction. However, not much overlapping between the predicted binding sites (by 3DLigandSite) and the ligand docking position was observed for these proteins. The multidrug transporters of the MFS superfamily are polyspecific and can extrude a remarkably diverse range of substrates. However, discussions pertaining to multi-substrate recognition and transport by members of the MFS are still open and it is not clear if the same amino acid residues are involved in substrate recognition and binding in varying conformations of the protein [62]. Biochemical studies on the Escherichia coli MFS drug/H+ antiporter concluded that the structural basis of substrate promiscuity is governed by a large, flexible and complex substrate recognition cavity within the protein, which enables different substrates to interact with different amino acid residues of the cavity, and to form different interactions with MFS transporter [63, 64]. The putative correlation between the influence of genetic polymorphisms on the structure and function of MFS transporters and chromium tolerance in A. flavus suggested the importance of efflux mechanism in microbial chromium tolerance. Our results supported previous reports of heavy metal efflux as one of the primary mechanisms of tolerance in microbial systems [65, 66]. Furthermore, ligand docking was observed in four proteins (g8975, g685, g6212, g9525) and their homologs coded by the test and the reference strains respectively. The non-synonymous amino acid changes in these cases seemed to have no influence on protein-ligand interaction.
In a recent study four populations of yeast, exposed to arsenic in its most toxic form, As (III), accumulated changes in DNA, adapted faster and went from poor to optimal performance for fitness components (length of lag phase, population doubling time and efficiency of growth) within just a few mitotic divisions. The study concluded that fitness component enhancements in yeast populations were adaptive responses to arsenic and not to other selective pressures [35]. The observed high rate of variations in the DNA of A. flavus strain TERIBR1 in our study, especially nsSNP polymorphisms, highlights the scope for additional research on genetic mechanisms operating in A. flavus in order to conclude on the role of stress mediated alterations in DNA on adaptation in micro-organisms.
Conclusions
Changes in DNA, guided by extreme environmental conditions, could influence the structure of proteins important in chromium stress tolerance in Aspergillus flavus. The structural changes in transporter proteins and enzymes are expected to have potential influence on their functional efficacy. Our study provided insights into the genetic factors governing heavy metal tolerance, which may aid in the development of future heavy metal bio-remediation technologies. Further, to ensure that the genes presenting nsSNPs are involved in the tolerance to chromium of the TERIBR1 strain, the results obtained in the present study demand cross validation by a proteome analysis.
Additional files
Table S1: Representation of candidate genes for chromium tolerance in A. flavus. (XLSX 97 kb)
Table S2: 16 genes coded by A. flavus strain TERIBR1 with high frequency of non-synonymous substitutions. (DOCX 15 kb)
Table S3: Phyre2 prediction and analysis of secondary structure. (DOCX 18 kb)
Figure S1. Protein-ligand interaction observed with homologous pairs of protein of A. flavus strains TERIBR1 and NRRL3357. (PDF 414 kb)
Acknowledgements
The authors are thankful to the TERI-Deakin Nanobiotechnology Research Centre, India for providing necessary infrastructure to carry out the required research work.
Funding
The manuscript consists of research work carried out in-house at the TERI-Deakin Nanobiotechnology Research Centre, India and was not supported by a particular funding source.
Availability of data and materials
The whole genome project has been deposited at https://submit.ncbi.nlm.nih.gov/subs/wgs/under Bioproject PRJNA362980.
Abbreviations
- ACE
Atomic Contact Energy
- BacMetdbs
Antibacterial Biocide & Metal Resistance Genes Database
- CHR
Chromate Transport Protein
- MFS
Major Facilitator Superfamily
- NrBlast
Non Redundant Basic Local Alignment Search Tool
- nsSNPs
Non Synonymous Single Nucleotide Polymorphisms
- Phyre2
Protein Homology/AnalogY Recognition server
- RCSB PDB
Research Collaboratory for Structural Bioinformatics Protein Data Bank
- sSNPs
Synonymous Single Nucleotide Polymorphisms
Authors’ contributions
All authors have read and approved the final manuscript. AJ was involved in bio-informatics and proteomics data analyses, data compilation and manuscript writing. DV supported genomics data analysis. AA isolated the A. flavus strain TERIBR1 and supervised wet lab assays for chromium resistance. PP was the coordinator of the project, involved in conceptualization of the project, study design, data analyses, data compilation, manuscript writing, critical inputs and finalization of the manuscript.
Ethics approval and consent to participate
Not Applicable.
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Akanksha Jaiswar, Email: akanksha.jaiswar@teri.res.in.
Deepti Varshney, Email: deepti.varshney12@gmail.com.
Alok Adholeya, Email: aloka@teri.res.in.
Pushplata Prasad, Phone: +91 11 24682100, Email: pushplata.singh@teri.res.in.
References
- 1.Pérez Silva RM, Ábalos Rodríguez A, Gómez Montes De Oca JM, Cantero Moreno D. Biosorption of chromium, copper, manganese and zinc by Pseudomonas aeruginosa AT18 isolated from a site contaminated with petroleum. Bioresour Technol. 2009;100:1533–1538. doi: 10.1016/j.biortech.2008.06.057. [DOI] [PubMed] [Google Scholar]
- 2.Shumate ES, Strandberg WG. Accumulation of metals by microbial cells. Comprehensive Biotechnology. 1985;13:235–247. [Google Scholar]
- 3.Andres Y, MacCordick HJ, Hubert JC. Bacterial biosorption and retention of thorium and uranyl cations by mycobacterium smegmatis. J Radioanal Nucl Chem. 1992;166:431–440. doi: 10.1007/BF02167788. [DOI] [Google Scholar]
- 4.Fourest E, Roux CJ. Heavy metal biosorption by fungal mycilial by- products: mechanisms and influence of pH. Appl Microbiol Biotechnol. 1992;37:399–403. doi: 10.1007/BF00211001. [DOI] [Google Scholar]
- 5.Hussein H, Krull R, SI AEL-ELA, Hempel DC. Conference Proceedings. Berlin, Germany: International Water Association World Water Conference; 2001. Interaction of the different heavy metal ions with immobilized bacterial culture degrading xenobiotic wastewater compounds. [Google Scholar]
- 6.Hussein H, Farag S, Moawad H. Isolation and characterisation of Pseudomonas resistant to heavy metals contaminants. Arab Journal of Biotechnology. 2003;7:13–22. [Google Scholar]
- 7.Hansda A, Kumar V, Anshumali A comparative review towards potential of microbial cells for heavy metal removal with emphasis on biosorption and bioaccumulation. World J Microbiol Biotechnol. 2016;32(10):170. doi: 10.1007/s11274-016-2117-1. [DOI] [PubMed] [Google Scholar]
- 8.Lacina C, Germain G, Spiros NA. Utilization of fungi for bio treatment of raw wastewaters. Afr J Biotechnol. 2003;2(12):620–630. doi: 10.5897/AJB2003.000-1116. [DOI] [Google Scholar]
- 9.Volesky B, Holan ZR. Biosorption of heavy metals. Biotechnol. Progr. 1995;11:235–250. doi: 10.1021/bp00033a001. [DOI] [PubMed] [Google Scholar]
- 10.Huang CP, Huang CP. Application of Aspergillus oryzae and Rhizopus oryzae. Wat. Res. 1996;30:1985–1990. doi: 10.1016/0043-1354(96)00020-6. [DOI] [Google Scholar]
- 11.Zafar S, Aqil F, Ahmad I. Metal tolerance and biosorption potential of filamentous fungi isolated from metal contaminated agriculture soil. Biores Technol. 2007;98:2557–2561. doi: 10.1016/j.biortech.2006.09.051. [DOI] [PubMed] [Google Scholar]
- 12.Das S, Santra SC. Bio-detoxification of chromium from industrial wastewater by fungal strains. BIOLOGIJA. 2012;58:1–6. doi: 10.6001/biologija.v58i1.2316. [DOI] [Google Scholar]
- 13.Li ZH, Li P, Randak T. Evaluating the toxicity of environmental concentrations of waterborne chromium (VI) to a model teleost, Oncorhynchus mykiss: A comparative study of in vivo and in vitro. Comp Biochem Physiol - C Toxicol Pharmacol. 2011;153:402–407. doi: 10.1016/j.cbpc.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 14.Bansal M, Singh D, Garg VK. A comparative study for the removal of hexavalent chromium from aqueous solution by agriculture wastes’ carbons. J Hazard Mater. 2009;171:83–92. doi: 10.1016/j.jhazmat.2009.05.124. [DOI] [PubMed] [Google Scholar]
- 15.Baral A, Engelken RD. Chromium-based regulations and greening in metal finishing industries in the USA. Environ Sci Pol. 2002;5:121–133. doi: 10.1016/S1462-9011(02)00028-X. [DOI] [Google Scholar]
- 16.Viti C, Marchi E, Decorosi F, Giovannetti L. Molecular mechanisms of Cr (VI) resistance in bacteria and fungi. FEMS Microbiol Rev. 2014;38:633–659. doi: 10.1111/1574-6976.12051. [DOI] [PubMed] [Google Scholar]
- 17.Levina A, Codd R, Dillon CT, Lay PA. Chromium in Biology: Toxicology and Nutritional Aspects. Prog Inorg Chem. 2003;51:145–250. [Google Scholar]
- 18.Samuel MS, Abigail ME, Ramalingam C. Biosorption of Cr (VI) by Ceratocystis paradoxa MSR2 Using isotherm modelling, kinetic study and optimization of batch parameters using response surface methodology. PLoS One. 2015;10:1–23. doi: 10.1371/journal.pone.0118999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Memon JR, Memon SQ, Bhanger MI, El-Turki A, Hallam KR, Allen GC. Banana peel. A green and economical sorbent for the selective removal of Cr (VI) from industrial wastewater. Colloids Surfaces B Biointerfaces. 2009;70:232–237. doi: 10.1016/j.colsurfb.2008.12.032. [DOI] [PubMed] [Google Scholar]
- 20.Moussavi G, Mahmoudi M. Removal of azo and anthraquinone reactive dyes from industrial wastewaters using MgO nanoparticles. J Hazard Mater. 2009;168:806–812. doi: 10.1016/j.jhazmat.2009.02.097. [DOI] [PubMed] [Google Scholar]
- 21.Anwar J, Shafique U, Waheed-uz-Zaman, Salman M, Dar A, Anwar S. Removal of Pb (II) and Cd (II) from water by adsorption on peels of banana. Bioresour Technol. 2010;101:1752–1755. doi: 10.1016/j.biortech.2009.10.021. [DOI] [PubMed] [Google Scholar]
- 22.Gupta VK, Rastogi A, Nayak A. Adsorption studies on the removal of hexavalent chromium from aqueous solution using a low cost fertilizer industry waste material. J Colloid Interface Sci. 2010;342:135–141. doi: 10.1016/j.jcis.2009.09.065. [DOI] [PubMed] [Google Scholar]
- 23.Wahab MA, Jellali S, Jedidi N. Ammonium biosorption onto sawdust. FTIR analysis, kinetics and adsorption isotherms modeling. Bioresour Technol. 2010;101:5070–5075. doi: 10.1016/j.biortech.2010.01.121. [DOI] [PubMed] [Google Scholar]
- 24.Hamdi Karaoğlu M, Zor Ş, Uğurlu M. Biosorption of Cr (III) from solutions using vineyard pruning waste. Chem Eng. 2010;159(1–3):98–106. doi: 10.1016/j.cej.2010.02.047. [DOI] [Google Scholar]
- 25.Gutiérrez-Corona JF, Romo-Rodríguez P, Santos-Escobar F, Espino-Saldaña AE, Hernández-Escoto H. Microbial interactions with chromium: basic biological processes and applications in environmental biotechnology. World J Microbiol Biotechnol. 2016;32:191. doi: 10.1007/s11274-016-2150-0. [DOI] [PubMed] [Google Scholar]
- 26.Cervantes C, Campos-García J, Devars S, Gutiérrez-Corona F, Loza-Tavera H, Torres-Guzmán JC, Moreno-Sánchez R. Interactions of chromium with microorganisms and plants. FEMS Microbiol Rev. 2001;25(3):335–347. doi: 10.1111/j.1574-6976.2001.tb00581.x. [DOI] [PubMed] [Google Scholar]
- 27.Srivastava S, Thakur IS. Isolation and process parameter optimization of Aspergillus sp. for removal of chromium from tannery effluent. Bioresour. Technol. 2006;97:1167–1173. doi: 10.1016/j.biortech.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 28.Gadd GM. Interactions of fungi with toxic metals. New Phytol. 1993;124:25–60. doi: 10.1111/j.1469-8137.1993.tb03796.x. [DOI] [Google Scholar]
- 29.Gadd GM, Mowll JL. Copper uptake by yeast-like cells, hyphae and chlamydospores of Aureobasidium pullulans. Exp Mycol. 1985;9:230–240. doi: 10.1016/0147-5975(85)90019-2. [DOI] [Google Scholar]
- 30.Gadd GM. Biosorption: critical review of scientific rationale, environmental importance and significance for pollution treatment. J Chem Technol Biotechnol. 2009;84:13–28. doi: 10.1002/jctb.1999. [DOI] [Google Scholar]
- 31.Shugaba A, Buba F. Uptake and Reduction of Hexavalent Chromium by Aspergillus Niger and Aspergillus Parasiticus. J Phylogenetics Evol Biol. 2012;3:119. [Google Scholar]
- 32.Pal C, Bengtsson-Palme J, Rensing C, Kristiansson E, Larsson DG. BacMet: antibacterial biocide and metal resistance genes database. Nucleic Acids Res. 2014;42(Database issue):D737–D743. doi: 10.1093/nar/gkt1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roberts RK, Marzluf AG. The specific interaction of chromate with the dual sulfate permease systems of Neurospora crassa. Arch Biochem Biophys. 1971;142:651–659. doi: 10.1016/0003-9861(71)90531-5. [DOI] [PubMed] [Google Scholar]
- 34.Flores-Alvarez LJ, Corrales-Escobosa AR, Cortés-Penagos C, Martínez-Pacheco M, Wrobel-Zasada K, Wrobel-Kaczmarczyk K, Cervantes C, Gutiérrez-Corona F. The Neurospora crassa chr-1 gene is up-regulated by chromate and its encoded CHR-1 protein causes chromate sensitivity and chromium accumulation. Curr Genet. 2012;58(5–6):281–290. doi: 10.1007/s00294-012-0383-5. [DOI] [PubMed] [Google Scholar]
- 35.Gjuvsland AB, Zörgö E, Samy JK, Stenberg S, Demirsoy IH, Roque F, Maciaszczyk Dziubinska E, Migocka M, Alonso Perez E, Zackrisson M, Wysocki R, Tamás MJ, Jonassen I, Omholt SW, Warringer J. Disentangling genetic and epigenetic determinants of ultrafast adaptation. Mol Syst Biol. 2016;12:892. doi: 10.15252/msb.20166951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Richards EJ. Inherited epigenetic variation-revisiting soft inheritance. Mycorrhiza. 2006;7:395–401. doi: 10.1038/nrg1834. [DOI] [PubMed] [Google Scholar]
- 37.Sharma S, Adholeya A. Detoxification and accumulation of chromium from tannery effluent and spent chrome effluent by Paecilomyces lilacinus fungi. Int Biodeterior Biodegrad. 2011;65:309–317. doi: 10.1016/j.ibiod.2010.12.003. [DOI] [Google Scholar]
- 38.White TJ, Bruns TD, Lee SB, Taylor JW. PCR - Protocols and Applications - A Laboratory Manual. 1990. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. [Google Scholar]
- 39.Prasad P, Varshney D, Adholeya A. Whole genome annotation and comparative genomic analyses of bio-control fungus Purpureocillium lilacinum. BMC Genomics. 2015;16:1004. doi: 10.1186/s12864-015-2229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allpaths LG: High quality genome assembly from low cost data:http://www.broadinstitute.org/software/allpaths-lg/blog/) Acceesed 9-oct-2016.
- 41.MAKER: http://www.yandell-lab.org/software/maker.html. Accessed 20-oct-2016.
- 42.InterProScan: protein sequence analysis & classification. (http://www.ebi.ac.uk/interpro/). Accessed 16-Nov-2016.
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kelley LA, Mezulis S, Yates CM, Wass MN, MJE S. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laskowski RA, MA MA, Moss DS, Thornton JM. PROCHECK - a program to check the stereochemical quality of protein structures. J. App. Cryst. 1993;26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 46.ERRAT: http://services.mbi.ucla.edu/ERRAT/ Accessed 10-Jan-2018.
- 47.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 48.Wass MN, Kelley LA, MJE S. 3DLigandSite: Predicting ligand-binding sites using similar structures. Nucleic Acids Res. 2010;38(2):W469–W473. doi: 10.1093/nar/gkq406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33:W363–W7.D. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Prabhu S, Rajeswari VD. In silico docking analysis of bioactive compounds from Chinese medicine Jinqi Jiangtang tablet (JQJTT) using patch dock. J Chem Pharm Res. 2016;5(8):15–21. [Google Scholar]
- 51.Berman J, Westbrook Z, Feng G, Gilliland TN, Bhat H, Weissig IN, PEB S. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.PyMOL Molecular Graphics System: https://www.pymol.org/. Accessed: 21-Jan-2018.
- 53.Xue M, Yang J, Li Z, Hu S, Yao N, Dean RA, Zhao W, Shen M, Zhang H, Li C, Liu L, Cao L, Xu X, Xing Y, Hsiang T, Zhang Z, Xu JR, Peng YL. Comparative analysis of the genomes of two field isolates of the rice blast fungus Magnaporthe oryzae. PLoS Genet. 2012;8(8):e1002869. doi: 10.1371/journal.pgen.1002869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shabalina SA, Spiridonov NA, Kashina A. Sounds of silence: synonymous nucleotides as a key to biological regulation and complexity. Nucleic Acids Res. 2013;41(4):2073–2094. doi: 10.1093/nar/gks1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Romanova NA, Wolffs PF, Brovko LY, Griffiths MW. Role of efflux pumps in adaptation and resistance of listeria monocytogenes to benzalkonium chloride. Appl Environ Microbiol. 2006;72(5):3498–3503. doi: 10.1128/AEM.72.5.3498-3503.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McGlynn P, Lloyd RG. Rescue of stalled replication forks by RecG: simultaneous translocation on the leading and lagging strand templates supports an active DNA unwinding model of fork reversal and Holliday junction formation. Proc Natl Acad Sci U S A. 2001;98:8227–8234. doi: 10.1073/pnas.111008698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Plaper A, Jenko-Brinovec S, Premzl A, Kos J, Raspor P. Genotoxicity of trivalent chromium in bacterial cells: Possible effects on DNA topology. Chem Res Toxicol. 2002;15:943–949. doi: 10.1021/tx010096q. [DOI] [PubMed] [Google Scholar]
- 58.Decorosi F, Tatti E, Mini A, Giovannetti L, Viti C. Characterization of two genes involved in chromate resistance in a Cr (VI)-hyper-resistant bacterium. Extremophiles. 2009;13:917–923. doi: 10.1007/s00792-009-0279-6. [DOI] [PubMed] [Google Scholar]
- 59.Chihomvu P, Stegmann P, Pillay M. Characterization and structure prediction of partial length protein sequences of pcoA, pcoR and chrB genes from heavy metal resistant bacteria from the Klip River, South Africa. Int J Mol Sci. 2015;16:7352–7374. doi: 10.3390/ijms16047352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu J, Jiang R. Prediction of deleterious nonsynonymous single-nucleotide polymorphism for human diseases. Sci World J. 2013;2013:675851. doi: 10.1155/2013/675851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhao N, Han JG, Shyu C-R, Korkin D. Determining effects of non-synonymous SNPs on protein-protein interactions using supervised and semi-supervised learning. PLoS Comput Biol. 2014;10:e1003592. doi: 10.1371/journal.pcbi.1003592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lewinson O, Adler J, Sigal N, Bibi E. Promiscuity in multidrug recognition and transport: the bacterial MFS Mdr transporters. Mol Microbiol. 2006;61(2):277–284. doi: 10.1111/j.1365-2958.2006.05254.x. [DOI] [PubMed] [Google Scholar]
- 63.Neyfakh AA. Mystery of multidrug transporters: the answer can be simple. Mol Microbiol. 2002;44(5):1123–1130. doi: 10.1046/j.1365-2958.2002.02965.x. [DOI] [PubMed] [Google Scholar]
- 64.Adler J, Bibi E. Determinants of Substrate Recognition by the Escherichia coli Multidrug Transporter MdfA Identified on Both Sides of the Membrane. J Biol Chem. 2004;279:8957–8965. doi: 10.1074/jbc.M313422200. [DOI] [PubMed] [Google Scholar]
- 65.Ianeva OD. Mechanisms of bacteria resistance to heavy metals. Mikrobiol. 2009;71(6):54–65. [PubMed] [Google Scholar]
- 66.Kaur S, Kamli MR, Ali A. Role of arsenic and its resistance in nature. Can J Microbiol. 2011;57(10):769–774. doi: 10.1139/w11-062. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Representation of candidate genes for chromium tolerance in A. flavus. (XLSX 97 kb)
Table S2: 16 genes coded by A. flavus strain TERIBR1 with high frequency of non-synonymous substitutions. (DOCX 15 kb)
Table S3: Phyre2 prediction and analysis of secondary structure. (DOCX 18 kb)
Figure S1. Protein-ligand interaction observed with homologous pairs of protein of A. flavus strains TERIBR1 and NRRL3357. (PDF 414 kb)
Data Availability Statement
The whole genome project has been deposited at https://submit.ncbi.nlm.nih.gov/subs/wgs/under Bioproject PRJNA362980.


